Abstract
Rhizosphere, the thin layer of soil under the direct influence of plant root, is a nutrient-rich microhabitat. The numbers of bacteria colonizing this niche are 100–1000 times more than the surrounding non-rhizosphere soil. More than 25 bacterial genera have been characterized as plant growth-promoting rhizobacteria, among which Bacillus sp. play a predominant role. Because of the non-fastidious nature and quick colonization of rhizosphere, these gram-positive rods are relatively abundant in the rhizosphere and can exert its plant growth-promoting benefits on the plant involved. Brevibacillus, Lysinibacillus, Bacillus subtilis, Bacillus cereus, and Bacillus amyloliquefaciens are some of the species that can act as plant growth-promoting rhizobacteria (PGPR). Bacillus sp. readily qualify as a PGPR owing to its phytohormone production, nitrogen fixation, siderophore production, hydrogen cyanide production, antagonism against plant pathogens, and production of certain allelochemicals. Some strains of Bacillus sp. show extreme tolerance to heavy metals and can be coupled with phytoremediating plants to remove heavy metal pollutants from contaminated soils. Bacillus sp. isolated from degraded mine soils also show plant growth-promoting effects and can be potentially used as a bioinoculant during the revegetation process of reclamation of mine ecosystem.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Plant growth-promoting rhizobacteria (PGPR) are a diverse group of bacterial genera that actively colonize the rhizosphere region of the plants. The plant and the PGPR species enjoy a mutually benefitting and non-obligatory relationship with each other. These free-living bacteria dwell in the rhizosphere, the region under the direct influence of the plant. Though the numbers of bacteria are 10–100 fold higher in the rhizosphere region, only 2–5% of rhizobacteria qualify to be a PGPR species (Antoun and Pre’vost 2005). Some of the PGPR so far identified include Alcaligenes, Arthrobacter, Azospirillum, Azotobacter, Bacillus, Bradyrhizobium, Burkholderia, Enterobacter, Flavobacterium, Klebsiella, Mesorhizobium, Pseudomonas, Rhodococcus, Streptomyces, Serratia, etc. (Tariq et al. 2017).
3.2 Root Colonization Traits of Bacillus
PGPR are effective colonizers of the root niches. PGPR display strong growth—competitive and colonizing skills over existing rhizosphere communities. While the colonizing traits of gram-negative PGPR is well understood, the underlying mechanisms and dynamics of colonization by gram positive bacteria are continued to be explored. Some of the competing colonizing traits of all genera of PGPR include chemotactic motility, higher growth rate, better adhesion to the roots, and synthesis of certain amino acids, vitamins, and some nucleotide bases such as uracil and biofilm formation (Lugtenberg and Kamilova 2009). There are many molecular mechanisms, operons, and sensory systems that are being discovered to explain the colonization behavior of PGPR.
3.2.1 Chemotactic Motility
Chemotactic motility is a colonization trait of PGPR genera Chemotaxis can be defined as a directed movement of an organism toward chemical stimuli. It allows bacterial cells to advance toward a chemical attractant or move away from a repellant. PGPR can respond to the secretion of a variety of substances termed chemoeffectors (either an attractant or repellant) in root exudates. The event of chemotactic response is a crucial step in root colonization. Chemoeffectors found in root exudates include organic acids amino acids, polyamines, sugars, alcohols, fatty acids, purines, phytohormones, flavonoids, terpenoids, and benzoxazinoids (Pétriacq et al. 2017) Some of the components that can trigger chemotactic response in Bacillus sp. are listed in the Table 3.1.
Among these, amino acids, sugars, and organic acids are the major chemoeffectors that attract PGPR. Chemotaxis is initiated through chemosensory system. Chemosensory system consists of a ternary complex made up of chemoreceptors (also known as methyl-accepting chemotaxis proteins [MCPs]) and transducer-like proteins or Tlps which consists of the CheA histidine kinase and the CheW coupling protein which can couple the CheA activity to the receptor control (Ortega et al. 2017). Figure 3.1 describes the chemosensory pathway in bacteria.
Chemoreceptors are located on the surface of bacteria. The structure of chemoreceptors can vary among bacteria. Most of the chemoreceptors have a transmembrane topology, while some of them are entirely cytosolic. Chemoreceptors are known to contain two domains or modules—an input module and an output module. The input module/domain is found extracellularly, which is also referred to as sensory domains or more commonly the ligand-binding domain (LBD). The output module is called cytoplasmic signaling domain which is a conserved structure known to be composed of four-helix bundle in antiparallel coil. Approximately 80% of the ligand-binding domain (LBD) belongs to any one of the three categories such as 4HB type (four-helix bundle), cache type and the PAS (Per-Arnt-Sim proteins) type.
In Bacillus subtilis, as many as ten chemoreceptors (MCPs) have been identified, which includes Mcp A, Mcp B, Mcp C, Tlp A, Tlp B, Tlp C, YfmS, YoaH, YvaQ, and Hem AT (Ortega et al. 2017; Feng et al. 2018). McpC is a broad range receptor that can bind to almost all amino acids except L asparagine. While McpB is identified as the receptor for L-asparagine. Mcp R is unique to Bacillus amyloliquefaciens which can specifically bind the amino acid arginine (Feng et al. 2018). HemAT is a myoglobin like receptor which aids in aerotaxis.
Chemoreceptors can recognize signals when chemoeffectors bind to their ligand-binding domain (LBD). This triggers a canonical signal transduction system in which CheA histidine kinase phosphorylates the CheY receptor control protein. This receptor signaling system displays a kinase on-off state. In either state, the CheY protein modifies or controls the flagellar motor rotation to switch from tumbling to swimming motility or vice versa. In the presence of attractant stimuli, the receptor signaling system shifts to the kinase-off state where the flux of phosphoryl groups from CheA to CheY slows down, forcing the flagellar motor to rotate in CCW (counterclockwise) direction resulting in a run. Conversely, kinase-on state phosphorylates the CheY protein which turns the flagellar motor to CW (clockwise) direction resulting in a tumble. Based on the chemoreceptor attractant-repellant signaling, bacterial cells respond to chemoeffectors by suppressing the CW signals during runs to take them in their favorable direction (Parkinson et al. 2015).
3.2.2 Biofilm Formation
Biofilms are structured communities of cells that are adherent to a surface, an interface, or each other and are encased in a self-produced polymeric matrix (Stanley et al. 2003). Bacterial biofilms established on plant roots by PGPR can protect the colonization sites and act as a sink for the nutrients in the rhizosphere. It prevents access to the root exudates by the plant pathogens which will otherwise colonize the root niche and establish infection. Biofilm formation is a root-colonizing trait of PGPR. It is stated that an efficient PGPR will be competent in biofilm formation since many genes involved in biofilm formation are also engaged in root colonization process. In fact, the eps and Tas A mutants (eps and Tas A are genes responsible for biofilm formation in B. subtilis) failed to colonize the roots (Beauregard et al. 2013). Hence, matrix formation is an essential feature for root colonization.
The extracellular matrix (ECM) that is produced by B. subtilis is mainly composed of an exopolysaccharide (EPS) and the protein TasA (Branda et al. 2006) both of which are extremely essential for root colonization. The EPS and the Tas A protein polymerize into amyloid-like fibers. Three master transcriptional regulators are identified in Bacillus sp. which are responsible for switching from motile individual cells to biofilm forming state. Those are DegU for exoprotease secretion, ComA for competence and surfactin production, and Spo0A for matrix production and, eventually, spore formation (Blake et al. 2021).
The eps A-O operon and tas A operon are under the direct control of the master transcriptional regulator spoOA which controls the transcription of two regulatory genes sinI and sinR. The product of the gene sinR (sin R) acts as a repressor that binds to the promoter region of the eps operon. This represses the transcription of the matrix genes responsible for biofilm formation. Mutants of sinR always produce rugose colonies consisting of chains of nonmotile cells in robust multicellular structures. In contrast, the product of sinI (sin I) is an antagonist to sin R. It counteracts the sinR-mediated repression of the eps operon and activates the genes responsible for biofilm formation. Mutants of sinI always produce motile cells that do not form chains. Phosphorylated form of spoOA stimulates the synthesis of sin I protein which, in turn, can activate the matrix genes. Unphosphorylated form of spoOA activates sin R which is a repressor that shuts off the production of matrix genes. Bacillus subtilis exists in two mutually exclusive physiological states where cells grow either in bundled chains or motile single cells. It is speculated that sinI/sinR plays the role of switch between these two physiological states that determine biofilm production by the species (Kearns et al. 2005).
DegU helps in biofilm formation by way of repressing motility genes and by regulating the synthesis of poly-γ-glutamic acids (PGA), which are needed for a stable biofilm. ComA is indirectly involved in biofilm formation by regulating the production of surfactin which is a signaling molecule in biofilm formation (Nakano et al. 1991). Bacillus sp. are efficient biofilm producers which makes them an effective PGPR.
3.3 Plant Growth-Promoting Traits of Bacillus
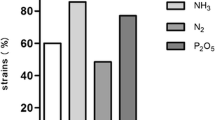
Bacillus sp. readily qualifies as a PGPR since it possesses all PGPR traits such as phytohormone production, mobilization of nutrients, bioactivity against plant pathogens, and reduction of biotic and abiotic stress of the plants (Fig. 3.2).
3.3.1 Phytohormone Production
Plant growth hormones such as auxins, gibberellins, cytokinins, ethylene, and abscisic acid are implicated in all aspects of plant growth and development.
Indole acetic acid (IAA) is the predominant type of auxin produced by plants. Plant cell division, enlargement, response of plants to light and gravity, and tissue differentiation are few processes that are controlled by auxin (Teale et al. 2006). It is also involved in initiation of lateral and adventitious roots, mediates growth events like florescence and photosynthesis, and is known to influence metabolite productions and affect responses to biotic and abiotic stresses. Bacteria use five different pathways using tryptophan as the precursor molecule for the synthesis of IAA, viz., indole-3-acetamide pathway, indole-3-pyruvate pathway, tryptamine pathway, tryptophan side-chain oxidase pathway, and indole-3-acetonitrile pathway (Spaepen and Vanderleyden 2011). Bacillus sp. are known to produce IAA in quantities up to 330 μg mL−1 (Wagi and Ahmed 2019). While the role of IAA is clear in plants, its function in bacterial growth is fairly ambiguous. It plays an important role in plant microbe interaction as in the pathogenesis by tumorigenic bacteria, root proliferation by PGPR, legume-Rhizobium symbiosis, and auxin signaling in plant defense.
IAA is involved in phytopathogenesis as evident from gall/tumor formation by Agrobacterium sp. where the genes responsible for IAA production are transferred by the bacterium into the plants. IAA synthesis is also involved in the pathogenesis of other bacteria such as Ag. rhizogenes, Ps. savastanoi, and Pa. agglomerans pv. gypsophilae. IAA production by rhizobacteria is a significant PGPR trait which results in plant growth promotion. About 80% of rhizosphere bacteria are able to synthesize IAA. In the root microniche, secretion of IAA by bacteria induces root proliferation, faster emergence of seedling, and improved seedling vigor. IAA synthesis by bacteria can actually be a detoxifying mechanism since tryptophan, the precursor of IAA, is toxic at higher levels (Bar and Okon 1992). The role of IAA in legume-Rhizobium synthesis is also well documented. IAA is also known to be involved in plant defense against pathogens. Plant defense genes are repressed by auxin signaling, thereby enhancing the vulnerability of plants to bacterial infection (Navarro et al. 2006). There is a link between auxin signaling and plant defense which the bacteria can employ as a colonization strategy. IAA can act as a signaling molecule in microorganisms regulating their IAA biosynthesis, vir gene expression in Agrobacterium, virulence, and infection by pathogens (Spaepen et al. 2007).
Gibberellins are tetracyclic diterpenoid acids consisting of 6-5-6-5 tetracyclic ent-gibberellane backbone and produced by plants, fungi, and bacteria (Bottini et al. 2004). Gibberellins are found to influence various physiological events in plants that involve seed germination, development of seedling, elongation of stem and internodes, induction of flowering, and ripening of fruit (King and Evans 2003). The bioactive gibberellins belong to GA1, GA 3, GA4, and GA7 classes. There is no apparent role of gibberellins in bacteria except that it can act as a signaling molecule toward the host. Besides this, gibberellins can also regulate auxin-related genes and can significantly influence other plant growth regulatory hormones (Castro-Camba et al. 2022). Bacillus methylotrophicus, Brevibacillus, Lysinibacillus, Bacillus subtilis, Bacillus cereus, Bacillus amyloliquefaciens, Bacillus siamensis, Bacillus pumilus, and Bacillus licheniformis are some of the species of Bacillus that can synthesize gibberellins (Gohil et al. 2022; Radhakrishnan and Lee 2016; Naveena and Gowrie 2019).
3.3.2 Nutrient Mobilization in the Rhizosphere
Plants require macronutrients like nitrogen, phosphorus, and potassium and micronutrients such as zinc, iron, magnesium, manganese, etc. for their growth. Most of them are insoluble or unavailable to the plants. PGPR can mobilize these nutrients and supply them to plants. The presence of nif gene cluster in Bacillus enables efficient nitrogen fixation. Paenibacillus sp. consists of nine genes (nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA, nifV) (Shi et al. 2016). Several species such as Bacillus tequilensis, B. megaterium, B. cereus, B. amyloliquefaciens, B. aryabhattai, B. safensis, B. aerophilus, B. subtilis, B. rhizosphaerae, B. pumilus, B. fluminensis and B. indica, and B. mycoides B. aerophilus, B. oceanisediminis, B. flexus, B. aquimaris, B. vietnamensis, and B. subterraneaus, B. marisflavi, and Paenibacillus massiliensis can fix atmospheric nitrogen for plant uptake (Singh et al. 2020; Ding et al. 2005). It is reported that B. megaterium could demonstrate high nitrogen fixing potential (210.05 ± 7.0 nmol C2H4/mg protein/day (Yousuf et al. 2017).
Iron acquisition by Bacillus is mediated through siderophore, a low molecular weight metal-chelating compound produced under iron-deficient conditions. Siderophores are octahedral structures that can bind to Fe (III) atom coordinated with oxygen atoms. Gram-positive bacteria chelate ferric ion and transport it across cell membrane. Depending on the chemical nature, siderophores are classified into hydroxamate, catecholate, carboxylate, and mixed-type siderophores.
Hydroxamate siderophores consist of C(=O)N-(OH)R structure, with R being an amino acid or its derivative. It forms a bidentate ligand with Fe ions and oxygen atoms. Each siderophore is capable of forming octahedral complex compounds with Fe(III) ions. Rhizobactin, shizokinen, and vicibactin are some of the examples of hydroxamate type. Bacillus megaterium is known to produce two hydroxamate siderophores.
In catecholate-type siderophores, the Fe(III) ion is bound to hydroxyl or catecholate groups. When it chelates with Fe(III) atom, a hexadentate-octahedral complex is formed along with two oxygen atoms from each catecholate group. Catecholate siderophores are derivatives of salicylic or 2,3-dihydroxybenzoic acid. Bacillibactin, aminocholine, spirilobactin, nitrocholine, protochelin, 2,3-DHBA, and agrobactin are some catecholate-type siderophores produced by different bacteria.
In carboxylate type of siderophores, the Fe-chelating groups are the carboxyl and hydroxyl groups. Staphyloferrin A and B, rhizobactin, and rhizoferrin are some of the carboxylate siderophores produced by Staphylococcus, Rhizobium sp., and Mucorales respectively. Mycobactin, Pyoverdine, and azobactin are some of the other types of siderophores characterized (Singh et al. 2022). Bacterial siderophores provide plants with iron and promote their growth when the bioavailability of iron is low. Bacterial siderophores complexed with iron are reduced to give Fe(II) back to the plant transport system. In this mechanism, Fe(III) siderophores from bacteria are first transported to the plant root apoplast, where siderophore reduction occurs resulting in higher concentration of iron near the root surface. The second mechanism is mediated through ligand exchange between bacterial siderophores and phytosiderophores (Timofeeva et al. 2022). Siderophores can also bind with toxic metals chromium Cr3+, copper Cu3+, Cu2+, lead Pb2+, vanadium V4+, and aluminum Al3+ and reduces the toxicity rendered to plants by these metals. Siderophore-producing Bacillus species are also capable of removing heavy metals and are bioremediating in nature in heavy metal polluted sites.
Mobilization of phosphorus is yet another feat of Bacillus sp. Phosphate-solubilizing Bacillus sp. employ one of the many strategies such as production of phosphatase enzyme, secretion of organic acids, chelation, and ion exchange to solubilize phosphorus to be made available to plants. B. mucilagenosus and B. edaphicus are also known to solubilize potassium rock through secretion of organic acids (Han and Lee 2005).
3.3.3 Bioactive Potential Against Pathogens
Members of the Bacillus genus exhibit strong activity against plant pathogens. As an efficient colonizer of plant roots, Bacillus sp. target the pathogens through synthesis and secretion of antagonistic substances, induced systemic resistance (ISR), and competitive occupation of the root niches.
About 5–8% of the genome of Bacillus sp. is devoted for synthesis of bioactive secondary metabolites that are antagonistic substances directed to inhibit pathogens A wide variety of antagonistic substances like non-ribosomally synthesized peptides and lipopeptides, polyketide compounds, bacteriocins, and siderophores are employed by Bacillus sp. to inhibit pathogens in the root environment (Fira et al. 2018). Lipopeptides are classified into three families, namely, the iturin, fengycin, and surfactin lipopeptide families. Bacillomycin D and mycosubtilin are compounds of the iturin family produced by Bacillus sp., which are potent antifungal compounds. Fengycin is also a strong antifungal compound. Surfactin compounds are implicated in biofilm formation which is a root-colonizing trait of the bacterium. Zwittermicin A is an aminopolyol antibiotic which demonstrates broad spectrum activity against other microbes. Volatile organic compounds (VOCs), such as 2,3-butanediol, are produced by B. subtilis and B. amyloliquefaciens. These are implicated on biocontrol of pathogens and elicit induced systemic resistance (ISR) of plants (Saraf et al. 2014). Research revealed benzaldehyde, 1,2-benzisothiazol-3(2 H)-one, and 1,3-butadiene are VOCs that are produced by Bacillus sp. which had strong inhibitory activity against bacterial wilt disease pathogen, Ralstonia solanacearum (Tahir et al. 2017a). Bacillus sp. produce extracellular enzymes such as chitinase, β-1,3-glucanase and β-1,4-glucanase, laminarinase, oxalate oxidase, cellulases, proteases, phytases, and lipases. These enzymes can degrade the cell wall components of fungi which can be seen as a mechanism of warding off of fungal pathogens (Kumar et al. 2012).
Induced systemic resistance (ISR) is a mechanism of plant defense in which plants are preconditioned by appropriate stimulation or interaction to resist subsequent infection by pathogens. Plants can recognize and distinguish between invading pathogenic and nonpathogenic microorganisms and activate appropriate defense-related mechanisms. Microbial- or pathogen-associated molecular patterns (MAMPs or PAMPs) such as flagellin or fungal chitin can be easily recognized by plants through their transmembrane pattern recognition receptors (PRRs) to direct the first level of immune defense called pattern-triggered immunity (PTI). PTI mechanism fails when pathogens secrete virulence effectors into plant cells. In response to these effectors, induction of second layer of immunity called effector-triggered immunity (ETI) is sequestered. Pathogens often evade both PTI and ETI. Induced systemic resistance come into play when colonization of beneficial bacteria “prime” the host plants against future infections by pathogenic bacteria. This phenomenon of induced systemic resistance was originally thought to be routed through activation of jasmonic acid (JA) and ethylene (ET) pathway rather than salicylic acid (SA) and PR-related defense proteins which is a feature of systemic acquired resistance (SAR) but now found to be a complex interplay of both pathways (Yu et al. 2022).
ISR by Bacillus sp. against plant diseases is listed in Table 3.2. ISR in plants commence when plants can significantly recognize MAMPs from beneficial bacteria through receptors and elicit PTI. MAMPs may include flagellin, LPS, and oligo and polysaccharides. Flagellin sensing (FLS2) receptors are some proteins found in plants to sense flagellin from Bacillus subtilis. PTI triggered from beneficial bacteria causes less cellular damage than pathogen-induced PTI which helps in easy colonization of plant roots by beneficial bacteria. Once ISR is initiated, early ISR events will begin to unfold in plants which include expression of PR genes and production of ROS, volatiles, phenyl alanine ammonia lyase, polyphenol oxidase, peroxidase, β-1, 3 glucanase, and chitinase depending on the nature of the invading plant pathogen. These events can phenomenally protect the plants against the attack of the plants by pathogens by exerting certain specific mechanisms such as antagonistic and antimicrobial effects, reduction of oxidative stress, promotion of plant growth, inhibition of spore germination, hyphal growth, stomatal regulation, etc. In a study of mechanism of ISR by PGPR B. pumilus strain SE34 in pea plants, primed roots limited the spread of Fusarium oxysporum f. sp. pisi in epidermis and outer cortex, while the non-primed roots had extensive invasion of cortex, endodermis, up to vascular stele. There are several reports of Bacillus species involved in ISR against diseases like stem blight, crown rot, leaf spot, systemic viral infections, blue mold and wilts (Kloepper et al. 2004).
3.3.4 Reduction in Plant Stress
Stress, both biotic and abiotic, can lead to adverse effects on plant growth. Abiotic stresses such as low or high temperature, high salinity, drought, UV radiation, heavy metal, nutrient deficiency, etc. can hinder plant growth. PGPR-seeded plants can combat these stresses efficiently.
Salinity tolerance is imparted to plants by the production of exopolysaccharide (EPS) by the root-colonizing PGPR. Salinity levels, low pH, and high metal ions are some environmental triggers that activate the gene eptA coding for the enzyme phosphoethanolamine-lipid-A transferase (EptA) which regulates the synthesis of EPS. Hence, under high salinity levels, EPS accumulation covers the root surface and prevents the influx of Na + ions into the roots. Inoculation of plants with Bacillus subtilis GB03 is known to downregulate the HKT (high-affinity K+ transporter) expression, which reduces Na+ uptake, thus providing salt tolerance to plants (Zhang et al. 2008). Production of ACC deaminase lowers the level of ABA in plants which can limit accumulation of Na + imparting salt tolerance. ACC deaminase production also enhances salt tolerance by increasing the activity of antioxidant enzymes, such as superoxide dismutase, peroxidase, and catalase, and upregulation of ROS pathway genes (Habib et al. 2016).
Under water-deficit stress, some Bacillus sp. that are ABA (abscisic acid) producers confer salt and water tolerance. ABA increases the accumulation of ROS which controls the Ca2+ channels that are involved in stomatal conductance and stomatal closure, thereby reducing water loss (Goswami and Deka 2020; Grover et al. 2021). Another possible mechanism of water-deficit stress management could be the increase in the soluble sugar content mediated by PGPR. Accumulation of soluble sugars results in drought resistance since sugars are known to function as osmoprotective agents (Sánchez et al. 1998). Cold stress tolerance is also imparted on plants when Bacillus sp. modulates the expression of the ABA-related genes.
The signaling pathways of stress hormones such as salicylic acid (SA) and abscisic acid (ABA), which are the central regulators of biotic and abiotic stresses in plants, are known to be modified by PGPR. This reprogramming of expression of SA and ABA by PGPR opens up a myriad of alteration of expression of other stress-related genes converging in reduced stress. Furthermore, PGPR-treated plants show decreased antioxidant enzyme levels indicating minimal occurrence of oxidative damage by ROS accumulation when compared to the stressed plants.
3.4 Bacillus as PGPR in Post-mined Lands
Bacillus sp. isolated from post-mined and revegetated mine soil and identified as PGPR has proven to be an efficient bioinoculant during the revegetation process. Open cast mining employs manual, semi-mechanized, and fully mechanized methods to remove the overburden present on the upper layers of soil and is mainly used to extract energy resources like coal and lignite and other minerals. The excavated soil which is termed as overburden is dumped near pre-excavated sites to be subjected to reclamation processes. This overburden poses a huge threat to ecosystem owing to the presence of elevated levels of heavy metals and increased compaction and sand content with low organic matter content and nutrient availability and less microbial population (Sheoran et al. 2008). Extraction of deep-seated metals and minerals from the earth generates huge amount of overburden and is responsible for biodiversity loss, soil erosion, loss of ecosystem balance, soil, air and water pollution, and loss of livelihood (Wong 2003).
Revegetation has become one of the predominant and traditional methods of ecological restoration of mine land ecosystem. During revegetation, successful establishment of plant or tree species is very important in stabilizing the disordered mined land not only through providing a good ground cover but also reengineer the disturbed soil ecosystem back to near normal health. Seeding the plants intended for revegetation can be a huge advantage since they supply the plants with the proven benefits that are tagged with PGPR. It can reestablish the biogeochemical cycling of nutrients; restore the altered physicochemical properties of the soil such as pH, bulk density, CEC, and NPK levels; remove heavy metals and other toxicities; and improve the biological properties of the soil. When applied along with organic amendments, PGPR can readily colonize the rhizosphere and exert its benefits. Soil pH can be brought to near neutral, compaction of soil indicated by high bulk density (which is detrimental to plants and microbes) is lowered to less than 1.5 g/cc, the numbers of microbiota is increased, soil texture and structure is improved, concentration of heavy metals can be reduced, and levels of soil NPK can be significantly raised to productive levels. Several research works indicate the successful use of PGPR on the reclamation of post-mined soils. Bacillus paramycoides, Bacillus subtilis, and Brevibacterium were successfully employed in restoration of phosphate mined soils (Mghazli et al. 2022). Bacillus subtilis, Lysinibacillus fusiformis, Bacillus cereus, Brevibacillus borstelensis, and Bacillus amyloliquefaciens isolated from lignite mine soils have proven PGPR abilities (Naveena and Gowrie 2018).
References
Antoun H, Pre’vost D (2005) Ecology of plant growth promoting rhizobacteria. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 1–38
Bar T, Okon Y (1992) Induction of indole-3-acetic acid synthesis and possible toxicity of tryptophan in Azospirillum brasilense Sp7. Symbiosis 13:191–198
Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolte R (2013) Bacillus subtilis biofilm induction by plant polysaccharides. PNAS 110(17):E1621–E1630
Blake C, Christensen MN, Kov ́acs AT (2021) Molecular aspects of plant growth promotion and protection by Bacillus subtilis. Mol Plant Microbe Interact 34(1):15–25
Bottini R, Cassán F, Piccoli P (2004) Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol 65:497–503
Branda SS, Chu F, Kearns DB, Losick R, Kolter R (2006) A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238
Castro-Camba R, Sánchez C, Vidal N, Vielba JM (2022) Interactions of gibberellins with phytohormones and their role in stress responses. Horticulturae 8:241
Chakraborty U, Chakraborty B, Basnet M (2006) Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. J Basic MicrobiolJ Basic Microbiol 46:186–195
Chen L, Wang X, Ma Q, Bian L, Liu X, Xu Y, Zhang H, Shao J, Liu Y (2020) Bacillus velezensis CLA178-induced systemic resistance of Rosa multiflora against crown gall disease. Front MicrobiolFront Microbiol 11:587667
Chowdappa P, Kumar SPM, Lakshmi MJK, Upreti K (2013) Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol ControlBiol Control 65:109–117
Chung EJ, Hossai MT, Khan A, Kim KH, Jeon CO, Chung YR (2015) Bacillus oryzicola sp. nov., an endophytic bacterium isolated from the roots of Rice with antimicrobial, plant growth promoting, and systemic resistance inducing activities in Rice. Plant Pathol JPlant Pathol J 31(2):152–164
Ding Y, Wang J, Liu Y, Chen S (2005) Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region. J Appl Microbiol 99(5):1271–1281
Feng H, Zhang N, Du W, Zhang H, Liu Y, Fu R, Shao J, Zhang G, Shen Q, Zhang R (2018) Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol Plant-Microbe Interact 31(10):995–1005
Fira D, Dimkić I, Berić T, Lozo J, Stanković S (2018) Biological control of plant pathogens by bacillus species. J BiotechnolJ Biotechnol 10(285):44–55
Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW (2012) The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol ChemJ Biol Chem 287:39412–39418
Gohil RB, Raval VH, Panchal RR, Rajput KN (2022) Plant growth-promoting activity of bacillus sp. PG-8 isolated from fermented Panchagavya and its effect on the growth of Arachis hypogea. Front Agron 4:805454
Goswami M, Deka S (2020) Plant growth-promoting rhizobacteria—alleviators of abiotic stresses in soil: a review. Pedosphere 30:40–61
Grover M, Bodhankar S, Sharma A, Sharma P, Singh J, Nain L (2021) PGPR mediated alterations in root traits: way toward sustainable crop production. Front Sustain Food Syst 4:1–28
Habib SH, Kausar H, Saud HM (2016) Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed Res IntBiomed Res Int 2016:6284547
Han HS, Lee KD (2005) Phosphate and potassium solubilizing bacteria effect on mineral uptake, soil availability, and growth of egg plant. Res J Agric Biol Sci 1:176–180
Hanlon DW, Ordal GW (1994) Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol ChemJ Biol Chem 269:14038–14046
Jiang C, Li Z-J, Tang S-Y et al (2022) Plant growth-promoting rhizobacterium AR156 - induced systemic resistance against multiple pathogens by priming of phytoalexin synthesis and secretion. Authorea
Kearns DB, Chu F, Branda SS, Kolter R, Losick R (2005) A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55(3):739–749
King RW, Evans LT (2003) Gibberellins and flowering of grasses and cereals: prising open the lid of the “florigen” black box. Annu Rev Plant Physiol Plant Mol Biol 54:307–328
Kloepper JW, Ryu C-M, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Kumar P, Dubey RC, Maheshwari DK (2012) Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res 167:493–499
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annual reviews of. Microbiology 63:541–556
Mghazli N, Bruneel O, Zouagui R, Hakkou R, Sbabou L (2022) Characterization of plant growth promoting activities of indigenous bacteria of phosphate mine wastes, a first step toward revegetation. Front MicrobiolFront Microbiol 13:1026991
Nakano MM, Xia LA, Zuber P (1991) Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol 173:5487–5493
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Naveena ML, Gowrie U (2018) Identification of plant growth promoting rhizobacteria (PGPR) from revegetated soils of lignite mines of Neyveli, Tamilnadu. Int J Pharm Bio Sci 9(1):121–129
Naveena ML, Gowrie U (2019) Potential metabolites of Bacillus subtilis strain isolated from Rhizospheric soils of revegetated mine spoil dumps. Indian J Environ Prot 39(10):938–944
Ortega Á, Zhulin IB, Krell T (2017) Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol RevMicrobiol Mol Biol Rev 81:e00033–e00017
Parkinson JS, Hazelbauer GL, Falke JJ (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends MicrobiolTrends Microbiol 23(5):257–266
Pétriacq P, Williams A, Cotton A, McFarlane AE, Rolfe SA, Ton J (2017) Metabolite profiling of non-sterile rhizosphere soil. Plant JPlant J 92:147–162
Radhakrishnan R, Lee IJ (2016) Gibberellins producing bacillus methylotrophicus KE2 supports plant growth and enhances nutritional metabolites and food values of lettuce. Plant Physiol BiochemPlant Physiol Biochem 109:181–189
Rajkumar K, Naik MK, Amaresh YS, Chennappa G (2018) Induction of systemic resistance by Bacillus subtilis isolates against fusarium wilt of chilli. Int J Curr Microbiol App Sci 7(7):2669–2680
Sánchez FJ, Manzanares MA, de Andres EF, Tenorio JL, Ayerbe L (1998) Turgor maintenance, osmotic adjustment and soluble sugar and proline accumulation in 49 pea cultivars in response to water stress. Field Crop Res 59:225–235
Saraf M, Pandya U, Thakkar A (2014) Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol Res 169:18–29
Sheoran AS, Sheoran V, Poonia P (2008) Rehabilitation of mine degraded land by metallophytes. Min Eng J 10(3):11–16
Shi HW, Wang LY, Li XX, Liu XM, Hao TY, He XJ, Chen SF (2016) Genomewide transcriptome profiling of nitrogen fixation in Paenibacillus sp. WLY78. BMC MicrobiolBMC Microbiol 16:25
Singh RK, Singh P, Li HB, Song QQ, Guo DJ, Solanki MK, Verma KK, Malviya MK, Song XP, Lakshmanan P, Yang LT, Li YR (2020) Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: a comprehensive study of plant-microbe interactions for growth enhancement in saccharum spp. BMC Plant BiolBMC Plant Biol 18; 20(1):220
Singh P, Chauhan PK, Upadhyay SK, Singh RK, Dwivedi P, Wang J, Jain D, Jiang M (2022) Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded land. Front MicrobiolFront Microbiol 13:898979
Spaepen S, Vanderleyden J (2011) Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol 3:a001438
Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plants signalling. FEMS Microbiol Rev 31:425–448
Stanley NR, Britton RA, Grossman AD, Lazazzera B (2003) Identification of catabolite repression as a phys-iological regulator of biofilm formation by bacillus subtilisusing DNA microarrays. J Bacteriol 185:1951–1957
Tahir HAS, Gu Q, Wu H et al (2017) Effect of volatile compounds produced by Ralstonia solanacearum on plant growth promoting and systemic resistance inducing potential of bacillus volatiles. BMC Plant Biol 17:133
Tahir HA, Gu Q, Wu H, Niu Y, Huo R, Gao X (2017a) Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci RepSci Rep 7:40481
Tariq M, Noman M, Ahmed T, Hameed A, Manzoor N, Zafar M (2017) Antagonistic features displayed by plant growth promoting rhizobacteria (PGPR): a review. J Plant Sci Phytopathol 1:038–043
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859
Timofeeva AM, Galyamova MR, Sedykh SE (2022) Bacterial siderophores: classification, biosynthesis, perspectives of use in agriculture. Plants 11(22):3065
Tohidifar P, Bodhankar GA, Pei S, Cassidy CK, Walukiewicz HE, Ordal GW, Stansfeld PJ, Rao CV (2020) The unconventional cytoplasmic sensing mechanism for ethanol chemotaxis in Bacillus subtilis. MBio 11:e02177–e02120
Wagi S, Ahmed A (2019) Bacillus spp.: potent microfactories of bacterial IAA. PeerJ 7:e7258. https://doi.org/10.7717/peerj.7258. PMID: 31372316
Wong MH (2003) Ecological restoration of mine degraded soils, with emphasis on metal contaminated soils. Chemosphere 50:775–780
Xie S, Jiang H, Ding T, Xu Q, Chai W, Cheng B (2018) Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol Plant PatholMol Plant Pathol 19(7):1612–1623
Xing Z, Wu X, Zhao J et al (2020) Isolation and identification of induced systemic resistance determinants from Bacillus simplex sneb 545 against Heterodera glycines. Sci Rep 10:11586
Yousuf J, Thajudeen J, Rahiman M, Krishnankutty S, Alikunj P, A, A Abdulla MH. (2017) Nitrogen fixing potential of various heterotrophic bacillus strains from a tropical estuary and adjacent coastal regions. J Basic MicrobiolJ Basic Microbiol 57(11):922–932
Yu Y, Gui Y, Li Z, Jiang C, Guo J, Niu D (2022) Induced systemic resistance for improving plant immunity by beneficial microbes. Plan Theory 11:386
Zhang H, Kim MS, Sun Y, Dowd SE, Shi H, Paré PW (2008) Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol Plant-Microbe InteractMol Plant-Microbe Interact 21:737–744. https://doi.org/10.1094/MPMI-21-6-0737
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Linnet Naveena, M. (2024). Plant Growth-Promoting Traits of Bacillus and Related Genera. In: Mageshwaran, V., Singh, U.B., Saxena, A.K., Singh, H.B. (eds) Applications of Bacillus and Bacillus Derived Genera in Agriculture, Biotechnology and Beyond. Microorganisms for Sustainability, vol 51. Springer, Singapore. https://doi.org/10.1007/978-981-99-8195-3_3
Download citation
DOI: https://doi.org/10.1007/978-981-99-8195-3_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-8194-6
Online ISBN: 978-981-99-8195-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)