Abstract
Magnetic resonance imaging (MRI) is a vital and universally recognized medium to assess brain neoplasms. This paper presents a study on brain tumor segmentation based on the random walk algorithm which is a graph-based method in which pixels of a brain MR image are treated as nodes. Segmentation is performed by interactively labeling certain nodes as foreground and background seeds, followed by computing the probability of each unlabeled node to reach all the labeled nodes using random paths. The method is applied on two different MR modalities viz. T2-weighted MRI with fluid attenuated inversion recovery (FLAIR), and T2 MRI to segment complete tumor, and tumor core regions, respectively, by utilizing visual traits of MRI images and identifying local and global brain tissues information. Efficacy is validated quantitatively as well as qualitatively through performing the experiments on publicly available brain tumor segmentation challenge (BRATS-2013) dataset. Results demonstrate that the proposed method performs favorable as compared to several existing methods.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
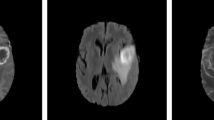
Automated MRI image analysis plays a significant role in the diagnosis and assessment of various brain tumor types and for sophisticated treatment planning. Glioblastoma multiforme (GBM), a malignant grade-IV tumor type, is one of the most affected and dangerous brain diseases in the world [3, 17]. Automation, accuracy, and time efficiency for a method of brain tumor segmentation are the three major needs of the hour because of the following reasons, (i) manual brain image analysis is quite time taking and depends upon the expertise of the radiologist, (ii) variable and amorphous structure of tumor affects the robustness of any tumor segmentation method. Except to the healthy or normal brain tissues, i.e., white matter, gray matter, and cerebro-spinal fluid (CSF), brain lesion is subdivided into four major components, edema, non-enhancing solid core, enhancing tumor, and necrosis. MRI in the form of its various modalities expresses extensive global and local level information of brain’s healthy and neoplastic tissues. Community of worldwide researchers involved in BRATS challenge is playing a significant role by creating and maintaining benchmarks specially in terms of providing a dataset [17] and defining evaluation measures. BRATS dataset contains MR images in four MRI modalities, i.e., FLAIR, T1-weighted, T2-weighted, and post-contrast T1-weighted MRI as displayed in Fig. 1. Annotation of dataset is performed by different experts from various institutions based on the visual characteristics of MR image modalities. T2-weighted MR modality images are helpful to extract active tumor also known as tumor core without edema. Similarly, FLAIR images are suitable to identify whole tumor region by observing the hyper-intense lesion [8].
Axial view of MRI modalities of a selected subject from BRATS dataset [14]. From left to right: T1-weighted, T2-weighted, T1c, FLAIR followed by the corresponding annotated ground truth
State-of-the-art methods in the literature of brain lesion detection and segmentation can be categorized in four major ways as follows: atlas-based methods, region-based methods, gradient and edge-based methods, clustering and classification-based methods. Atlas-based methods utilize the anatomical structure of healthy brain tissues followed by registering the same with tumorous brain image in order to capture and analyze the affected brain region [1, 17, 23]. As the anatomy of human brain tissues possesses significant variation case by case, atlas-based methods are only useful for the rough estimation of lesion area, and hence, they are generally used as a preprocessing step in any lesion segmentation method. Region-based methods perform segmentation by first investigating features of different pixels (or voxels) and further finding the affinity among them [15, 22]. Feature extraction is the major step in region-based segmentation followed by a similarity measurement criteria which often leads to oversegmentation problem. Edges and corner regions are the group of pixels which represent the significant information whenever there is an abrupt change in the intensity or appearance in an image. Edges and corners are the joint gradient information in both horizontal and vertical directions which can be exploited to recognize salient objects specially abnormal lesion in brain MR images [11, 20]. However, disadvantages of such approach include user interaction for initial region selection and appearance of lesion in low contrast images. Combining the hypo-intense and hyper-intense appearance of multiple MR modalities together is been a solution of such problems. Moreover, clustering and classification-based methods are currently the most popular due to the significant growth of machine learning paradigm. classification-based methods usually rely on certain features of different MR modalities such as local intensity of pixels, texture information around the neighborhood, statistical information, and spatial distribution of pixels. Thus, extracted features are exploited to segment the image using either supervised or unsupervised (also known as clustering) classification algorithms [2, 3, 12, 13, 18, 24].
Agn et al. [1] proposed an atlas-based lesion segmentation method in which the initial lesion area is detected by registering the input brain MR images with the healthy atlas-based brain tumor prior. Tumor sub-compartments have been further segmented by implementing a Convolutional Restricted Boltzmann Machines classifier (RBMs). Another generative method is suggested by Menze et al. [17] to segment brain tumor in multi-modality MR images by deriving a tumor estimation algorithm. However, atlas-based segmentation methods prone to false segmentation since the segmentation quality depends on accurate registration procedure. Working toward the similar objective, Letterboer et al. [15] proposed a region-based method which gives radiologists an initial estimation of brain abnormality towards providing a better treatment planning. Recently, Tong et al. [22] presented a method using texture feature extraction and kernel dictionary learning. Dictionary coding for normal and abnormal brain voxels is performed which in order to further classify the tumor region using linear discrimination function. Sachdeva et al. [19] proposed a whole tumor region segmentation method based on statistical and texture features and active contour model. Content-based active contour is a deformable model which is employed to evolve a user-initialized curve around the specified object boundary under the influence of internal and external forces. Exploiting the classification-based method, Bauer et al. [2] suggested joint architecture of hierarchical conditional random field (CRF) and support vector machine (SVM) classifiers for efficient segmentation of whole tumor volume. Corso et al. [3] proposed a method to segment tumor and edema regions based on Bayesian classification in which weighted aggregation is used to compute the model-aware affinities. Moreover, advancements of classification-based methods have been carried out using convolution operation-based machine learning methods in order to improve the performance of brain lesion segmentation. Havaei et al. [12] proposed a two-path way convolutional neural network (CNN)-based segmentation method which utilizes local as well as global features of brain tissues. Likewise, Kamnitsas et al. [13] presented a three-dimensional version of CNN architecture with 11 layers in its pipeline. A three-dimensional CRF is also exploited in the end to refine the performance of lesion segmentation. Pei et al. [18] suggested another classification-based method utilizing tumor cell density and textures features of brain tissues in association with random forest (RF) classifier. The method also implements joint-label fusion approach with another segmentation method to improve the performance. In a recent study, Zhao et al. [24] trained the RF classifier by extracting gradient and circular context-sensitive features. The total extracted features are reduced by using minimum redundancy maximum relevance (MRMR) approach.
We propose a semi-automated tumor segmentation method with following major contributions:
-
1.
Random walks algorithm is implemented in semi-automated manner to detect and segment complete tumor and tumor core regions from FLAIR and T2-weighted MR images, respectively.
-
2.
By utilizing the prior visual constraints of different MR modalities, multiple seed points representing foreground and background are chosen interactively in order to significantly improve the segmentation performance.
-
3.
The proposed method segments complete tumor and tumor core regions in 0.2 s for each set of T2-weighted and FLAIR MR images which illustrates the computational efficiency of the method.
Remaining of the paper is structured in the following way: Sect. 2 describes the proposed brain lesion segmentation method in detail. Section 3 demonstrates the experimental setup and results implemented on the benchmark datasets along with the comparison with other existing methods. Lastly, conclusion presents the significant outcomes of this work with future works in Sect. 4.
2 Proposed Method
In this study, a semi-automated method is proposed to recognize and segment two different affected regions of brain tumor by exploiting random walks method. The segmentation procedure of random walks algorithm is shown in Fig. 2 which illustrates that how a node (pixel in an image) is labeled based on the interactively selected foreground and background seed points. Probability shown at each node represents that how easy or difficult it is to reach a selected seed point starting from that particular node by following random walks. The proposed method is implemented on FLAIR and T2 MR images to obtain complete tumor and tumor core regions, respectively. Segmentation in various classes can be done based on the chosen seed points. However, we have chosen seeds corresponding to two labels (for foreground and background) by utilizing the visual constraints and hyper-intense regions of FLAIR and T2-weighted MRI modalities.
Example showing segmentation using random walks. a Two seeds points \(L_1\) and \(L_2\) as shown in blue and red colors. b Calculation of probabilities to reach seed \(L_1\) starting from each node through random walks. c Calculation of probabilities to reach seed \(L_2\) starting from each node through random walks. d Assigning labels to each node of the graph based on the calculated probabilities corresponding to initially selected seed points
2.1 Random Walks Segmentation
Image segmentation using random walks is a graph-based semi-automated method in which a 2-dim input image represents a graph and each pixel is treated as a node (vertex) of the graph [10]. A few nodes (seed points) of the graph are interactively chosen as labels. In our case, seeds points are chosen corresponding to two labels, i.e., foreground and background for each image. A probability corresponding to each unmarked node is computed which shows that how easy or how difficult it is to reach a particular label (background or foreground) starting from each unmarked node through a randomly chosen walk. Thus, two probabilities are calculated for each unmarked node based on which the node is segmented in either of the class.
The mathematical formulation and description of the random walks method is given as follows [10]: Let \(G=(V,E)\) be the graphical representation of an image where V and E represent the set of vertices (pixels) and set of edges between two vertices, respectively. Each edge \(e\in E\) between vertices \(v_p\) and \(v_q\) has a weight \(w_{pq}\). Let G be an undirected graph and \(d_p=\sum _q w_{pq}\) be the degree of a vertex \(v_p\) for the edges \(e_{pq}\) which incident on the vertex \(v_p\). Value of the weight \(w_{pq}\) is defined based on the Gaussian function applied over the difference between the local level features (pixel’s intensity) of the two vertices \(v_p\) and \(v_q\) as given below:
where \(f_p\) and \(f_q\) represent the local features of pixels \(v_p\) and \(v_q\) respectively. \(\beta \) is the additional parameter whose value can be set empirically depending on the application and image type. The random walks segmentation method is inspired by the flow of electrons through random paths in an electrical circuit network. \(\beta \) indicates the conductance phenomenon over the network by guiding how easy or difficult it is to reach a marked node from each unmarked node following through random paths. It becomes very easy to move from one node \(v_p\) to another node \(v_q\) if the value of \(w_{pq}\) is very close to 1. Similarly, it is very difficult to move from one node to another in case the value of \(w_{pq}\) is close to 0.
The solution of random walks segmentation problem can be solved through electrical current and voltage laws. However, another alternate solution of the same is given in [10] by computing the probabilities of random walks analytically in order to improve the efficiency of the method. Computing the probabilities of random walks can be represented in terms of combinatorial Dirichlet problem which is solved over a Laplacian equation derived in context of random walks problem. The functional of combinatorial Dirichlet integral [10] is given as below:
where \(\mu \) and \(\varPsi \) represent field and regions, respectively [4]. The Dirichlet problem is defined on the basis of a harmonic function with its corresponding boundary constraints. A harmonic function compatible to the boundary constraints also optimizes the value of Dirichlet integral due to the Euler-Lagrange property of the Laplacian equation [4]. The combinatorial Laplacian matrix [7] is defined as follows:
where \(L_{pq}\) is indexed by nodes \(v_p\) and \(v_q\).
Let \(V_M\) be the set of marked nodes (seed points) corresponding to the foreground and background and \(V_U\) be the set of unmarked nodes (except to the chosen seed points); the probabilities of the set of nodes \(V_U\) can be calculated by considering it as a type of Laplacian function which is compatible to the predefined boundary conditions of the marked nodes \(V_M\). Based on this consideration, the probability estimation of each node of image graph is equivalent to the minimization criterion of the above stated Dirichlet integral:
where \(x_p\) and \(x_q\) represent the probabilities at vertices \(v_p\) and \(v_q\), respectively. The critical points of Dirichlet integral D[x] become minima due to the semi-definite and positive property of the Laplacian matrix L. The above stated equation can be rewritten in terms of set of marked and unmarked node \(V_M\) and \(V_U\) as follows:
where \(x_M\) and \(x_U\) represent the potential or the computed probabilities of marked seed points and unmarked vertices, respectively. The following equation is obtained by performing differentiation of \(D[x_U]\) with respect to \(x_U\) as the solution of random walks problem in order to calculate the potential or probabilities of unmarked vertices:
Segmented complete tumor and tumor core regions on the selected subjects of BRATS2013 high-grade (HG) dataset. Each row delineates the subject id slice sequence number. a FLAIR MR image, b T2 MR image, c segmented complete tumor region, d ground truth corresponding to complete tumor region, e segmented tumor core region, f ground truth corresponding to tumor core region
Practically, number of unmarked nodes in an image graph is much greater than that of marked seed nodes which makes this solution computationally complex due to the complex calculation of \(L_U^{-1}\) while solving the equation \(x_{U} = - L_U^{-1} B^T x_M\). However, user interaction for choosing initial foreground and background seed points makes this method efficient in terms of producing accurate and fast segmentation results.
3 Experimental Results
Efficacy of the method is validated qualitatively as well as quantitatively through performing the experiments on real glioma images from publicly available benchmark datasetFootnote 1 (BRATS-2013) as shown in Fig. 3 and Table 1. Experimental work has been carried out on the Dell desktop having 64-bit Windows 10 OS, with Intel(R) i-7 processor and 8 GB of random access memory (RAM).
3.1 Dataset
BRATS-2013 dataset contains numerous real Glioma images of low-grade (LG) and high-grade (HG) brain tumor type. Each 2-dim slice of dataset is skull stripped in order to maintain the anonymity of patients and available in the form of four different modalities such as T1-weighted images, T1c images, T-2 weighted images, and FLAIR images. However, only T2-weighted and FLAIR MR image are utilized in this work. Real image data consists of the images of 20 HG and 10 LG Glioma subjects.
As per the BRATS challenge guidelines, Glioma brain tumor type can be classified into edema, non-enhanced solid core, enhancing tumor, and necrosis [16]. Edema can be identified as the swelling around other brain tumor tissues. Therefore, edema tissues are predominantly curable as compared to other tumor tissue types. Performance of a method is evaluated by effective segmentation of the following regions:
-
1.
Complete tumor region as the fusion of edema, non-enhanced solid core, enhancing tumor, and necrotic core.
-
2.
Tumor core region also known as complete tumor region without edema as the fusion of non-enhanced solid core, enhancing tumor, and necrotic core.
3.2 Qualitative Evaluation
Segmented complete tumor region as well as tumor core region from few of the selected 2-dim slices of HG subjects is shown in Fig. 3. Identification of subject and its corresponding slice is shown at the extreme left of each row. Each row shows the input MR images as well as the segmented brain tumor region along with the ground truth. The visual appearance of the shown images illustrates that the proposed approach produces favorable results for brain tumor segmentation. Moreover, the method lags while detecting tumor core regions as it produces many false positive cases due to the similar range of intensity local feature of lesion and CSF.
3.3 Quantitative Evaluation
The segmentation performance is also validated quantitatively with the available ground truth using the relevant evaluation measures such as dice similarity coefficient (DSC), sensitivity or true positive rate, and positive predictive value (PPV). DSC [6] also known as the dice score represents an overall agreement between the predicted result (output) and the actual result (ground truth) by measuring the overlapped region. Sensitivity represents the rate with which our prediction is true positive (TP) means the predicted result is positive (for tumorous brain tissues) and actual result is also positive. Positive predictive value (PPV) refers to the number of true positive predictions over all the positive predictions (either positive or negative) predicted by the proposed method.
where TP, TN, FP, and FN represent true positive, true negative, false positive, and false negative, respectively. Average quantitative results for all 20 HG as well as 10 LG subjects in terms of aforementioned performance measures are given in Table 1. Each subject’s 3-dim data size is \(216\times 176\times 176\) which means total 176, 2-dim slices are available of \(216\times 176\) size each. Out of the total 176 slices, tumorous brain tissues information is present only in few of the slices only. Typically, most of the brain tissues information is present in the slices which belong to the middle position. Due to the same fact, 20 middle slices of each subject’s brain slices are considered for our experiments.
The segmentation results of the proposed method are also compared with the works of Demirhan et al. [5], Geremia et al. [9], and Taylor et al. [21] after reproducing the segmentation results on the same settings. Segmentation results of Demirhan et al. [5] are obtained by training the corresponding model on BRATS-2013 training dataset. As BRATS dataset provides us the real Glioma images which are skull stripped and registered with T1c MRI, these preprocessing steps were skipped while implementing this model. While reproducing the results of Taylor et al. [21], a Hidden Markov Model (HMM) is trained on 80% of BRATS 2013 training dataset. Pixel-wise classification is performed for both low-grade and high-grade Glioma dataset images. As the dataset is highly unbalanced, down-sampling is done for pixels representing healthy brain tissues before training the model. The detailed segmentation scores corresponding to the selected performance measures are shown in Table 1. We achieved the DSC of 0.67 for HG subjects and 0.66 for LG subjects for segmenting complete tumor region. The DSC for tumor core region is achieved as 0.49 and 0.52 for HG and LG subjects, respectively. Performance in segmenting tumor core region is due to the complex appearance of T2-weighted MR images where not only abnormal brain tissues but also CSF appear hyper-intensive. Still the result of tumor core segmentation is competitive as reported in Table 1. The performance of our method in terms of sensitivity and PPV is also favorable with the compared methods for both complete tumor and tumor core regions. However, our method outperforms in all the measures for segmenting LG subjects data for both complete tumor and tumor core regions. The performance score obtained indicates than the proposed brain tumor segmentation approach gives significant contribution.
3.4 Discussion
The quantitative and qualitative results shown above suggest that the proposed brain tumor segmentation approach is suitable and efficient for real time brain tumor segmentation. The proposed approach detects and segments the most significant tumor regions, i.e., complete tumor and tumor core regions in order to assist radiologists and provide better treatment planning. However, enhancing brain tumor region cannot be detected separately due to the visual constrains of FLAIR and T2-weighted MRI. All the experiments are conducted on the middle axial slices of each subject of the dataset to ensure that the significant amount of tumorous and healthy tissues present in each slice. Pixels were marked for both background and foreground interactively based on which probability of all unmarked pixels was computed in order to segment each node in the specified labels. The proposed segmentation approach is based on random walks algorithm in which \(\beta \) is the only free parameter in Eq. 1 in order to perform accurate segmentation. For all the experiments, value of parameter \(\beta \) is kept constant as 350. The proposed method is also time efficient as it segments both complete tumor and tumor core regions for each set of T2-weighted and FLAIR MR images in 0.2 s only.
4 Conclusion
In this paper, an efficient segmentation approach is proposed to detect and segment tumor in two significant complete tumor and tumor core regions. A probability-based random walks segmentation approach is exploited slice-by-slice manner to divide each MR image in two segments based on the pre-selected (marked nodes) foreground and background seed points interactively. Each MR image is treated as a graph in which probability of each unmarked node is computed by finding its accessibility against marked nodes through considering random walks in its four-connected neighborhood. All four tumor labels, i.e., edema, non-enhanced solid core, enhancing tumor, and necrotic core are extracted and combined as complete tumor region by implementing the proposed method in FLAIR MR image slices of each subject. Similarly, tumor core region (all tumor labels without edema) is extracted from T2-weighted MR image slices. The evaluation results demonstrated the high performance of the proposed brain tumor segmentation approach when compared to other existing methods. The proposed approach can assist radiologists because of its computational efficiency. In future, the proposed brain tumor segmentation approach can be extended by selecting foreground and background seed points automatically using feature descriptors like scale invariant feature transform (SIFT) and Harris edge and corner detector.
References
Agn, M., Puonti, O., Law, I., af Rosenschöld, P., van Leemput, K.: Brain tumor segmentation by a generative model with a prior on tumor shape. In: Proceeding of the Multimodal Brain Tumor Image Segmentation Challenge, pp. 1–4. Springer (2015)
Bauer, S., Nolte, L.P., Reyes, M.: Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 354–361. Springer (2011)
Corso, J.J., Sharon, E., Dube, S., El-Saden, S., Sinha, U., Yuille, A.: Efficient multilevel brain tumor segmentation with integrated Bayesian model classification. IEEE Trans. Medi. Imag. 27(5), 629–640 (2008)
Courant, R., Hilbert, D.: Methods of mathematical physics, vol. 2. Wiley (1989)
Demirhan, A., Törü, M., Güler, I.: Segmentation of tumor and edema along with healthy tissues of brain using wavelets and neural networks. IEEE J. Biomed. Health Inform. 19(4), 1451–1458 (2014)
Dice, L.R.: Measures of the amount of ecologic association between species. Ecology 26(3), 297–302 (1945)
Dodziuk, J.: Difference equations, isoperimetric inequality and transience of certain random walks. Trans. Am. Math. Soc. 284(2), 787–794 (1984)
Drevelegas, A.: Imaging of brain tumors with histological correlations. Springer Science & Business Media (2010)
Geremia, E., Menze, B.H., Ayache, N., et al.: Spatial decision forests for glioma segmentation in multi-channel mr images. In: MICCAI Challenge on Multimodal Brain Tumor Segmentation, vol. 34, pp. 14–18. Citeseer (2012)
Grady, L.: Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 11, 1768–1783 (2006)
Harris, C., Stephens, M.: A combined corner and edge detector. In: Alvey Vision Conference, vol. 15, pp. 147–151. Citeseer (1988)
Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., Pal, C., Jodoin, P.M., Larochelle, H.: Brain tumor segmentation with deep neural networks. Med. Image Anal. 35, 18–31 (2017)
Kamnitsas, K., Ledig, C., Newcombe, V.F., Simpson, J.P., Kane, A.D., Menon, D.K., Rueckert, D., Glocker, B.: Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Kistler, M., Bonaretti, S., Pfahrer, M., Niklaus, R., Büchler, P.: The virtual skeleton database: an open access repository for biomedical research and collaboration. J. Med. Internet Res. 15(11), e245 (2013)
Letteboer, M.M., Olsen, O.F., Dam, E.B., Willems, P.W., Viergever, M.A., Niessen, W.J.: Segmentation of tumors in magnetic resonance brain images using an interactive multiscale watershed algorithm1. Acad. Radiol. 11(10), 1125–1138 (2004)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R., et al.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imag. 34(10), 1993 (2015)
Menze, B.H., Van Leemput, K., Lashkari, D., Weber, M.A., Ayache, N., Golland, P.: A generative model for brain tumor segmentation in multi-modal images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 151–159. Springer (2010)
Pei, L., Bakas, S., Vossough, A., Reza, S.M., Davatzikos, C., Iftekharuddin, K.M.: Longitudinal brain tumor segmentation prediction in mri using feature and label fusion. Biomed. Signal Process. Control 55, 101,648 (2020)
Sachdeva, J., Kumar, V., Gupta, I., Khandelwal, N., Ahuja, C.K.: A novel content-based active contour model for brain tumor segmentation. Magn. Resonance Imag. 30(5), 694–715 (2012)
Shivhare, S.N., Kumar, N., Singh, N.: A hybrid of active contour model and convex hull for automated brain tumor segmentation in multimodal mri. In: Multimedia Tools and Applications, pp. 1–23 (2019)
Taylor, T., John, N., Buendia, P., Ryan, M.: Map-reduce enabled hidden Markov models for high throughput multimodal brain tumor segmentation. In: Multimodal Brain Tumor Segmentation, vol. 43 (2013)
Tong, J., Zhao, Y., Zhang, P., Chen, L., Jiang, L.: Mri brain tumor segmentation based on texture features and kernel sparse coding. Biomed. Signal Process. Control 47, 387–392 (2019)
Ullmann, J.F., Janke, A.L., Reutens, D., Watson, C.: Development of mri-based atlases of non-human brains. J. Comp. Neurol. 523(3), 391–405 (2015)
Zhao, J., Meng, Z., Wei, L., Sun, C., Zou, Q., Su, R.: Supervised brain tumor segmentation based on gradient and context-sensitive features. Front. Neurosci. 13 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Shivhare, S.N., Kumar, N. (2021). Brain Tumor Segmentation Using Random Walks from MRI Images. In: Panigrahi, C.R., Pati, B., Pattanayak, B.K., Amic, S., Li, KC. (eds) Progress in Advanced Computing and Intelligent Engineering. Advances in Intelligent Systems and Computing, vol 1299. Springer, Singapore. https://doi.org/10.1007/978-981-33-4299-6_3
Download citation
DOI: https://doi.org/10.1007/978-981-33-4299-6_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-4298-9
Online ISBN: 978-981-33-4299-6
eBook Packages: EngineeringEngineering (R0)