Abstract
This study was conducted to determine the antimicrobial resistance profiles of Escherichia coli isolated from food samples received at the National Food Safety Agency in Burkina Faso. A total of 155 isolates from dairy foods (29), fish-based dishes (7), mango juices (4), lettuces (35), RTE salads (21), sandwiches (32), and sesames (27) were included for testing with the Kirby-Bauer disk diffusion method. PCR targeting ampicillin (blaTEM, blaSHV, temA, and temB), tetracyclines [tet(A) and tet(B)], sulfamethoxazole (sul1 and sul2), aminoglycosides (StrA and aadA) and quinolones (GyrA) resistance genes were performed to elucidate the genotypic resistance mechanism. Of the 155 isolates, 105 (67.7%) were resistant to at least one antimicrobial agent. Resistances to tetracycline (33.5%), ampicillin (32.9%), cefoxitin (18.7%), gentamycin (15.5%), amoxicillin-clavulanate acid (15.5%), nalidixic acid (12.9%), chloramphenicol (11.6%), trimethoprim-sulfamethoxazole (11.6%), and ciprofloxacin (8.4%) were observed. Multidrug resistance was recorded in 26.5% of the isolates. Antimicrobial resistance genes including blaTEM (19/51, 37.3%), blaSHV (19/51, 37.3%), temB (17/51, 33.3%), tet(A) (24/52, 46.2%), tet(B) (9/52, 17.3%), sul1 (8/18, 44.4%), sul2 (4/18, 22.2%), aadA (11/24, 45.83%) and GyrA (31/36, 86.1%) were detected. All E. coli isolates resistant to at least 2 antibiotics were positive for the class 1 integron gene (intI1). These findings raise concerns about food safety and public health and demonstrate the need for strict government control and continuous monitoring.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Antimicrobial resistance (AMR) is a growing global concern and poses a grave threat to public health (Robinson et al. 2016). The void in the development of new therapeutic substances has exacerbated the problem, and as a result, very few antimicrobials are available to treat infections caused by multidrug-resistant (MDR) pathogens (Robinson et al. 2016). Numerous reports have described the presence of large amounts of antimicrobial resistant bacteria and antimicrobial resistant genes (ARGs) in various food products, such as meat, fish, vegetables, ready-to-eat (RTE) foods, and fruits (Rajaei et al. 2021; Vuthy et al. 2017; Amer et al. 2018). This presence of MDR bacteria in the food chain is a rising public health concern. Indeed, besides contributing to the spread of foodborne pathogens, an additional concern is that food represents a vehicle for the transmission of AMR determinants to humans, which may occur through contact or consumption of contaminated food products (Amer et al. 2018).
Escherichia coli (E. coli) is a ubiquitous rod-shaped Gram-negative bacterium abundant in the gastrointestinal tract of humans and animals, where it plays a crucial role in digestion and absorption (Kaushik et al. 2018). However, E. coli can infect animals and humans, causing diseases ranging from opportunistic wound infections to severe systemic infections. Furthermore, E. coli can easily spread through the food chain and function as ARG carriers/reservoirs, sharing them with other bacteria of the same or other species (Kaushik et al. 2018). As a result, E. coli strains are frequently used as an appropriate indicator bacterium in AMR surveillance, as changes in these species can serve as an early warning of the emergence of resistance in similar pathogenic bacteria (Guo et al. 2019). Resistant E. coli strains containing various ARGs have been extensively investigated, mainly in industrialized nations (Balbin et al. 2020; Li et al. 2020a). Most of the AMR research in Burkina Faso has focused on clinical samples from humans (Ouedraogo et al. 2016; Dissinviel et al. 2017; Kpoda et al. 2018; Dembélé et al. 2020). There is limited information on the antimicrobial resistance and molecular characteristics of E. coli strains isolated from food sources throughout our country. Accordingly, in this study, we performed antimicrobial susceptibility and molecular characterization of resistance genes from E. coli strains obtained from various foods controlled by Burkina Faso’s National Food Safety Agency. All isolates were tested for antimicrobial susceptibility using the disk diffusion method, followed by a polymerase chain reaction (PCR) assay to screen 11 ARGs and integrons. To our knowledge, this report is the first to investigate the role of E. coli in the propagation of AMR in the food chain in Burkina Faso. The results call for strengthening the monitoring of foodborne E. coli in the Burkina Faso food chain to protect human health.
2 Material and methods
2.1 Bacterial isolates and storage
This study included 155 E. coli isolates from food samples obtained from the Burkina Faso National Food Safety Agency surveillance activities between 2018 and 2021. All samples received were processed for isolation and identification of E. coli on MacConkey agar (HiMedia, Mumbai, India) and Eosin methylene blue (EMB) agar (HiMedia, Mumbai, India) followed by biochemical characterization by standard procedures. Isolated strains were stored at -80 °C in brain heart infusion (BHI), (HiMedia, Mumbai, India) medium containing 15% (v/v) glycerol (HiMedia, Mumbai, India). For the purpose of this study, all frozen E. coli isolates were reactivated from cryovials on Nutrient Agar (HiMedia, Mumbai, India) at 37 °C for 18–24 h. The identification of E. coli was confirmed by Gram staining and biochemical characterization using API 20E kits (BioMérieux, Marcy l’Etoile, France). The isolates were recovered from 7 types of food samples, including dairy foods (29), fish-based dishes (7), mango juices (4), lettuces (35), RTE salads (21), sandwiches (32) and sesame (27) (Table 1).
2.2 Antimicrobial susceptibility tests
The disk diffusion method on Mueller-Hinton (MH) agar (HiMedia, Mumbai, India) was used for antimicrobial susceptibility testing, as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2017). In brief, a single bacterial colony from nutrient agar was randomly selected and suspended in 0.85% sterile saline (HiMedia, Mumbai, India) to achieve an inoculum equivalent to 0.5 McFarland (HiMedia, Mumbai, India). A sterile cotton swab (HiMedia, Mumbai, India) was immersed in the 0.5 McFarland suspension and spread on MH plate. The plate was then allowed to dry completely at room temperature (no more than 15 min). Antimicrobial disks (Condalab, Madrid, Spain) were then applied to the MH (HiMedia, Mumbai, India) agar surface with sterile fine curved stainless-steel splinter forceps at a distance to avoid overlapping of the inhibition zones. The following antibiotics were used: cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 µg), cefepime (FEP, 30 µg), aztreonam (ATM, 30 µg ), amoxicillin/clavulanic acid (AMC, 30 µg), ampicillin (AMP, 10 µg), cefoxitin (FOX, 30 µg), chloramphenicol (C, 30 µg), trimethoprim-sulfamethoxazole (STX, 1.25/23.75 µg), nalidixic acid (NA, 30 µg), tetracycline (TE, 30 µg), imipenem (IMP, 10 µg ), meropenem (MEM, 10 µg), gentamicin (CN, 10 µg), amikacin (AK, 30 µg), and ciprofloxacin (CIP, 10 µg). The inoculated plates were incubated at 37 °C for 24 h and the inhibition zones were measured in millimeters across the diameters. The standard strain of E. coli ATCC 35218 was used as a quality check. Isolates exhibiting resistance to 3 or more classes were considered multidrug-resistant (MDR) (Magiorakos et al. 2012). Multiple antimicrobial resistance (MAR) indexes were calculated using Krumperman’s approach (Krumperman 1983).
2.3 DNA extraction
The heat lysis method (Yang et al. 2019) was used to extract genomic DNA from resistant bacteria, which was then used as a PCR template for all assays. Briefly, a single pure bacterial colony from a Nutrient agar (HiMedia, Mumbai, India) plate was subcultured in 1 ml of BHI (HiMedia, Mumbai, India) overnight. The next day, each culture broth was centrifuged at 13,000 rpm for 5 min at 4 °C, and the cell pellets were resuspended in 500 µl of sterile distilled water and heated at 100 °C in a water bath for 10 min. The cell suspension was immediately frozen at -20 °C for 10 min and then centrifuged at 13,000 rpm for 5 min at 4 °C. The supernatant was transferred to a new Eppendorf tube and conserved at -20 °C until use.
2.4 Identification of resistance genes and integrons
The presence of antimicrobial resistance genes among the resistant isolates was determined using the uniplex PCR tests to amplify ARGs. 11 pairs of oligonucleotide primers (Genecust, Paris, France) were used to target antimicrobial resistance genes that confer resistance to antimicrobial agents, including ampicillin (blaTEMblaSHV, temA, temB), tetracycline, [tet(A), tet(B)], aminoglycosides (StrA, aadA), quinolone (GyrA) and trimethoprim-sulfamethoxazole (sul1, sul2). The PCR cycling conditions for reactions were set at 94 °C for 3 min, followed by 30 cycles of 1 min at 94 °C, 30 s at annealing temperature and 1 min at 72 °C and final step for 7 min at 72 °C. Furthermore, uniplex PCR was conducted to detect the presence of intI1 integrase genes in the 65 isolates resistant to at least 2 antimicrobials, to detect the presence of Class 1 integrons. The thermal cycling conditions included preincubation at 94 °C for 1 min, followed by 35 cycles of denaturation at 98 °C for 30 s, annealing at 60 °C for 30 s, and polymerization at 72 °C for 30 s, and final extension at 72 °C for 10 min. Subsequently, the variable regions (VRs) of the strains that were positive for the intI1 gene were evaluated by uniplex PCR using the primers hep58 and hep59 with the following cycling conditions: preincubation at 94 °C for 1 min, followed by 35 cycles of denaturation at 98 °C for 30 s, annealing at 55 °C for 30 s, and polymerization at 72 °C for 4 min and final extension at 72 °C for 10 min. The primer (Genecust, Paris, France) sequences, the annealing temperatures, expected product sizes, and references are presented in Table 2. The PCR was done using a TECHNE Prime Elite thermal cycler (Bibby scientific, Staffordshire, United Kingdom). Appropriate positive and negative controls for amplifications were used in each PCR procedure. All PCR amplifications were performed in a reaction mixture comprising of 4 µl FIREPol® Master Mix (Solis BioDyne, Tartu, Estonia), 0.4 µl of each primer, 3 µl of DNA template and nuclease-free water to make volume up to 20 µl. Amplified PCR products were run on a 1.5% agarose gel containing 0.5 µg/ml of ethidium bromide (ThermoFisher, Waltham, United States) for 60 min at 100 V, then visualized using the gel documentation system E-BOX VX5 (VILBER, Marne-la-Vallée, France).
3 Results and discussion
In this study, we characterized the antimicrobial resistance profiles and ARGs of a collection of 155 E. coli isolates recovered from sandwich, dairy food, fish, sesame, lettuce and RTE salad at the Burkina Faso National Food Safety Agency. To our knowledge, this is the first time a study was conducted with many E. coli isolates from various food sources in our country.
3.1 Antimicrobial susceptibility of E. coli isolates
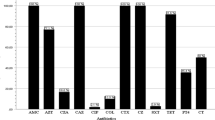
Among the E. coli isolates used in the present study, 105 (67.7%) were resistant to at least 1 of the 16 antimicrobials used (Table 1). A similarly high level of resistance (74.2%) was reported in isolates collected from RTE foods in 2017 in Shaanxi Province, Republic of China (Baloch et al. 2017). In our study, resistances to tetracycline, ampicillin, gentamycin, amoxicillin-clavulanate acid, nalidixic acid, chloramphenicol, and trimethoprim-sulfamethoxazole were generally observed. These findings are consistent with previously published studies showing that E. coli isolates from foods were resistant to several older antimicrobials, including tetracycline, streptomycin, sulfonamides, ampicillin, gentamicin, and nalidixic acid. Vuthy et al. (2017), reported high resistances to tetracycline, amoxicillin, and sulfamethoxazole (63.1–76.1%) of E. coli isolated from chicken food chains in Phnom Penh (China). In addition, E. coli isolated from RTE foods (Singapore) during a study of Guo et al. (2019) showed resistances to tetracycline (17.2%), ampicillin (15.2%) and chloramphenicol (10.1%). In a study conducted by Ayamah et al. (2021) in Ghana, all isolates obtained from kebabs (beef, chevon, and gizzard) were resistant to ampicillin, tetracycline, and gentamicin. Furthermore, Somda et al. (2018) found that E. coli isolated from RTE chickens in Burkina Faso were resistant to tetracycline (64.3%), ampicillin (42.8%), amoxicillin + clavulanic acid (46.3%), and trimethoprim-sulphamethoxazole (32.14%). These traditional antimicrobials have been overused and abused throughout the years by consumers, veterinarians, farmers, physicians, and patients due to their low prices and easy availability over the counter, rendering them empirically ineffective (Guo et al. 2019). Interestingly, our study did not find an E. coli isolate resistant to cefotaxime, ceftazidime, cefepime, and aztreonam (ESBL-producing strain). On the contrary, resistance to these antimicrobials was observed among E. coli isolated from street foods in India (Sivakumar et al. 2021). Additionally, a study of street food in Tamale (Ghana) showed that 55.4% (41/74) of the isolated E. coli were ESBL producers (Karikari et al. 2022). This disparity may be attributed to the sample sources, that is, foods of plant origin in our case and street foods in the others. In addition, bacteria found in meat (especially meat poultry origin) can be resistant to a wide range of antibiotics, including quinolones and the third generation of cephalosporins (Guo et al. 2019). It is important to note that none of the isolates in this study were resistant to carbapenems (imipenem and meropenem). Until recently, carbapenems, such as meropenem, imipenem, ertapenem, and doripenem, were the antibi-otics of last choice for treatments of MDR pathogenic bacteria (Osei et al. 2016). As a result, prescribers should not abuse these antibiotics to avoid the emergence and development of resistances. In our study, the E. coli isolates also showed low resistance to ciprofloxacin (8.4%). This finding is of public health significance, as they are critically important for the treatment of serious human infections. In developed countries, the use of fluoroquinolones has been banned for use in animal husbandry; therefore, E. coli isolated from this source showed very little resistance to these antibiotics (Boerlin et al. 2005).
3.2 Profile of MDR E. coli isolates
Our study revealed a moderate percentage of MDR strains among E. coli (26.5%). MDR strains are isolates resistant to 3 or more different classes of antibiotics (Magiorakos et al. 2012). Previous studies have reported that multidrug resistance is common in poultry and other food animals (Li et al. 2020a; Abatcha et al. 2018; Adzitey et al. 2020). The presence of MDR bacteria in foods (meat, vegetables, fruit and RTE foods) is a public health concern since they can be transmitted to humans directly or indirectly by eating contaminated RTE foods. The MDR bacteria identified in the present study may result from environmental contamination or transmission by food handlers due to poor personal hygiene and inadequate sanitation in the processing plant. Therefore, the microbiological quality of food should be monitored to ensure public health safety. As shown in Table 3, this study also revealed that the multiple antibiotic resistance (MAR) index ranged from 0.13 (resistant to 2 antibiotics) to 0.47 (resistant to 7 antibiotics). 19 resistant isolates (18.7%) had a MAR index > 0.2. Bacteria that have a MAR index > 0.2 originate from a high-risk source of contamination where antibiotics or growth promoters are frequently used (Krumperman 1983). Overall, the extensive use by humans and livestock production, particularly in poultry, may have led to the rise of resistance (Rajaei et al. 2021). Therefore, One Health strategies which require multidisciplinary collaborations, adequate surveillance systems, and strong laboratory capacity are needed in order to reduce antibiotic misuse.
3.3 PCR-based detection of antimicrobial resistance genes
The result of PCR screening for the presence of resistance genes among phenotypically resistant E. coli isolates is shown in Table 4. Among drug resistant isolates, PCR tests detected 9 genes (blaTEM, blaSHV, temB, tet(A), tet(B), aadA, GyrA, sul1, and sul2). In the present study, detection of beta-lactamase genes among ampicillin resistant isolates revealed the predominance of blaTEM and blaSHV genes (37.3% each), followed by the temB genes (33.3%). Beta-lactamase, which deactivates beta-lactam antibiotics by hydrolysis of the beta-lactam ring, is one of the most significant enzymes implicated in bacterial resistance to antibiotics (Shaikh et al. 2015). Although numerous beta-lactamases have been identified, TEM and SHV are reported to be the main causes of bacterial resistance to beta-lactam antibiotics (Hassani et al. 2022). All beta-lactamase genes detected during our work have been widely documented in the community, clinical isolates, and food-producing animals (Guo et al. 2019). The tetracycline resistance genes tet(A) and tet(B) were found in 46.2% and 17.3% of the isolates tested in this study, respectively. This finding supports previous research that identified tet(A) genes as the most common tetracycline resistance genes (Vuthy et al. 2017; Guo et al. 2019; Dissinviel et al. 2017; Abdelwahab et al. 2022). Currently, 30 different tetracycline resistance genes (tet genes) have been identified (Eliopoulos et al. 2003). All tet efflux genes encode membrane-associated energy-dependent proteins, which export tetracycline out of the cell. Among our isolates resistant to trimethoprim-sulfamethoxazole, 44.4% carried sul1 genes, while 22.2% harbored sul2 genes. These 2 genes that express highly resistant dihydropteroate synthases to sulfamethoxazole are carried by transposons and plasmids (Roberts 1996). However, almost 20 different resistance genes that express drug-insensitive dihydrofolate reductase are spread as cassettes in integrons, transposons, and plasmids (Nelson et al. 2019). In this study, 86% of E. coli resistant to quinolones carried the GyrA gene. Overexpression of naturally occurring efflux pumps, resistance encoded by the chromosome, mutations in molecular target DNA gyrase and topoisomerase IV, and resistance mediated by plasmids are molecular mechanisms of bacterial resistance to quinolone drugs (Abdelwahab et al. 2022). However, the most common mechanism of resistance to fluoroquinolones are mutations of target genes such as GyrA, parE, and parC that encode type II topoisomerases (Guo et al. 2019; O’Bryan et al. 2018).
In summary, it is worth noting that several genes can be involved in AMR. In this study, we did not evaluate the entire suite of resistance genes. Therefore, some resistant isolates did not carry any of the AMR genes tested, indicating that other AMR genes may be present or there are other novel genetic resistant determinants (Abdelwahab et al. 2022). Therefore, isolates that were phenotypically resistant but negative for PCR should be screened using novel techniques such as metagenomic analysis based on high-throughput sequencing (HTS) to provide more comprehensive information on their antibiotic resistomes (Li et al. 2020b).
3.4 PCR-based detection of class 1 integrons genes
The present study has shown the presence of MDR among E. coli isolates, as mentioned above, suggesting the existence of mobile genetic elements such as integrons, plasmids, and transposons (Deng et al. 2015). Of these, integrons play a central role in the spread of ARGs among gram-negative bacteria (Kaushik et al. 2018). Integrons capture gene cassettes using an integron-integrase and then express cassettes using an integron encoded promoter. There are many types of integrons based on the integrase gene (intI), but most of the integrons found in clinical isolates belong to Class 1 (Deng et al. 2015). Table 5 shows the distribution of antimicrobial resistance, class-1 integrons in the 65 isolates resistant to at least 2 antimicrobials. All the isolates were positive for class 1 integrons, which is consistent with previous findings (Zhang et al. 2019; Malek et al. 2015). Particularly in Burkina Faso, class 1 integrons were found in 65% of Salmonella isolated from environment and diarrhea stool samples (Somda et al. 2021). Subsequently, strains harboring intI genes were elaborated for the amplification of variable regions (VRs). The occurrence of VRs within class 1, integron positive isolates were 32.3% (21/65 strains). The integrons that did not carry gene cassettes are called empty integrons and can have the potential to rapidly convert themselves into MDR strains (Mohamed et al. 2020; Fang et al. 2019).
3.5 Limitations
This study has some limitations due to a lack of financial resources. Firstly, appropriate tests were not done to confirm the PCR product identity. The confirmation tests could be: (i) DNA sequencing of the PCR product; (ii) hybridization of the PCR product with specific DNA probes; (iii) or restriction analysis of the PCR product. Secondly, we have focused exclusively on the antimicrobial resistance in E. coli isolates, and the pathogenicity of the collected E. coli was not assessed.
4 Conclusion
This study generated preliminary data on the resistance of E. coli identified in the food chain of Burkina Faso. Phenotypic characterization revealed resistance to tetracycline, ampicillin, cefoxitin, gentamicin, clavulanic acid, and sulfamethoxazole/trimethoprim. In addition, isolates displayed multidrug resistance characteristics and revealed possible molecular resistance mechanisms. Therefore, a prospective study with a well-designed geographic distribution of the samples is required.
Data availability
All data generated or analyzed during this study are available from the corresponding authors upon request.
References
Abatcha MG, Effarizah ME, Rusul G (2018) Prevalence, antimicrobial resistance, resistance genes and class 1 integrons of Salmonella serovars in leafy vegetables, chicken carcasses and related processing environments in Malaysian fresh food markets. Food Control 91:170–180. https://doi.org/10.1016/j.foodcont.2018.02.039
Abdelwahab GE, Ishag HZA, Al Hammadi ZM, Al Yammahi SMS, Mohd Yusof MFB, Al Yassi MSY, Al Neyadi SSA, Al Mansoori AMA, Al Hamadi FHA, Al Hamadi IAS, Hosani MAAA, Mohammed Al Muhairi SS (2022) Antibiotics Resistance in Escherichia coli Isolated from Livestock in the Emirate of Abu Dhabi, UAE, 2014–2019. Int J Microbiol 2022:e3411560. https://doi.org/10.1155/2022/3411560
Adzitey F (2020) Incidence and antimicrobial susceptibility of Escherichia coli isolated from beef (meat muscle, liver and kidney) samples in Wa Abattoir, Ghana. Cogent Food Agric 6:1718269. https://doi.org/10.1080/23311932.2020.1718269
Amer MM, Mekky HM, Amer AM, Fedawy HS (2018) Antimicrobial resistance genes in pathogenic Escherichia coli isolated from diseased broiler chickens in Egypt and their relationship with the phenotypic resistance characteristics. Vet World 11:1082–1088. https://doi.org/10.14202/vetworld.2018.1082-1088
Ayamah A, Sylverken AA, Ofori LA (2021) Microbial load and antibiotic resistance of Escherichia coli and Staphylococcus aureus isolated from ready-to-eat (RTE) Khebab Sold on a University Campus and its environs in Ghana. J Food Qual 2021:1–9. https://doi.org/10.1155/2021/8622903
Balbin MM, Hull D, Guest C, Nichols L, Dunn R, Hull D, Thakur S (2020) Antimicrobial resistance and virulence factors profile of Salmonella spp. and Escherichia coli isolated from different environments exposed to anthropogenic activity. J Glob Antimicrob Resist 22:578–583. https://doi.org/10.1016/j.jgar.2020.05.016
Baloch AB, Yang H, Feng Y, Xi M, Wu Q, Yang Q, Tang J, He X, Xiao Y, Xia X (2017) Presence and Antimicrobial Resistance of Escherichia coli in Ready-to-eat Foods in Shaanxi, China. J Food Prot 80:420–424. https://doi.org/10.4315/0362-028X.JFP-16-175
Boerlin P, Travis R, Gyles CL, Reid-Smith R, Janecko N, Lim H, Nicholson V, McEwen SA, Friendship R, Archambault M (2005) Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol 71:6753–6761. https://doi.org/10.1128/AEM.71.11.6753-6761.2005
CLSI (2017) Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. file:///C:/Users/Biochimie/Desktop/telechargement/2017_CLSI_M100_PerformanceStandardsforAntimicrobialSusceptibilityTesting_27thed.pdf
Dembélé R, Konaté A, Traoré O, Kaboré WAD, Soulama I, Kagambèga A, Traoré AS, Guessennd NK, Aidara-Kane A, Gassama-Sow A, Barro N (2020) Extended spectrum beta-lactamase and fluoroquinolone resistance genes among Escherichia coli and Salmonella isolates from children with diarrhea, Burkina Faso. BMC Pediatr 20:459. https://doi.org/10.1186/s12887-020-02342-z
Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, Chen D, Bian H, Li Y, Yu G (2015) Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob 14:45. https://doi.org/10.1186/s12941-015-0100-6
Dissinviel SK, Nathalie G, Juste IB, Mohamed BO, Fernique K, Abraham A, Jacques S, Rasmata O, Koiné MD, Lassana S, Mireille D, Alfred T (2017) Prevalence and resistance profile of extended-spectrum β-lactamases-producing Enterobacteriaceae in Ouagadougou, Burkina Faso. Afr J Microbiol Res 11:1120–1126. https://doi.org/10.5897/AJMR2017.8598
Eliopoulos GM, Eliopoulos GM, Roberts MC (2003) Tetracycline therapy: update. Clin Infect Dis 36:462–467. https://doi.org/10.1086/367622
Fang J, Shen Y, Qu D, Han J (2019) Antimicrobial resistance profiles and characteristics of integrons in Escherichia coli strains isolated from a large-scale centralized swine slaughterhouse and its downstream markets in Zhejiang, China. Food Control 95:215–222. https://doi.org/10.1016/j.foodcont.2018.08.003
Guo S, Tay MYF, Aung KT, Seow KLG, Ng LC, Purbojati RW, Drautz-Moses DI, Schuster SC, Schlundt J (2019) Phenotypic and genotypic characterization of antimicrobial resistant Escherichia coli isolated from ready-to-eat food in Singapore using disk diffusion, broth microdilution and whole genome sequencing methods. Food Control 99:89–97. https://doi.org/10.1016/j.foodcont.2018.12.043
Hassani S, Moosavy M-H, Gharajalar SN, Khatibi SA, Hajibemani A, Barabadi Z (2022) High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci Rep 12:3878. https://doi.org/10.1038/s41598-022-07845-6
Karikari AB, Kpordze SW, Yamik DY, Saba CKS (2022) Ready-to-eat food as sources of extended-spectrum β-Lactamase-producing Salmonella and E. coli in Tamale, Ghana. Front Trop Dis 3:2673–7515. https://doi.org/10.3389/fitd.2022.834048
Kaushik M, Kumar S, Kapoor RK, Virdi JS, Gulati P (2018) Integrons in Enterobacteriaceae: diversity, distribution and epidemiology. Int J Antimicrob Agents 51:167–176. https://doi.org/10.1016/j.ijantimicag.2017.10.004
Kpoda DS, Ajayi A, Somda M, Traore O, Guessennd N, Ouattara AS, Sangare L, Traore AS, Dosso M (2018) Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Res Notes 11:471. https://doi.org/10.1186/s13104-018-3581-5
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170. https://doi.org/10.1128/aem.46.1.165-170.1983
Li Y, Zhang M, Luo J, Chen J, Wang Q, Lu S, Ji H (2020) Antimicrobial resistance of Escherichia coli isolated from retail foods in northern Xinjiang, China. Food Sci Nutr 8:2035–2051. https://doi.org/10.1002/fsn3.1491
Li Y, Cao W, Liang S, Yamasaki S, Chen X, Shi L, Ye L (2020a) Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci Rep 10:15175. https://doi.org/10.1038/s41598-020-72620-4
Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Malek M, Amer FA, Allam A, El-Sokkary R, Gheith T, Arafa M (2015) Occurrence of classes I and II integrons in Enterobacteriaceae collected from Zagazig University Hospitals, Egypt. Front Microbiol 6:601. https://doi.org/10.3389/fmicb.2015.00601
Mohamed SA, Ardiyati T, Rifa’i M, Widodo (2020) Detection of class 1 integron-associated gene cassettes and tetracycline resistance genes in Escherichia coli isolated from ready to eat vegetables. Ann Med Surg 55:327–331. https://doi.org/10.1016/j.amsu.2020.04.044
Nelson DW, Moore JE, Rao JR (2019) Antimicrobial resistance (AMR): significance to food quality and safety. Food Qual Saf 3:15–22. https://doi.org/10.1093/fqsafe/fyz003
O’Bryan CA, Crandall PG, Ricke SC (2018) Chap. 6 - Antimicrobial Resistance in Foodborne Pathogens. In: Ricke SC, Atungulu GG, Rainwater CE, Park SH (eds) Food and Feed Safety Systems and Analysis. Academic Press, pp 99–115. https://doi.org/10.1016/B978-0-12-811835-1.00006-3
Osei Sekyere J (2016) Current state of resistance to Antibiotics of Last-Resort in South Africa: a review from a Public Health Perspective. Front Public Health 4:209. https://doi.org/10.3389/fpubh.2016.00209
Ouedraogo A-S, Sanou M, Kissou A, Sanou S, Solaré H, Kaboré F, Poda A, Aberkane S, Bouzinbi N, Sano I, Nacro B, Sangaré L, Carrière C, Decré D, Ouégraogo R, Jean-Pierre H, Godreuil S (2016) High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect Dis 16:326. https://doi.org/10.1186/s12879-016-1655-3
Rajaei M, Moosavy M-H, Gharajalar SN, Khatibi SA (2021) Antibiotic resistance in the pathogenic foodborne bacteria isolated from raw kebab and hamburger: phenotypic and genotypic study. BMC Microbiol 21:272. https://doi.org/10.1186/s12866-021-02326-8
Roberts MC (1996) Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev 19:1–24. https://doi.org/10.1111/j.1574-6976.1996.tb00251.x
Robinson TP, Bu DP, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, Hay SI, Jiwakanon J, Kakkar M, Kariuki S, Laxminarayan R, Lubroth J, Magnusson U, Thi Ngoc P, Van Boeckel TP, Woolhouse MEJ (2016) Antibiotic resistance is the quintessential one health issue. Trans R Soc Trop Med Hyg 110:377–380. https://doi.org/10.1093/trstmh/trw048
Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal MA (2015) Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci 22:90–101. https://doi.org/10.1016/j.sjbs.2014.08.002
Sivakumar M, Abass G, Vivekanandhan R, Anukampa, Singh DK, Bhilegaonkar K, Kumar S, Grace MR, Dubal Z (2021) Extended-spectrum beta-lactamase (ESBL) producing and multidrug-resistant Escherichia coli in street foods: a public health concern. J Food Sci Technol 58:1247–1261. https://doi.org/10.1007/s13197-020-04634-9
Somda NS, Bonkoungou OJI, Zongo C, Kagambèga A, Bassolé IHN, Traoré Y, Mahillon J, Scippo M-L, Hounhouigan JD, Savadogo A (2018) Safety of ready-to-eat chicken in Burkina Faso: microbiological quality, antibiotic resistance, and virulence genes in Escherichia coli isolated from chicken samples of Ouagadougou. Food Sci Nutr 6:1077–1084. https://doi.org/10.1002/fsn3.650
Somda NS, Bonkoungou IJO, Sambe-Ba B, Drabo MS, Wane AA, Sawadogo-Lingani H, Savadogo A (2021) Diversity and antimicrobial drug resistance of non-typhoid Salmonella serotypes isolated in lettuce, irrigation water and clinical samples in Burkina Faso. J Agric Food Res 5:100167. https://doi.org/10.1016/j.jafr.2021.100167
Vuthy Y, Lay KS, Seiha H, Kerleguer A, Aidara-Kane A (2017) Antibiotic susceptibility and molecular characterization of resistance genes among Escherichia coli and among Salmonella subsp. in chicken food chains. Asian Pac J Trop Biomed 7:670–674. https://doi.org/10.1016/j.apjtb.2017.07.002
Yang X, Wu Q, Zhang J, Huang J, Chen L, Wu S, Zeng H, Wang J, Chen M, Wu H, Gu Q, Wei X (2019) Prevalence, bacterial load, and Antimicrobial Resistance of Salmonella Serovars isolated from Retail Meat and Meat products in China. Front Microbiol 10:2121. https://doi.org/10.3389/fmicb.2019.02121
Zhang S, Yang H, Rehman MU, Yang K, Dong M, Yang J, Wang M, Jia R, Chen S, Liu M, Zhu D, Zhao X, Yang Q, Wu Y, Zhang L, Liu Y, Yu Y, Tian B, Pan L, Chen X, Cheng A (2019) Class 1 integrons as predominant carriers in Escherichia coli isolates from waterfowls in Hainan, China. Ecotoxicol Environ Saf 183:109514. https://doi.org/10.1016/j.ecoenv.2019.109514
Acknowledgements
The authors are grateful to the Laboratoire National de Santé Publique (LNSP) of Burkina Faso for providing E. coli isolates, laboratory space, and reagents.
Funding
This study submission does not include any research funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Not required.
Competing interests
The authors declare no conflict of interest with this research.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soubeiga, A.P., Kpoda, D.S., Sawadogo, A. et al. Antimicrobial susceptibility and genotypic characterization of Escherichia coli isolated from foods controlled by the National Food Safety Agency in Burkina Faso. J Consum Prot Food Saf 19, 213–223 (2024). https://doi.org/10.1007/s00003-024-01493-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00003-024-01493-w