Abstract
Recently emerging evidence has shown that epigenetic mechanisms are involved in initiation and progression of various diseases, including kidney diseases. In the present article, we review the current data regarding the role of epigenetic modulation in chronic kidney disease (CKD) and kidney fibrosis, including DNA methylation and histone modification. Especially we focused on the role of transcription factors in epigenetic modulation and the possibility of therapeutic target of CKD. We have recently reported that transcription factor Kruppel-like factor 4 (also known as gut-enriched Kruppel-like factor) is expressed in kidney podocytes (visceral epithelial cells) and modulates podocyte phenotype by gene-selective epigenetic control. Targeting transcription factors for epigenetic modification may be a good candidate for remission and regression of CKD. It is necessary for the therapy of CKD with an epigenetic-based approach to investigate organ-, tissue-, or gene-specific treatment methods for reduction of side effects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Epigenetic Modulation in Onset and Progression of Kidney Diseases
Epigenetics refers to modifications in gene expression which are not explained by changes in DNA sequence. Epigenetic signals are responsible for the establishment, maintenance, and reversal of metastable transcriptional states that are fundamental for the cell’s ability to remember past events, such as changes in the external environment or developmental cues (Bonasio et al. 2010).
Recently it has been revealed that epigenetic mechanisms are involved in initiation and progression of various diseases, such as cancer and metabolic diseases. Concerning chronic kidney disease (CKD), Kottgen et al. performed the genome-wide association study and showed that the identified 13 new loci affecting renal function and CKD are associated with 1.4 % of the variation in eGFRcrea (Kottgen et al. 2010). However, it is not enough to be elucidated underlying risk factors, and recently epigenetic modulations are recognized as important factors of disease risks. This review provides an overview of epigenetic modifications in CKD, especially focused on DNA methylation and histone modification, and implication of therapeutic approaches.
Epigenetic Modification in CKD
DNA methylation-dependent gene silencing is one of the most characterized mechanisms in epigenetic regulation of gene expression. In CKD, exposure of uremic toxins may exert a crucial impact on epigenetic gene regulation and may thus lead to CKD-associated arteriosclerosis and cardiovascular diseases. For instance, homocysteine levels are elevated in CKD, which causes increased S-adenosylhomocysteine (SAH) levels. SAH inhibits transmethylation reactions that S-adenosylmethionine converts to SAH with supplying methyl groups to other methyl acceptors. Ingrosso et al. reported lower DNA methylation levels in hemodialysis patients with hyperhomocysteinemia (Ingrosso et al. 2003). However, Nanayakkara et al. (2008) showed no significant correlations between global DNA methylation levels and homocysteine or arteriosclerosis levels in CKD stage 2–4 patients. Although these studies performed in CKD patients of different stages and the possibility that it affected the results of the studies cannot be ruled out, these results indicate that homocysteine may cause epigenetic changes in gene-specific manner rather than global DNA methylation or demethylation. It is also reported that uremic toxins lead to DNA methylation of promoter in Klotho genes, which causes down-regulation of the gene expression in renal tubular cells (Azuma et al. 2012; Sun et al. 2012). These results suggest that uremic toxins may not cause global DNA methylation or demethylation in kidney with CKD, but may regulate particular gene expression related to the pathophysiology of CKD. Recent investigation of methylome-wide loci for association with CKD suggested that particular pattern of DNA hypermethylation and hypomethylation is present at different loci in CKD (Smyth et al. 2014).
Concerning histone modifications, Van Beneden et al. (2011) reported that acetylated histone H3K9 was decreased in glomeruli of adriamycin nephropathy model in mice and histone deacetylase (HDAC) inhibitor valproic acid attenuates proteinuria in these mice. HDAC inhibitor is reported to be effective in recovery after AKI (Cianciolo Cosentino et al. 2013), in gentamycin-induced nephrotoxicity (Sun et al. 2013), and in obstructive nephropathy (Kinugasa et al. 2010; Marumo et al. 2010; Pang et al. 2009), and have additional renoprotective effect when combined with angiotensin-converting enzyme inhibitor (Zhong et al. 2013). These results suggest that global histone deacetylation may be a common mechanism in the process of kidney injury and might be a therapeutic target of CKD. Histone methylation is also suggested to be involved in pathogenesis of CKD. Lefevre et al. showed that methylation of histone H3K4 in podocytes promoted CKD (Lefevre et al. 2010). Phosphorylation of histone H3 contributes to the incapacity of tubular epithelial cells to regenerate and proliferate in CKD (Yang et al. 2010). These results highlight the crucial role of various histone modifications in kidney health and diseases.
Epigenetic Modification in Renal Fibrosis
Renal fibrosis is a final common pathway towards end-stage renal disease in CKD caused by various mechanisms. Recently several reports have suggested the important correlation between renal fibrosis and epigenetic regulation of related genes. Bechtel et al. (2010) demonstrated that DNA hypermethylation of RASAL1 activates fibroblasts and induces fibrogenesis in the kidney. Recently they also showed that administration of BMP7 caused normalization of RASAL1 promoter hypermethylation and inhibited experimental kidney fibrosis (Tampe et al. 2014).
Histone modification is also implicated in kidney fibrosis especially in the epigenetic control of transforming growth factor (TGF)-β signaling pathway. Histone H3K4 methylation increases in TGF-β1-mediated ECM gene expression in mesangial cells (Sun et al. 2010). Blocking the class I HDAC by MS-275 ameliorates renal fibrosis and inhibits renal fibroblast activation via modulating TGF-beta and EGFR signaling (Liu et al. 2013).
KLF4 and Epigenetic Modulation in Kidney Podocytes
We have recently reported that transcription factor Kruppel-like factor 4 (KLF4; also known as gut-enriched Kruppel-like factor) is expressed in kidney podocytes (visceral epithelial cells) (Hayashi et al. 2014). The importance of KLF4 in cell biology was underscored by the recent finding as one of the factors to drive the reprogramming of somatic cells into induced pluripotent stem (iPS) cells (Aoi et al. 2008; Maherali et al. 2008; Takahashi and Yamanaka 2006; Takahashi et al. 2007). KLF4 expression in podocytes was decreased in proteinuric kidney diseases in several mouse models and humans. Restoration of KLF4 by podocyte-specific transgenic mice or gene transfer induced recovery of a podocyte epithelial marker, nephrin expression with demethylation of the nephrin promoter region and amelioration of albuminuria. These results suggest that epigenetic change in podocyte genes through decreased KLF4 expression is a common mechanism in exacerbation in proteinuric kidney diseases. Kidney glomerular diseases with proteinuria occurs by various causes, however, decreased KLF4 expression accompanied with epigenetic changes could be a common downstream regulator of podocyte function to numerous upstream mechanisms. This suggests the possibility that transcription factors such as KLF4 may be a target of treatment of proteinuria in various kidney diseases via epigenetic modulations.
As exemplified by the generation of iPS cells from mouse embryonic fibroblasts by transient overexpression of cocktails of transcription factors, an epigenetic state can transit to a different one, given the appropriate combination of transcription factors. It is a very suggestive result for epigenetic modulation as disease therapy, because treatment with appropriate transcription factors is enough to change or reset epigenetic status without addition of direct epigenetic modulator such as histone modifier or DNA methyltransferases.
Role of Transcription Factors in Coordinate and Selective Epigenetic Regulation
Epigenetic modulations, such as histone modifications and DNA methylation, occur through histone acetyl transferases, HDAC, DNA methyltransferases (DNMT) and DNA demethylases in organ-, tissue- and gene-specific manners. These reactions are usually accompanied with complex of transcription factors, and it is probable that these transcription factors may contribute to the determination of the specificity of epigenetic modulations. UHRF1 (ubiquitin-like, containing plant homeo domain and really interesting new gene finger domains 1) binds to methylated CpG and recruits DNMT1 or HDAC1 (Bostick et al. 2007; Sharif et al. 2007). It is reported that transcription factors directly recruit chromatin-modifying enzymes to their genomic targets (Hervouet et al. 2009).
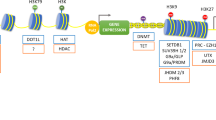
In our recent findings in kidney podocytes, DNA methylation profiling and bisulfite genomic sequencing revealed that KLF4 expression reduced methylation at the nephrin promoter and the promoters of other epithelial markers, whereas methylation was increased at the promoter regions of mesenchymal genes, suggesting selective epigenetic control of KLF4 in podocytes (Hayashi et al. 2014). These results highlight the possibility that KLF4 or a combination of appropriate transcription factors can cause more effective or selective epigenetic changes in podocytes, which may be a therapeutic target for proteinuria. A scheme of changes in transcription factors and epigenetic modifications in CKD was shown in Fig. 1.
Scheme of changes in transcription factors and epigenetic modulations in CKD. Various renal insults cause changes in expression of transcription factors, which lead to coordinate and gene-selective epigenetic modifications. The epigenetic modifications change various gene expressions and induce CKD onset and progression
Therapeutic Approach in the Future
Recently several epigenetic modification drugs are clinically used for the treatment of hematological malignancy. DNA methyltransferase inhibitor Azacitidine and Decitabine were approved by US Food and Drug Administration (FDA) for the treatment of myelodysplastic syndrome in 2004 and 2006, respectively. HDAC inhibitor Vorinostat and Romidepsine were approved by FDA for treatment of cutaneous T cell lymphoma (CTCL) in 2006 and 2009, respectively. Although HDAC inhibition is shown to be effective in CKD of mice model, it is reported that transient serum creatinine elevation and proteinuria were detected in Vorinostat-treated patients of CTCL (Mann et al. 2007). It is unclear why the effect of HDAC inhibitors in humans is different from the results reported in animal models (Cianciolo Cosentino et al. 2013; Kinugasa et al. 2010; Marumo et al. 2010; Pang et al. 2009; Sun et al. 2013; Van Beneden et al. 2011), and whether it is due to differences between characteristics of each HDAC inhibitors, doses, species, or disease states. In the future, these epigenetic modification drugs may become good candidates for treatment of CKD, however further investigation is needed to determine susceptible disease types and minimize the side effects.
To reduce side effects caused by systemic epigenetic treatment, new approaches targeted to specific genes and organs are expected. For example, Deelman et al. (2010) showed the possibility of microbubbles and ultrasound technique which could enhance the cellular uptake of drugs (including gene constructs) into the kidney. Shimizu et al. (2010) reported the delivery method of targeting glomeruli using siRNA/nanocarrier complex. Targeting kidney podocytes, Hauser et al. (2010) used a new delivery technique named shamporter (sheep-anti-mouse transporter), involving a monovalent sheep anti-mouse IgG directed against an unknown podocyte antigen, to specifically target siRNA to the podocyte.
Induction of transcriptional factors such as KLF4 is one of the good candidates for a new epigenetic therapy for CKD. It is well expected that modification of the transcriptional factors involving in epigenetic modulations at specific genes and tissues, may be a therapeutic target with fewer side effects. There may be several ways to induce particular transcription factors. One is direct gene therapy, which is targeted to the kidney. Another way is administration of agents or drugs inducing the expression of appropriate transcription factors in the kidney. Further studies are needed to find these agents for epigenetic treatment of CKD in the future.
Conclusion
There has been great progress in epigenetic regulation of disease onset and progression. A better understanding of epigenetic signals in various research areas, including stem cell function and differentiation may benefit research progression. Epigenetic modification in kidney gene expression via transcription factors may be a good therapeutic target of CKD, which is progressive and difficult to obtain remission or regression with present therapies. Investigation of more efficient and specific treatment with epigenetic modifiers is necessary in the future.
References
Aoi T, Yae K, Nakagawa M et al (2008) Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321:699–702
Azuma M, Koyama D, Kikuchi J et al (2012) Promoter methylation confers kidney-specific expression of the Klotho gene. FASEB J 26:4264–4274
Bechtel W, McGoohan S, Zeisberg EM et al (2010) Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 16:544–550
Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330:612–616
Bostick M, Kim JK, Esteve PO et al (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764
Cianciolo Cosentino C, Skrypnyk NI, Brilli LL et al (2013) Histone deacetylase inhibitor enhances recovery after AKI. J Am Soc Nephrol 24:943–953
Deelman LE, Decleves AE, Rychak JJ et al (2010) Targeted renal therapies through microbubbles and ultrasound. Adv Drug Deliv Rev 62:1369–1377
Hauser PV, Pippin JW, Kaiser C et al (2010) Novel siRNA delivery system to target podocytes in vivo. PLoS One 5:e9463
Hayashi K, Sasamura H, Nakamura M et al (2014) KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J Clin Invest 124:2523–2537
Hervouet E, Vallette FM, Cartron PF (2009) Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 4:487–499
Ingrosso D, Cimmino A, Perna AF et al (2003) Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet 361:1693–1699
Kinugasa F, Noto T, Matsuoka H et al (2010) Prevention of renal interstitial fibrosis via histone deacetylase inhibition in rats with unilateral ureteral obstruction. Transpl Immunol 23:18–23
Kottgen A, Pattaro C, Böger CA et al (2010) New loci associated with kidney function and chronic kidney disease. Nat Genet 42:376–384
Lefevre GM, Patel SR, Kim D et al (2010) Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet 6:e1001142
Liu N, He S, Ma L et al (2013) Blocking the class I histone deacetylase ameliorates renal fibrosis and inhibits renal fibroblast activation via modulating TGF-beta and EGFR signaling. PLoS One 8:e54001
Maherali N, Ahfeldt T, Rigamonti A et al (2008) A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3:340–345
Mann BS, Johnson JR, Cohen MH et al (2007) FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 12:1247–1252
Marumo T, Hishikawa K, Yoshikawa M et al (2010) Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol 298:F133–F141
Nanayakkara PW, Kiefte-de Jong JC, Stehouwer CD et al (2008) Association between global leukocyte DNA methylation, renal function, carotid intima-media thickness and plasma homocysteine in patients with stage 2–4 chronic kidney disease. Nephrol Dial Transplant 23:2586–2592
Pang M, Kothapally J, Mao H et al (2009) Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am J Physiol Renal Physiol 297:F996–F1005
Sharif J, Muto M, Takebayashi S et al (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908–912
Shimizu H, Hori Y, Kaname S et al (2010) siRNA-based therapy ameliorates glomerulonephritis. J Am Soc Nephrol 21:622–633
Smyth LJ, McKay GJ, Maxwell AP et al (2014) DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics 9:366–376
Sun G, Reddy MA, Yuan H et al (2010) Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 21:2069–2080
Sun CY, Chang SC, Wu MS (2012) Suppression of Klotho expression by protein-bound uremic toxins is associated with increased DNA methyltransferase expression and DNA hypermethylation. Kidney Int 81:640–650
Sun X, Zhang B, Hong X et al (2013) Histone deacetylase inhibitor, sodium butyrate, attenuates gentamicin-induced nephrotoxicity by increasing prohibitin protein expression in rats. Eur J Pharmacol 707:147–154
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Takahashi K, Tanabe K, Ohnuki M et al (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Tampe B, Tampe D, Müller CA et al (2014) Tet3-mediated hydroxymethylation of epigenetically silenced genes contributes to bone morphogenic protein 7-induced reversal of kidney fibrosis. J Am Soc Nephrol 25:905–912
Van Beneden K, Geers C, Pauwels M et al (2011) Valproic acid attenuates proteinuria and kidney injury. J Am Soc Nephrol 22:1863–1875
Yang L, Besschetnova TY, Brooks CR et al (2010) Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med 16:535–543
Zhong Y, Chen EY, Liu R et al (2013) Renoprotective effect of combined inhibition of Angiotensin-converting enzyme and histone deacetylase. J Am Soc Nephrol 24:801–811
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Hayashi, K., Itoh, H. Transcription Factors and Epigenetic Modulation: Its Therapeutic Implication in Chronic Kidney Disease. Arch. Immunol. Ther. Exp. 63, 193–196 (2015). https://doi.org/10.1007/s00005-014-0326-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00005-014-0326-6