Abstract
Classification is a supervised learning model where the class labels are accurately identified for future samples. Medical data is an important source for understanding and improving health outcomes and classification algorithms are often used to analyze these data. Learning models give significant experiences into the situational needs of patients. Various hypotheses have been carried out on different datasets yet it is truly challenging to track down which model is suitable. Proposed work compares the performance of classification models like LR, DT, SVM, NB, KNN, and RF on various datasets. SVM classifier yields accuracy of 0.59 for the Diabetic dataset as it considers individual model opinion, while RF classifier surpassed them both with accuracy 0.9974 for the breast cancer Wisconsin dataset since it is an ensemble approach that takes majority opinions. These findings highlight the need for careful consideration of the choice of classification model when analyzing medical data and provide valuable insights for researchers and practitioners working with these data.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Classification techniques are mostly used in the field of medical research to predict the livelihood of a patients having various disease or condition based on their medical history and other relevant features. These techniques have the potential to dramatically increase the accuracy as well as efficiency of diagnosis, treatment, and prognosis making them an important tool in the practice of medicine. Below are some of the classification techniques, their merits, and demerits.
1.1 Logistic Regression
Binary classification challenges are handled by the nominal/ordinal machine learning technique known as logistic regression. It is frequently used in the medical sector to forecast the possibility of a specific result, such as the likelihood that a patient would contract a specific disease or the likelihood that they will respond to a specific therapy. The chance that a patient has a specific disease, for instance, might be predicted using a logistic regression model based on symptoms, test findings, and other characteristics.
One of the main advantages of using logistic regression in the medical field is to implement and can handle huge amount of data types, including continuous and categorical variables. It can also provide insights into the relationships between different features (e.g., symptoms and test results) and the likelihood of a particular outcome. However, logistic regression is a linear model and can only capture linear correlation between the input features and the output which might be a limitation when the data is not linearly separable or when there are nonlinear relationships in the data that are important to consider. Logistic regression can also be sensitive to the presence of outliers in the data which can affect the model’s performance [1,2,3,4].
1.2 Decision Tree
A well-liked machine learning model called decision trees is utilized for both classification and regression problems. They function by building a tree-like structure, in which the leaf nodes indicate the predicted class or value and the inside nodes reflect judgments depending on the values of the input characteristics. Decision trees may be used for binary and multi-class classification tasks and can handle continuous and categorical information.
Decision trees have been used in the medical sector for a range of tasks, including determining the variables that affect the risk that a patient would contract a specific disease or forecasting the efficacy of a specific treatment for a specific patient. They have also been used to predict the likelihood of a patient being readmitted to the hospital. Decision trees are often chosen for these tasks because they are easy to interpret and implement and can often achieve good performance on many types of data. Decision trees may not generalize well to novel, untested data and might be prone to over fitting. They can also be computationally expensive to build and use which can be a drawback when working with larger datasets [5].
1.3 Support Vector Machine
The machine learning approach known as support vector machines (SVMs) is utilized for both classification and regression tasks. SVMs have been utilized in the medical profession for a number of purposes, including forecasting the chance that a patient would develop a certain disease, the likelihood that a patient will react to a specific therapy, and the risk that a patient will be readmitted to the hospital.
One of the main advantages of using SVMs in the medical field is that they can handle high-dimensional data and can find complex, nonlinear relationships in the data. They are also robust to noise and can handle large datasets efficiently. However, the choice of kernel and other hyperparameters, which might have an impact on the model's performance, is one SVM restriction. In addition, SVMs can be computationally expensive to train which can be a drawback when working with larger datasets. It is important for researchers to carefully evaluate the performance of SVM models and to consider alternative algorithms when appropriate [4,5,6,7].
1.4 Naive Bayes Classifier
Naive Bayes is an algorithm which is based on the concept of Bayes Theorem and is a probabilistic algorithm that makes predictions about the likelihood of an event based on prior knowledge and statistical data; Naive Bayes may be used in the medical industry to forecast a patient’s chance of having a certain disease based on their symptoms or their likelihood of benefiting from a particular therapy, among other things.
The Naive Bayes algorithm's relative simplicity and ease of use are two of its key features. It also performs well when dealing with large amounts of data and can be used to make predictions in real time. The Naive Bayes algorithm's fundamental drawback is that it assumes that all characteristics are independent of one another, which may not always be the case in real-world scenarios. This can lead to less accurate predictions compared to other algorithms that do not make this assumption [1].
1.5 K-Nearest Neighbors
The machine learning technique K-nearest neighbors (KNN) is utilized for both classification and regression applications. It has been used in the medical industry for a range of purposes, including forecasting the risk that a patient would contract a certain illness or the efficacy of a specific treatment for a specific patient. A prediction is made using the class labels or values of the K data points in the training set that are closest to the new data point in KNN.
One of the main advantages of using KNN in the medical field is that it is simple to implement, doesn’t involve a training phase, and can handle huge data types that includes continuous and categorical variables. KNN is also flexible and can be used for multi-class and binary classification tasks. However, one limitation of KNN is expensive to use, particularly when it is used with large datasets. The choice of K and the distance metric employed to gauge similarity between data points can both have an impact on how well KNN performs. It is important for researchers to carefully evaluate the performance of KNN models and to consider alternative algorithms when appropriate [8, 9].
1.6 Random Forests
An ensemble machine learning approach known as random forests is utilized for both classification and regression problems. They have been used in the medical industry for a range of purposes, including forecasting the risk that a patient would contract a certain illness or the efficacy of a specific treatment for a specific patient. The chance of a patient being readmitted to the hospital has also been predicted using them [10, 11].
One of the main advantages of using random forests in the medical field is that they can handle high-dimensional data and can find complex, nonlinear relationships in the data. They are also robust to noise and can handle large datasets efficiently. In addition, random forests can provide feature importance scores which can help researchers understand which features are most important for predicting the outcome. However, one limitation of random forests is that they can be difficult to interpret as the decision-making process is distributed across many different decision trees. They may also be sensitive to the selection of hyperparameters, which might impact the effectiveness of the model [12,13,14].
2 Related Work
Recent years have seen a significant amount of study on the use of classification algorithms to medical data. For example, Arwatki Chen et al. [15] have achieved their goal by predicting diabetes with a stable and high accuracy using various machine learning algorithms. The model's consistent accuracy was made possible by the use of several risk factors and cross-validation methods. The study was constrained by the fact that it was based on a single dataset, and more testing and validation on a bigger, more varied dataset may be required to fully assess the model's efficacy. The study could be expanded in the future to include more deep learning and deep learning methods and to test the model on a bigger dataset in order to increase precision and generalizability. The model's practical application and effectiveness in the early diagnosis of diabetes would also be further understood by using it in a real-world situation, such as a hospital or medical clinic.
Li et al. [10] included applications of machine learning classifiers like DT, LR, KNN, SVM, ANN, NB, employing feature selection methods including MRMR, Relief, LLBFS, and LASSO. The feature selection issue for heart disease diagnosis has also been addressed using the novel feature selection algorithm FCMIM. The models that use feature selection methods in addition to the LOSO cross-validation approach have given a good accuracy. Hence, they claimed to improve the performance of the diagnosis of cardiac problems by adding new feature selection methods and optimization approaches.
Li et al. [11] the preoperative identification and staging of pancreatic cancer, computed tomography (CT) images, and an ensemble learning-support vector machine (EL-SVM) were used. The Least Absolute Shrinkage and Selection Operator (LASSO) approach was employed for feature selection, and it achieved high accuracy at various points. The effectiveness of models with more feature selection and hyperparameter tuning techniques would be interesting to study.
Anthimopoulos et al. [2] have published a deep convolutional neural network (CNN) for categorizing lung computed tomography (CT) image patches into seven groups, including healthy tissue and six different interstitial lung illnesses (ILDs). The proposed network architecture, designed to capture the low-level textural features of the lung tissue, consisted of five convolutional layers and three dense layers. On a challenging dataset of 120 CT images from multiple hospitals and scanners, the proposed technique outperformed the state of the art and yielded encouraging results. Negative aspects of the recommended technique include its large number of parameters, delayed training period (often a few hours), and some variance in output for the same input due to the random initialization of the weights. In order to help in the differential diagnosis of ILDs, the authors intend to expand the approach to take into account three-dimensional data from multidetector CT volume scans.
Liu et al. [13], For the simultaneous classification of Alzheimer's disease and clinical score regression, a deep multi-task multi-channel learning (DM2L) architecture was built employing magnetic resonance imaging (MR) imaging data and demographic information (i.e., age, gender, and education of participants). The study found that the DM2L technique outperformed many cutting-edge algorithms in the tasks of illness classification and clinical score regression on four publically accessible datasets. Existing convolution neural networks trained on other sizable 3D medical image datasets were needed to fine-tune the proposed network. Other study limitations include discrepancies in data distributions between the training and testing data, independence of the proposed deep feature learning framework from the landmark identification procedure, and the need for more research. For further enhancing the performance of the DM2L approach, they have included a number of directions, such as researching model adaptation strategies, combining landmark detection and landmark-based classification/regression into a single deep learning framework, optimizing the convolution neural networks that have already been trained on other substantial 3D medical image datasets, and automatically learning weights for the tasks of disease classification and clinical score regression.
Tsanas et al. [17] used prolonged vowel phonations; a variety of traditional and cutting-edge speech signal processing methods were used to separate Parkinson's disease (PWP) patients from healthy controls. The accuracy of their categorization was improved from prior research’ 93% accuracy using a subset of 22 features to the authors reported 99% accuracy using ten dysphonic measures. From the initial 132 features, four different feature selection algorithms found a small subset of 10 features that were informative for the binary classification task. Although RELIEF offered the subset with the lowest classification error, the FS methods still performed rather well. Signal-to-noise ratio (SNR) and mel-frequency cepstral coefficients (MFCCs) measurements were discovered to be consistently chosen by the FS algorithms, demonstrating the significance of these measurements for the evaluation of vocal pathology in PWP. For the purpose of mapping characteristics to the response, the scientists also examined the effectiveness of nonlinear random forests (RFs) and support vector machines (SVMs) and discovered that RF classifier works well than the SVM classifier. The authors did observe that the RF classifier was more susceptible to the FS method and training set selection. They also emphasized the necessity for more investigation into how PWP affects vocal tract articulatory dysfunction as well as the need of employing a sizable and varied dataset for vocal pathology evaluation.
Liu et al. [14] have discussed multimodal neuroimaging data, and a deep learning system has been suggested for detection of Alzheimer's disease (AD). They showed the system could discriminate between several phases of AD development by combining unsupervised feature representation with deep learning techniques. The framework was evaluated using data from the ADNI repository, and it was discovered to perform better than existing deep learning frameworks including the most advanced SVM-based approach for classifying AD. The authors claim that the approach might be extended to additional unlabeled data for feature engineering and could yet use more training data. The technique may be applied to more efficiently depict multimodal neuroimaging biomarkers.
Tao et al. [21] had conducted a study on machine learning methods using Magnetocardiography (MCG) data, and an efficient and precise approach for the automated diagnosis and localization of ischemic heart disease was created. Following the extraction of 164 features from the T wave segmentation and comparison of multiple classifiers, the SVM-XGBoost model gives the good results for IHD identification, with 94.03% accuracy and an AUC of 0.98. The XGBoost model successfully localized in ischemia in the left circumflex, left anterior descending, and right coronary arteries with accuracy values of 0.68, 0.74 and 0.65, respectively. This approach may broaden the use of MCG data in clinical settings by giving doctors a valuable tool for interpreting the data. Furthermore, the possibility of noninvasive ischemia localization is suggested by the link between magnetic field patterns and stenosis location. To increase the localization accuracy and validate the findings using bigger datasets, further effort is still required.
Arwatki Chen et al. [15] employed a variety of machine learning methods, and it was able to predict diabetes with a steady and high level of accuracy. The model's consistent accuracy was made possible by the inclusion of several risk variables and cross-validation methods. The study was constrained by the fact that it was based on a single dataset, and further testing and validation on a bigger, more varied dataset may be required to fully assess the model's efficacy. To increase accuracy and generalizability, machine learning and deep learning models are applied to bigger dataset. Moreover, applying the model in a genuine environment, such as a hospital or medical facility, would give additional understanding of the usefulness for diabetes detection.
Kumari et al. [9] have suggested ensemble voting classifier has great accuracy in predicting diabetes mellitus and breast cancer, with 97.02% and 79.04% accuracy, respectively. Modern techniques and basic classifiers such as logistic regression, AdaBoost, support vector machine, Naive Bayes, random forest, Bagging, XGBoost, CatBoost, and GradientBoost were surpassed by ensemble approach. To properly assess the efficacy and robustness of the suggested strategy, however, more testing on a larger and more varied dataset may be required. Future advancements in the suggested method's accuracy might come from using deep learning models and investigating various ensemble methodologies.
Ambrish et al. [6] used logistic regression technique on the UCI dataset, and a prediction of cardiovascular disease with a high accuracy of 87.10% was made. The model's performance was enhanced by pre-processing the data, which involved cleaning, identifying missing values, and conducting feature selection. With more training data, the model's accuracy also rose. The study was only able to use the UCI dataset, but future research might expand to include other datasets for more reliable findings. The application of additional machine learning algorithms or for greater performance in the prediction of cardiovascular illness might also be investigated in the future study.
Astani et al. [3] a system for categorizing 13 kinds of tomato diseases in both lab and field settings was put forth and put to the test. On the Taiwan database, which includes photos with a variety of difficulties like shadow, background clutter, noise, low image quality, many leaves, varied textures, and brightness fluctuations, the approach employed ensemble classification and obtained an accuracy of 95.98%. The suggested approach was also discovered to be quicker than deep learning models and was very accurate in classifying photographs with low image quality, shadows, and cluttered backgrounds. To completely evaluate the approach's efficacy, more testing on additional plant species and illnesses is required. The method was only tested on two databases, though.
Piao et al. [18], Feature subset-based ensemble technique is suggested for leveraging miRNA expression data to categorize various tumors. In comparison with other widely used ensemble approaches, the method was able to produce good results and greater prediction accuracy. The capacity of this technique to take feature relevance and redundancy into account while creating numerous feature subsets, leading to more independent and informative models for the classification problem, is one of its key accomplishments. Additionally, performance was enhanced by the integration of various base classifiers and the average posterior probability. The suggested approach does have some limitations, though, one of which is that it cannot be used for low-dimensional data since there are only so many subsets that can be formed. This might potentially affect the algorithm's capacity to produce a base no of classifiers.
Sambasivam and Opiyo [19] used photos from a dataset amassed in Uganda with the goal of developing a machine learning model to precisely diagnose illnesses affecting cassava leaves. The majority of the photos belonged to the Cassava Mosaic Disease and Cassava Brown Streak Virus Disease categories, making up the limited and severely unbalanced dataset. The authors achieved an accuracy of 93% by using methods like class weight, SMOTE, and focused loss with deep convolution neural networks to overcome this class imbalance. The suggested approach performed well in real-world situations and was able to categorize the underrepresented groups appropriately. However, one drawback of the study is that it was only tested on one dataset; hence, more testing on a larger and more varied dataset may be required to corroborate the findings.
Hameed et al. [7], this study's proposed intelligent digital diagnosis strategy for skin disorders used deep learning to obtain a high classification accuracy of 96.47%. This is a noteworthy accomplishment since prompt and efficient skin disease treatment depends on precise diagnosis. A restriction of earlier research that only concentrated on a small number of illnesses has been addressed by the use of the multi-class multi-level (MCML) classification method, which is inspired by the “split and conquer” approach. Further research might be done to include more diseases in the categorization algorithm as the study only took a small number of skin conditions into account. Better testing could be carried out on actual patient cases to further confirm the suggested algorithm's efficacy as it was only tested using photos gathered from various sources. The suggested approach may be used to create a mobile-enabled expert system for usage in remote locations with sparse access to diagnostic resources. The availability and accessibility of skin disease diagnosis and treatment in these locations may significantly increase as a result [18, 20, 21].
Overall, the use of classification techniques on medical data has the potential to significantly improve the accuracy and efficiency of diagnosis, treatment, and prognosis, making it an important area of research in the field of medicine. However, it is crucial to carefully weigh the advantages and disadvantages of various categorization algorithms and to assess how well they perform on pertinent medical datasets [16, 17, 22,23,24,25,26,27].
3 Proposed Work
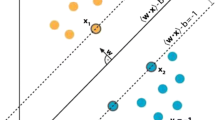
Empirical analysis is done on medical datasets by using various classification algorithms. Essential pre-processing methods are done on the datasets, such as feature scaling and missing value imputation, before building the models. Decision trees (DT), logistic regression (LR), support vector machine (SVM), k-nearest neighbors (KNN), random forests (RF), and Naive Bayes classifier (NBA) were some of the classification models used in this study. Figure 1 provides an information about building a classification models.
Thyroid Disease dataset [DS-1] includes a total of 3772 occurrences and 29 characteristics. The Chronic Kidney Disease dataset [DS-2] with the exception of the target variable, it had 400 instances in total and 25 characteristics. Diabetes 130-US hospitals for the years 1999–2008 dataset [DS-3] has 100,000 instances and 55 characteristics. Breast Cancer Wisconsin (Diagnostic) dataset [DS-4] contains 569 occurrences and 31 characteristics and Pima Indians Diabetes dataset [DS-5] which consist of 700 occurrences and 8 characteristics. Table 1 represents accuracy of different classification models on various datasets.
4 Results and Discussion
Figure 2 shows classification models accuracy on various datasets. Out of all the datasets, logistic regression, support vector machine, and random forest with an average of 99% accuracy for Breast Cancer Wisconsin, decision tree, and KNN with an average 98% for Thyroid Disease dataset performed well, whereas decision tree, KNN, and random forest with an average of 75% for Pima Indians Diabetes, logistic regression, and GNB with an average of 60% for diabetes 130-US hospitals datasets.
5 Conclusion
This work is helpful for researchers and practitioners making the choice of classification model when analyzing medical data. Out of all classification models random forest with accuracy 99.74, and logistic regression with 98.99 for Breast Cancer Wisconsin dataset as random forest is an ensemble method and logistic regression is good for classification problems performed well, Gaussian Naïve Bayes with 0.59 for diabetes dataset and support vector machine with 0.60 for Chronic Kidney Disease dataset does not performed well. In the future, researchers can get good accuracy of classification models by considering feature selection methods.
References
Aksoy S, Koperski K, Tusk C, Marchisio G, Tilton JC (2005) Learning Bayesian classifiers for scene classification with a visual grammar. IEEE Trans Geosci Remote Sens 43(3):581–589
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5):1207–1216
Astani M, Hasheminejad M, Vaghefi M (2022) A diverse ensemble classifier for tomato disease recognition. Comput Electron Agric 198:107054
Barakat N, Bradley AP, Barakat MNH (2010) Intelligible support vector machines for diagnosis of diabetes mellitus. IEEE Trans Inf Technol Biomed 14(4):1114–1120
Fayn J (2010) A classification tree approach for cardiac ischemia detection using spa-tiotemporal information from three standard ecg leads. IEEE Trans Biomed Eng 58(1):95–102
Ambrish G, Ganesh B, Ganesh A, Srinivas C, Dhanraj, Mensinkal K (2022) Logistic regression technique for prediction of cardiovascular disease. Glob Trans Proc 3(1):127–130. Int Conf Intell Eng Approach (ICIEA-2022)
Hameed N, Shabut AM, Ghosh MK, Hossain MA (2020) Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst Appl 141:112961
Hossain E, Hossain MF, Rahaman MA (2019) A color and texture based approach for the detection and classification of plant leaf disease using knn classifier. In: 2019 international conference on electrical, computer and communication engineering (ECCE). IEEE, pp 1–6
Kumari S, Kumar D, Mittal M (2021) An ensemble approach for clas-sification and prediction of diabetes mellitus using soft voting classifier. Int J Cognitive Comput Eng 2:40–46
Li JP, Ul Haq A, Ud Din S, Khan J, Khan A, Saboor A (2020) Heart disease identification method using machine learning classification in e-healthcare. IEEE Access 8:107562–107582
Li M, Nie X, Reheman Y, Huang P, Zhang S, Yuan Y, Chen C, Yan Z, Chen C, Lv X et al (2020) Computer-aided diagnosis and staging of pancreatic cancer based on ct images. IEEE Access 8:141705–141718
Lindner C, Thiagarajah S, Wilkinson JM, Wallis GA, Cootes TF, arcOGEN Consortium et al (2013) Fully automatic segmentation of the proximal femur using random forest regression voting. IEEE Trans Med Imag 32(8):1462–1472
Liu M, Zhang J, Adeli E, Shen D (2019) Joint classification and regression via deep multi-task multi-channel learning for alzheimer’s disease diagnosis. IEEE Trans Biomed Eng 66(5):1195–1206
Liu S, Liu S, Cai W, Che H, Pujol S, Kikinis R, Feng D, Fulham J, ADNI (2015) Multimodal neuroimaging feature learning for multi-class diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng 62(4):1132–1140
Lyngdoh AC, Choudhury NA, Moulik S (2021) Diabetes disease prediction using machine learning algorithms. In: 2020 IEEE-EMBS conference on biomedical engineering and sciences (IECBES), pp 517–521
Gunjan VK, Kumar S, Ansari MD, Vijayalata Y (2022) Prediction of agriculture yields using machine learning algorithms. In: Proceedings of the 2nd international conference on recent trends in machine learning, IoT, smart cities and applications: ICMISC 2021. Springer, Singapore, pp 17–26
Tsanas A, Little MA, McSharry PE, Spielman J, Ramig L-R (2012) Novel speech signal processing algorithms for high-accuracy classifica-tion of parkinson’s disease. IEEE Trans Biomed Eng 59(5):1264–1271
Piao Y, Piao M, Ryu KH (2017) Multiclass cancer classification using a feature subset-based ensemble from microrna expression profiles. Comput Biol Med 80:39–44
Sambasivam G, Opiyo GD (2021) A predictive machine learning application in agriculture: Cassava disease detection and classification with imbalanced dataset using convolutional neural networks. Egypt Inf J 22(1):27–34
Springer DB, Tarassenko L, Clifford GD (2015) Logistic regression-hsmm-based heart sound segmentation. IEEE Trans Biomed Eng 63(4):822–832
Tao R, Zhang S, Huang X, Tao M, Ma J, Ma S, Zhang C, Zhang T, Tang F, Jianping L, Shen C, Xie X (2019) Magnetocardiography-based ischemic heart disease detection and localization using machine learning methods. IEEE Trans Biomed Eng 66(6):1658–1667
Kumar S, Gunjan VK, Ansari MD, Pathak R (2022) Credit card fraud detection using support vector machine. In: Proceedings of the 2nd international conference on recent trends in machine learning, IoT, smart cities and applications: ICMISC 2021. Springer, Singapore, pp 27–37
Gaddam DKR, Ansari MD, Vuppala S, Gunjan VK, Sati MM (2022) A performance comparison of optimization algorithms on a generated dataset. In: ICDSMLA 2020: proceedings of the 2nd international conference on data science, machine learning and applications. Springer, Singapore, pp 1407–1415
Narayana GS, Ansari MD, Gunjan VK (2022) Instantaneous approach for evaluating the initial centers in the agricultural databases using K-means clustering algorithm. J Mob Multimedia 43–60
Kumar S, Ansari MD, Gunjan VK, Solanki VK (2020) On classification of BMD images using machine learning (ANN) algorithm. In: ICDSMLA 2019: proceedings of the 1st international conference on data science, machine learning and applications. Springer, Singapore, pp 1590–1599
Gunjan VK, Prasad PS, Pathak R, Kumar A (2020) Machine learning methods for extraction and classification for biometric authentication. In: ICDSMLA 2019: proceedings of the 1st international conference on data science, machine learning and applications. Springer, Singapore, pp 1984–1988
Kumar MR, Gunjan VK (2020) Review of machine learning models for credit scoring analysis. Ingeniería Solidaria 16(1)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Saketha Rama, B.V., Suryanarayana, G., Ansari, M.D., Begum, R. (2023). An Empirical Comparison of Classification Machine Learning Models Using Medical Datasets. In: Kumar, A., Gunjan, V.K., Hu, YC., Senatore, S. (eds) Proceedings of the 4th International Conference on Data Science, Machine Learning and Applications. ICDSMLA 2022. Lecture Notes in Electrical Engineering, vol 1038. Springer, Singapore. https://doi.org/10.1007/978-981-99-2058-7_29
Download citation
DOI: https://doi.org/10.1007/978-981-99-2058-7_29
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-2057-0
Online ISBN: 978-981-99-2058-7
eBook Packages: Computer ScienceComputer Science (R0)