Abstract
Biofilms are association of microorganisms that attach to each other to a surface enclosed in a self-generated extracellular matrix. Virtually (99.9%) all microorganisms have the competence to form biofilm. The formation of biofilm is a complex process, in which bacterial cells transform from planktonic cells to sessile mode of growth. The biofilm development results from the expression of specific genes. Biofilms have been developed as an adaptive strategy of bacterial species to survive in adverse environmental conditions as well as to establish antagonistic or beneficial interactions with their host. Molecular interaction and details of biofilm formation are not well-understood as bacteria in the biofilm have several orders of magnitude, more resistant to antibiotics compared to planktonic bacteria. Thus, the currently available drugs typically failed to target bacterial biofilms. Quorum sensing (QS) is a process of intercellular signalling or cell-cell communication and a vital regulatory mechanism for coordinating biofilm formation including common activities and physiological processes such as symbiosis, formation of spores or fruiting bodies, antibiotics synthesis, genetic competence, apoptosis and virulence in many bacterial species using extracellular QS signalling molecules, which is often referred to as autoinducers (AIs). Microorganisms produce a wide variety of QS signalling molecules that can be self-recognized in a concentration-dependent manner and subsequently induce or suppress expression of QS-controlled genes. Bacterial QS regulation is established through a wide range of signals such as oligopeptides, N-acyl homoserine lactones (AHLs), furanosyl borate, hydroxy palmitic acid methyl ester and methyldodecanoic acid. In this chapter, we highlight the current understanding of the processes that lead to bacterial biofilm formation, survival behaviours and mechanisms of antimicrobial resistance in bacteria.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Biofilm
- Quorum sensing
- Antibiotic resistance
- Quorum quenchers
- Acyl homoserine lactone
- Microbial communication
1 Introduction
Biofilms are ubiquitous in nature and can occur on both animate and inanimate surfaces. Biofilms have been recognized as aggregates of many bacteria and simple eukaryotes growth on natural aquatic surfaces, clinical, industrial and domestic domains (Irie and Parsek 2008). The natural and clinical biofilms are formed by different types of microbial species with wide range of structural characteristics, however, majority of the biofilms are encased in self-produced extracellular matrix or extracellular polysaccharide (EPS) layer (Sutherland 2001a; Irie and Parsek 2008). The composition of extracellular matrix can differ between organisms, but are commonly abundant in proteins (<1–2%) including enzymes, polysaccharides (1–2%), nucleic acids (<1%) including DNA and RNA and water (up to 97%) (Lu and Collins 2007). The phenotype of matrix generally depends on environmental stress such as temperature, pH, osmolarity, UV radiation, desiccation, oxygen tension and nutrient availability (Staley et al. 2014). Bacterial communities in biofilm can switch from planktonic form to sessile form. The planktonic bacteria have relatively high cell growth and reproduction rate but low EPS production (Rabin et al. 2015). However, the sessile form exhibits slower growth of bacteria yet increased EPS production which is useful to form biofilms (Chadha 2014). It should be noted that biofilm forming bacterial mutants are unable to produce the EPS (Watnick and Kolter 1999). The synthesis of EPS matrix intends the bacterial cells to attach to a surface. Biofilm forms a thin layer eventually building to a thick layer (>100 layers) establishes a ‘mushroom-’ or ‘tower’-shaped structure (Rabin et al. 2015). The bacterial arrangement in the biofilm depends on their metabolism and aero-tolerance; aerobic bacteria live in the upper layers, while anaerobic bacteria prefer to live in deeper layers of biofilm (Rabin et al. 2015).
Bacteria form biofilms for numerous advantages: enhance the tolerance of bacteria to harsh environmental conditions, avoid being washed away by water flow or blood stream and protection from antimicrobial agents and disinfectants (Jefferson 2004; Rabin et al. 2015). Further, the biofilm retards bacterial motility and increases cell density providing a suitable environment for plasmid exchange between bacterial communities by conjugation process. Some of these plasmids encode for antibiotic resistance (Hausner and Wuertz 1999; Rabin et al. 2015) and also enable them to overcome different environmental stresses (Chadha 2014). The bacteria in a biofilm may communicate their presence to each other using chemical communication known as quorum sensing (QS). QS is a mechanism by which bacteria apparently regulate collective behaviours in response to cell density (Lyon and Muir 2003; West et al. 2012). The bacteria produce and release small diffusible signal molecules for cell-cell signalling. Diffusing of these small molecules into cells regulates (autoinduction) totally different behaviours including the production of a variety of small molecules that are released from bacterial cells to help growth, mobility, competence, sporulation, bioluminescence emission, symbiosis, antibiotic production and biofilm formation (West et al. 2012; Li and Tian 2012). In addition, the diffusion of these small molecules also contributes to an increase in the production of the signal molecule itself (autoregulation). Therefore, the production of these signalling or autoinducing molecules leads to high cell densities, which afford a considerable increase in the production of signal and QS-controlled factors (West et al. 2012; Darch et al. 2012). Generally, in infectious diseases, the invading bacteria need to reach a critical cell density before they show virulence and defeat the host defence mechanisms (Costerton et al. 2003; Li and Tian 2012). It is now apprehended that quorum sensing mechanisms occur in both unicellular prokaryotic and single-celled eukaryotic organisms such as fungi (Miller and Bassler 2001; Waters and Bassler 2005; van Bodman et al. 2008; Sordi and Muhlschlegal 2009; Li and Tian 2012). Furthermore, the cell-cell communication is apparent between microbial human pathogens through QS having important implications in the infections.
2 Structure, Biofilm Formation, Survivability and Quorum Sensing
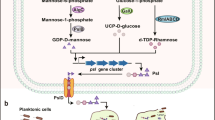
Microbial biofilm formation is a dynamic process that floating (planktonic) cells transform to immobile (sessile) form of growth (Okada et al. 2005). Still controversy exists in QS involved in biofilm formation (Parsek and Greenberg 2005). However, it is demonstrated that QS are influencing the biofilm formation in several species (Parsek and Greenberg 2005). Further, it has been suggested that biofilm formation depends on the specific gene expression (Okada et al. 2005; Sauer et al. 2004). Biofilm formation is generally considered to occur through series of stages (Fig. 3.1): (1) adsorption or accumulation of organisms on an aggregator surface, i.e. substrate (deposition); (2) attachment to a surface or the desegregation of the interface between organisms and aggregator for formation of polymer bridges; (3) cell proliferation or growth of organisms on the aggregator’s surface; (4) biofilm formation and maturation; and (5) detachment or dispersal (O’Toole et al. 2000; Garrett et al. 2008; Joo and Otto 2012).
In the accumulation step, microorganisms form initial conditioning layer composed of organic or inorganic molecules creating the foundation for biofilm growth. This layer provides a favourable environment for growth, nutrients and anchorage of the bacterial community (Characklis and Marshal 1990; Garrett et al. 2008). The attachment step is categorized as a two-stage process such as reversible attachment and irreversible attachment (Garrett et al. 2008; Renner and Weibel 2011). Bacteria should get closer enough to a surface for biofilm formation as bacterial cell meeting has both attractive and repulsive forces. When the bacterial cells have distance between 10 and 20 nm to a surface, the negative charges are repelled; however, van der Waals forces between the bacterial cells overcome the repulsion by attraction to a surface. Besides, fimbriae and flagella also provide the mechanical attachment to the surface (Palmer et al. 2007; Rabin et al. 2015). If the repulsive forces are higher than the attractive forces, the bacterial cells will disperse from the surface; this probably would occur before conditioning of a substrate (Garrett et al. 2008). In the early attachment, planktonic microbial cells are transferred from aqueous to the conditioned surface by either physical forces or bacterial flagella. Many environmental factors contribute to the reversible attachment of biofilm to a surface such as surface nature, temperature, pressure, available energy and bacterial orientation (Garrett et al. 2008). Besides, the reversibly attached cells persist immobilized and become irreversibly attached cells. The irreversibly attached biofilm can withstand greater physical or chemical forces (Sutherland 2001b; Liu et al. 2004; Rabin et al. 2015). The flagella and type IV pili play crucial role in irreversible attachment of cells to a surface and form microcolonies (Garrett et al. 2008; Rabin et al. 2015).
During the lag phase, the bacterial cells adapt to a surface or an environment for accumulation and attachment process. However, the rapid propagation in the population occurs in exponential phase or log phase. The bacterial rapid growth in the biofilm takes place with the sufficient nutrients accessible from the bulk fluid and the substrate surface depending on the nature of the environment (Garrett et al. 2008). During the cell division in the biofilm, daughter cells move outward and upward from the attachment point to form clusters, such interactions and growth provides mushroom-like structure (Hall-Stoodley and Stoodley 2002). It is believed that mushroom-like structure support the passage of nutrients to bacterial communities that lives in bottom of a biofilm. The secretions of extracellular matrix by bacteria in a biofilm aid to form bonding between cells due to interaction of polysaccharide, intercellular adhesion polymers and the presence of divalent cations (Dunne 2002; Garrett et al. 2008). Certain biofilm-related differential gene expression involved in the bacterial species for transforming planktonic to sessile form aids the cells for adhesion in the population. The motility of the sessile species is arrested and synthesis of external flagella is inhibited during this stage (Garrett et al. 2008). Meanwhile, the expression of number of genes in sessile species ameliorates the production of cell surface porin proteins and extracellular polysaccharides.
The cell surface porin proteins such as OprC and OprE provide the path of transportation of bacterial extracellular polysaccharides (homopolysaccharides or heteropolysaccharides) into the cell (Hancock et al. 1990; Sutherland 2001c). These polysaccharides play a key role in adhesion and cohesion of cells to form extracellular matrix. More than 50 cell surface proteins encoded for biofilm formation was found in the sessile cells which are absent in planktonic cells (Hall-Stoodley and Stoodley 2002). The fluid-filled matrix supports the distribution of nutrients consistently inside the biofilm (Parsek and Singh 2003). A cascade of cell signalling mechanisms are involved during high cell concentration in the biofilm. These signalling molecules or autoinducers (e.g. homoserine lactones and small peptides) are used to trigger gene expression by enzymatic process for developing and maturation of biofilm (Bassler 1999) (Fig. 3.2). The biofilm will break down during death phase by secreting of enzymes by the microbial community within the biofilm. The lytic enzymes produced by surface bacteria to break down the polysaccharides aggregate the biofilm for colonization of new substrates. Alginate lyase, N-acetyl-heparosan lyase and hyaluronidase are found to be generally used in the breakdown of the biofilm matrix in Pseudomonas spp., Escherichia coli and Streptococcus equi, respectively (Sutherland 1999). Concurrently, the gene coding proteins are up-regulated for organisms’ motility, pathogenicity, luminance and metabolites production (Garrett et al. 2008; Rabin et al. 2015) (Fig. 3.2).
3 Quorum Sensing in Bacteria
All microorganisms have the capability to form biofilm on any surface (Sekhar et al. 2009). During biofilm formation microbial communities including intraspecies and interspecies are able to communicate between them through a mechanism known as quorum sensing. There are well-known QS systems described in bacteria: acyl homoserine lactones (AHLs) are a major class of autoinducer signalling molecules used by Gram-negative species for quorum sensing. AHLs are composed of homoserine lactone (HSL) rings containing acyl chains of C4 to C18 in length (Ng and Bassler 2009). These side chains entertain occasional alteration, particularly at the C3 position, or unsaturated double bonds (Fig. 3.3a) (Ng and Bassler 2009). Gram-positive bacterial species predominantly use modified oligopeptides as autoinducers in quorum sensing-regulated gene expression systems (Fig. 3.3b). Furthermore, the autoinducer molecules called AI-2 and HAI-1 system were found in both Gram-positive and Gram-negative bacterial species (Fig. 3.3c) (Fuqua et al. 2001; Bassler 2002; Sturme et al. 2002).
Representative chemical structures of bacterial autoinducers and the responsible enzymes for their production. (a) Gram-negative N-acyl-homoserine lactone autoinducers. (b) Oligopeptide autoinducers and amino acid sequences of the peptide autoinducers produced by Gram-positive bacteria. The bolded tryptophan in Bacillus subtilis (ComX) is isoprenylated. (c) Autoinducer-2 family quorum sensing molecules. DPD (4,5-dihydroxy-2,3-pentanedione), the precursor to AI-2. In the presence of boron, AI-2 exists as S-THMF-borate ((2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate). In the absence of boron, AI-2 exists as R-THMF ((2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran)
These systems play the central role for the formation of biofilm. QS relies upon the intercommunication of a small diffusible signal molecule with a sensor or transcriptional activator to trigger gene expression for QS-coordinated activities (Li and Tian 2012) (Fig. 3.2). It is constituted that during QS bacteria concurrently regulate gene expression in response to changes in high cell population densities and complexity of microbial species (Ng and Bassler 2009). There are two types of gene expression systems on QS, i.e. low-cell density dependent for individual and non-social behaviours and high-cell density dependent for group and social behaviours (Parsek and Greenberg 2005; Waters and Bassler 2005; Williams et al. 2007; Novick and Geisinger 2008). The detecting and responding to variations in cell density is the essential phases of QS. The low molecular weight molecules of autoinducers are produced intracellularly and secreted outside the cells either passively or actively. The increase of concentration of autoinducer is proportional to increase in the number of cells of a population. When autoinducers accumulate to meet minimal threshold, QS-related receptors bind to the autoinducers and activate signal transduction cascades leading to gene expression in population-wide changes (Ng and Bassler 2009) (Fig. 3.2).
4 Quorum Sensing in Gram-Negative Bacteria
An archetypal Gram-negative bacterial quorum sensing circuit is shown in Fig. 3.4a. Acyl homoserine lactone (AHL)-mediated Lux-type QS is common in many Gram-negative bacterial species. The LuxR and LuxI homologs in Gram-negative bacterial species are responsible for production of autoinducers (Fuqua et al. 2001; Parsek and Greenberg 2005). The first AHL autoinducer identified in the marine bacterium Vibrio fischeri inhabits as an endosymbiont in the light organ of Hawaiian squid Euprymna scolopes (Ruby 1996). The luminescence produced by V. fischeri is utilized by host E. scolopes for its anti-predation mechanism (Ruby 1996). The LuxI and LuxR are important proteins for QS regulation of bioluminescence in V. fischeri. LuxI is an autoinducer of N-3-(oxo-hexanoyl) homoserine lactone of the QS synthases (Fig. 3.3a) (Engebrecht and Silverman 1984; Schaefer et al. 1996). The autoinducer diffuses passively through the bacterial membrane, and its concentration increases both intra- and extracellularly as the cell density of the population increases (Bassler 2002). The LuxR is the cytoplasmic receptor for autoinducer and also the transcriptional activator of the luciferase luxICDABE operon (Engebrecht and Silverman 1984). The autoinducer ligand is necessary for the stability of LuxR protein. When the QS autoinducer accumulates, it is bound by LuxR and the LuxRAHL complex identifying a unanimous binding system upstream of the luxICDABE operon and activates its expression (Stevens et al. 1994).
Pseudomonas aeruginosa is an opportunistic pathogen that primarily infects the immunocompromised individuals. It is a well-studied pathogen in terms of the regulation of virulence factors and the role the QS acts in pathogenicity. P. aeruginosa virulence characters are regulated by two different Lux-type QS systems such as las and rhl (Sifri 2008). This las and rhl system regulates the cascade of virulence regulators in P. aeruginosa including various virulence traits such as exoprotease secretion, toxin production, motility and biofilm formation (Van Delden and Iglewski 1998). The AHL quorum sensing in P. aeruginosa has been described by detection of las and rhl AHLs N-3-oxododecanoyl homoserine lactone and N-butanoyl-homoserine lactone in sputum samples collected from cystic fibrosis-infected patients (Singh et al. 2000; Erickson et al. 2002). The AHL autoinducer molecules are normally unique among which a specific AHL molecule is recognized only by the bacterial species that produces it. Therefore, the AHL-quorum sensing systems largely nurture intra-species cell-cell communication (Ng and Bassler 2009). In addition, the AHL especially N-3-oxododecanoyl homoserine lactone exhibits antibacterial activity exclusively against Gram-positive bacteria. Therefore, the production of AHLs enhance P. aeruginosa is more determined in a mixed bacterial population or in a biofilm formation (Sifri 2008). The third non-AHL-QS system that cell signalling occurs through quinolone compounds has been discovered in P. aeruginosa in addition to las and rhl systems (Sifri 2008). Production of quinolones (4-hydroxy-2-alkylquinolines) is regulated by the transcriptional regulator MvfR (Pesci et al. 1999). The MvfR regulates the expression of many genes through the trigger of PqsH in the production of anthranilic acid and its conversion to 4-hydroxy-2-heptylquinoline, and its further conversion to 3,4-dihydroxy-2-heptylquinoline is known as Pseudomonas quinolone signal (Gallagher et al. 2002; Deziel et al. 2004). Synthesis of MvfR and PqsH is regulated by LasR, thereby intertwining the mvfR signalling pathway with AHL quorum sensing (Deziel et al. 2004). Expression of QS-regulated genes is controlled by MvfR but is different from those regulated by AHL autoinducers (Deziel et al. 2005). The large network of QS-regulators such as las, rhl and mvfR are controlled by a wide range of cellular functions in P. aeruginosa.
5 Quorum Sensing in Gram-Positive Bacteria
Figure 3.4b displays the typical quorum sensing circuit of Gram-positive bacteria. Quorum sensing is a cell-cell communication and regulation of gene expression in Gram-positive bacterial species. In Gram-positive bacteria, the signalling molecules or autoinducers are mostly small post-translationally processed peptides called autoinducing peptides (AIPs) (Fig. 3.3b) (Monnet and Gardan 2015). The AIPs are impermeable to cell membranes; thereby secretion of QS small peptides is usually actively mediated by specialized transport proteins and secreted into the extracellular environment (Ng and Bassler 2009). Furthermore, in many cases, the initially produced small peptides are modified by processing and cyclization during secretion (Havarstein et al. 1995; Solomon et al. 1996; Ji et al. 1997; Ng and Bassler 2009). One of the major differences between Gram-positive and Gram-negative QS systems is the site of the cognate receptors. The Gram-negative species of LuxR-type receptors are cytoplasmic-bound, while the sensors for small oligopeptides in Gram-positive species are cell membrane-bound. Therefore, the signal transduction in Gram-positive species occurs through a series of phosphorylation cascade using membrane-bound two signalling proteins (Simon et al. 2007; Ng and Bassler 2009). The two-component signalling proteins such as membrane-bound histidine kinase receptor and a cognate cytoplasmic response regulator function as a transcriptional regulator (Simon et al. 2007; Ng and Bassler 2009). Similarly in AHL-QS systems, the concentration of secreted small oligopeptide autoinducers increases the cell density (Ng and Bassler 2009). A membrane-bound histidine kinase receptor activates its intrinsic autophosphorylation process by quorum sensing via detection of oligopeptide autoinducer accumulation and reaching a threshold concentration in the extracellular environment (Ji et al. 1995). This ATP-driven phosphorylation activity ensuring a conserved histidine residue (H) in the cytoplasm subsequently transfers the phosphate group to the conserved aspartate residue (D) of a cognate response regulator (Ng and Bassler 2009). Phosphorylation action triggers the regulators to employ as DNA-binding transcription factors to control expression of target genes (Ng and Bassler 2009).
Gram-positive peptide autoinducers are different from Gram-negative bacterial AHLs as they are genetically encoded, not showing similarity on a single core molecule. Therefore, Gram-positive bacterial species can produce a signal with a unique sequence (Fig. 3.3b) (Ng and Bassler 2009). The quorum sensing system of Staphylococcus aureus is a well-studied system, which is encoded by the accessory gene regulator (agr) locus (Sifri 2008). The agr system plays a crucial role in regulating the syntheses of a wide range of S. aureus virulence factors (Novick 2003) and complex association with biofilm formation (Sifri 2008). The agr locus consists of two different transcripts such as RNAII and RNAIII. The RNAII encoding agrB, agrD, agrC and agrA and RNAIII are instrumental in suppression of cell wall-related protein production and increase the production of exoprotein secretion in response to high cell concentration (Sifri 2008). The colonization facilitated by cell wall-associated adhesins however secreted products of S. aureus inevitably for invasion and dissemination. The four genes encoded by RNAII are involved in the production and sensation of the AIPs (Sifri 2008). The agrD encodes the precursor of the AIP; however, integral membrane protein AgrB controls its processing and excretion as a thiolactone-modified cyclic oligopeptide. The extracellular accumulation of the AIP is regulated by a two-component histidine kinase that constituted AgrA and AgrC, whereas the transcription of RNAII and RNAIII is induced by activation of AgrA-AgrC. Interestingly, RNAIII undergo self-transcription and acts as the regulatory effector molecule for the agr system, mainly by translational inhibition of the virulence gene repressor and possibly other gene regulators (Geisinger et al. 2006; Boisset et al. 2007; Sifri 2008). The autoinduction and signal transduction of agr system regulates the staphylococcal virulence (Novick 2003; Sifri 2008).
6 Quorum Sensing in Intraspecies or Cross-Species
The third type of quorum sensing known as autoinducer 2 (AI-2) has been reported in both Gram-negative and Gram-positive species (Miller and Bassler 2001; Federle and Bassler 2003; Waters and Bassler 2005). The AI-2 quorum sensing system is different from the other two quorum sensing systems that are particularly implemental in signalling in intraspecies or cross-species communication (Schauder and Bassler 2001; Federle and Bassler 2003). However, two autoinducers, namely HAI-1 and AI-2, are produced by a marine bacterium V. harveyi (Fig. 3.3c). Notably, HA-1 is archetypal to the Gram-negative QS metabolite AHL. However, HA-1 synthesis is not dependent on a LuxI-like enzyme. AI-2 is a furanosyl borate diester that regulates cell density-dependent bioluminescence in V. harveyi (Chen et al. 2002; Vendeville et al. 2005). Both the HAI-1 and AI-2 signal transductions occur through similar phosphorylation cascade of Gram-positive species. Later, Miller et al. (2004) also reported a chemically distinct form of the quorum sensing signalling molecule AI-2 from Salmonella typhimurium (Fig. 3.3c).
The model system of quorum sensing circuit controlling bioluminescence shown in Fig. 3.4c is that of the Gram-negative bacterium V. harveyi. A highly conserved catalytic enzyme LuxS plays a vital role in AI-2 synthesis. The AI-2 quorum sensing is predominant among prokaryotic organisms as luxS gene has been determined in several bacterial species (Li and Tian 2012). In AI-2 synthesis, LuxS primarily converts S-ribosylhomocysteine to homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) (Fig. 3.3c) where the DPD is a precursor molecule for synthesizing AI-2 (Irie and Parsek 2008). The LuxP protein acts as a cytoplasmic receptor and a transcriptional activator in the V. harveyi system (Vendeville et al. 2005). However, in Salmonella enterica sv. Typhimurium system, AI-2 is first transported inside the cell prior to initiating a signalling cascade (Taga et al. 2001; Irie and Parsek 2008). A number of diverse interspecies cell-cell communication have been reported by AI-2 that regulates specific target gene expression in V. harveyi, Vibrio cholerae, Escherichia coli, Salmonella typhimurium, Shigella flexneri, Streptococcus pyogenes, Clostridium perfringens, Porphyromonas gingivalis, Neisseria meningitidis, Borrelia burgdorferi and Actinobacillus actinomycetemcomitans (Bassler 2002).
7 Biofilm as a Mechanism of Resistance to Antibiotics
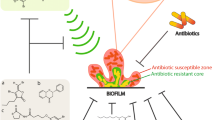
It is now confirmed that intra- and intercellular communications in microorganisms govern its microbial ecology (Penesyan et al. 2015). The complex cell-cell signalling in natural bacterial communities promotes the evolution and leads the bacterial species as super bugs that are extremely resistant antibiotics. More than 75% of bacterial infections involved biofilm formation and encompasses surface-attached bacterial colonies that are protected by an extracellular matrix (Musk and Hergenrother 2006). It is reported that the immobile or sessile bacterial species in a biofilm are 1000 times more resistant to antibiotic treatment than the same organism that were grown as free-floating planktonic cells, which rigorously complicates treatment choices (Rasmussen and Givskov 2006). The knowledge about the molecular mechanisms of antibiotic resistance in biofilms is meagre, despite the existence of a decade of research. However, some intrinsic and extrinsic resistance factors are documented to biofilm resistance to antibiotics (Anderson and O’Toole 2008).
The intrinsic factors also known as innate resistance are associated with biofilm development and life cycle (Paraje 2011). There are many different types of intrinsic biofilm factors that are influential of the antibiotic resistance in bacterial communities in biofilm. The factors being: (1) Biofilms can act as diffusion barriers to preclude antibiotics to reach their targets. Biofilm constitutes a rich exopolysaccharide, enzymes, DNA, protein, water channel and bacterial cells (Rabin et al. 2015). These physical and chemical properties of the matrix make the antibiotics liable to limited diffusion or render it ineffective against bacterial species in biofilm and make them more resistant against antibiotics. (2) The deficiency of nutrients and oxygen inside biofilms facilitates the bacterial communities to establish the microenvironment within the biofilm. This microenvironment might induce alternative metabolic activity resulting in slow growth of the bacteria (Paraje 2011). Furthermore, many studies are evidential to the oxygen limitation, hypoxic zones, restricted nutrient diffusion and slow or no growth of bacteria within biofilms (Patel 2005; Paraje 2011). The slow growth certainly leads resistance to killing of bacteria which are occupied within the biofilm (Costerton et al. 1999). However, the stationary planktonic cells are being killed due to slow growth in the microenvironments that undermine the activity of antibiotics by pH variations (Patel 2005; Høiby et al. 2010). (3) A small subpopulation of bacteria within biofilm possibly differentiates into persister cells (Paraje 2011). Generally, the non-growing or slow-growing bacteria in the biofilms that differentiate into dormant cells are considered as persister cells, which are highly resistant to antibiotic treatment (Lewis 2005). The persister cells undergo phenotypic variations by stable genetic changes for withstanding in the extreme antibiotic treatment environment (Keren et al. 2004). However, the association among planktonic persisters and biofilm resistance and the mechanisms of antibiotic tolerance are unclear (Paraje 2011). (4) Microbial communities in biofilms producing imbalanced or increased oxidants such as the free radicals, peroxide and nitric oxide lead to overproduction of reactive oxygen species (ROS) (Paraje 2011). Resulting in the detoxification of ROS by antioxidant defence enzymes particularly, superoxide dismutase (SOD) and catalase (CAT) are inadequate to eliminate the free radicals in the biofilms (Sardesai 1995). Cumulatively, ROS known as oxidative stress may result in significant damage to cell structures including the matrix, DNA, proteins and lipids (Becerra et al. 2006; Baronetti et al. 2011; Arce Miranda et al. 2011).
Furthermore, the amplified synthesis of oxidative stress stimulates specific variations in the physiology of bacteria (Paraje 2011). It is also important to note that biofilm development is determined by the balanced production of oxidants (ROS and NO) and antioxidant defences (SOD) which can be much affected by diverse environmental stress factors that would lead to cellular stress causing a reduction in the extracellular matrix of the biofilms (Arce Miranda et al. 2011). In addition, bacteria in biofilms can trigger oxidative stress mode through inducing the SOS response and activating DNA repair systems, such as methyl mismatch repair (MMR) or the DNA oxidative repair system (GO) (Jolivet-Gougeon and Bonnaure-Mallet 2014). Unexpectedly, the immense number of bacteria with mutations in DNA repair genes has been detected inside biofilms, contributing in a hypermutator phenotype with a mutation rate up to 1000-fold (Jolivet-Gougeon and Bonnaure-Mallet 2014). This phenotype grants a critical advantage for strong mutator species relating to adhesion capability (Le Bars et al. 2012), growth in biofilms (Lujan et al. 2011) and persistence (Mena et al. 2007). (5) Bacteria within biofilms produce a prophylactic shield against phagocytes through QS-regulated synthesis of virulence factors such as enzymes and cellular lysins (Paraje 2011). In addition, QS influence the biofilm development and regulate the tolerance of biofilms to antibiotic action (Bjarnsholt et al. 2005).
The extrinsic or induced resistance factors are induced transcriptionally in biofilm-growing bacteria against antibiotic treatment. The highest occurrence of mutation has been observed in sessile bacteria compared to free-floating planktonic bacteria residing in biofilm through horizontal gene transmission (Paraje 2011). Studying horizontal gene transfer in natural environments contributes emergence of multidrug-resistant bacteria and genetic diversity of microbial communities (Martínez 2009, 2012). Bacteria can accumulate high levels of enzymes in response to antibiotics. For example, Pseudomonas aeruginosa biofilm secrete high amount of beta-lactamase regulated by beta-lactamase gene (ampC) by expression of the green fluorescent protein (GFP) after exposure to high dose of ceftazidime (Bagge et al. 2004; Jolivet-Gougeon and Bonnaure-Mallet 2014). Therefore, the bacterial cells within biofilms might concurrently produce antibiotic degrading enzymes that affect the affinity of antibiotic target and over express efflux pumps that have a broad range of substrates (Paraje 2011; Jolivet-Gougeon and Bonnaure-Mallet 2014). The multidrug efflux pumps in biofilm-growing bacteria importantly contribute to biofilm formation, and this mechanism is arsenal in defeating various classes of antibiotics.
8 Conclusion
The bacterial physiological functions such as motility, development of antibiotic resistance and virulence factors and biofilm formation are regulated by quorum sensing mechanism. It is undeniable that increased antibiotic resistance in biofilm forming bacteria and further comprehensive studies of molecular mechanisms are needed to understand the regulatory system of QS.
References
Anderson GG, O’Toole GA (2008) Innate and induced resistance mechanisms of bacterial biofilms. In: Romeo T (ed) Bacterial biofilms, Current topics in microbiology and immunology, vol 322. Springer-Verlag, Berlin/Heidelberg, pp 85–105
Arce Miranda JE, Sotomayor CE, Albesa I, Paraje MG (2011) Oxidative and nitrosative stress in Staphylococcus aureus biofilm. FEMS Microbiol Lett 315:23–29
Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Hoiby N (2004) Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 48:1168–1174
Baronetti JL, Angel Villegas N, Paraje MG, Albesa I (2011) Nitric oxide-mediated apoptosis in rat macrophages subjected to Shiga toxin 2 from Escherichia coli. Microbiol Immunol 55:231–238
Bassler BL (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 2:582–587
Bassler BL (2002) Small talk: cell-to-cell communication in bacteria. Cell 109:421–424
Becerra MC, Paez PL, Larovere LE, Albesa I (2006) Lipids and DNA oxidation in Staphylococcus aureus as a con sequence of oxidative stress generated by ciprofloxacin. Mol Cell Biochem 285:29–34
Bjarnsholt T, Jensen PØ, Burmølle M, Hentzer M, Haagensen JAJ, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Høiby N, Givskov M (2005) Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383
Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator rot by an antisense mechanism. Genes Dev 21:1353–1366
Chadha T (2014) Bacterial biofilms: survival mechanisms and antibiotic resistance. J Bacteriol Parasitol 5:190
Characklis WG, Marshal KC (1990) Biofilms. Wiley, New York
Chen X, Schauder S, Potier N, Van Dorssealaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G (2003) The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112:1466–1477
Darch SE, West SA, Winzer K, Diggle SP (2012) Density-dependent fitness benefits in quorum sensing bacterial populations. Proc Natl Acad Sci U S A 109:8259–8263
Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1144
Deziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55:998–1014
Dunne WM (2002) Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166
Engebrecht J, Silverman M (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A 81:4154–4158
Erickson DL, Endersby R, Kirkham A, Stuber K, Vollman DD, Rabin HR, Mitchell I, Storey DG (2002) Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun 70:1783–1790
Federle MJ, Bassler BL (2003) Interspecies communication in bacteria. J Clin Invest 112:1291–1299
Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468
Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C (2002) Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184:6472–6480
Garrett TR, Bhakoo M, Zhang Z (2008) Bacterial adhesion and biofilms on surfaces. Prog Nat Sci 18:1049–1056
Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61:1038–1048
Hall-Stoodley L, Stoodley P (2002) Developmental regulation of microbial biofilms. Curr Opin Biotechnol 13:228–233
Hancock REW, Siehnel R, Martin N (1990) Outer membrane proteins of Pseudomonas. Mol Microbiol 4:1069–1075
Hausner M, Wuertz S (1999) High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol 65:3710–3713
Havarstein LS, Coomaraswamy G, Morrison DA (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144
Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O (2010) Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332
Irie Y, Parsek MR (2008) Bacterial biofilms. In: Romeo T (ed) Quorum sensing and microbial biofilms. Springer, Berlin/Heidelberg, pp 67–84
Jefferson KK (2004) What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173
Ji G, Beavis RC, Novick RP (1995) Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A 92:12055–12059
Ji G, Beavis R, Novick RP (1997) Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030
Jolivet-Gougeon A, Bonnaure-Mallet M (2014) Biofilms as a mechanism of bacterial resistance. Drug Discov Today Technol 11:49–56
Joo HS, Otto M (2012) Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol 19:1503–1513
Keren I, Kaldalu N, Spoering A, Wang YP, Lewis K (2004) Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18
Le Bars H, Le Gall-David S, Renoux VM, Bonnaure-Mallet M, Jolivet-Gougeon A, Bousarghin L (2012) Impact of a mutator phenotype on motility and cell adherence in Salmonella Heidelberg. Vet Microbiol 159:99–106
Lewis K (2005) Persister cells and the riddle of biofilm survival. Biochemistry 70:267–274
Li YH, Tian X (2012) Quorum sensing and bacterial social interactions in biofilms. Sensors 12:2519–2538
Liu Y, Yang SF, Li Y, Xu H, Qin L, Tay JH (2004) The influence of cell and substratum surface hydrophobicities on microbial attachment. J Biotechnol 110:251–256
Lu TK, Collins JJ (2007) Dispersing biofilms with engineered enzymatic bacteriophage. PNAS 104:11197–11202
Lujan AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM (2011) Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 6:e27842
Lyon GJ, Muir TW (2003) Chemical signaling among bacteria and its inhibition. Chem Biol 10:1007–1021
Martinez JL (2009) The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci 276:2521–2530
Martínez JL (2012) Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front Microbiol 3:1
Mena A, Macia MD, Borrell N, Moya B, de Francisco T, Perez JL, Oliver A (2007) Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J Bacteriol 189:3665–3668
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199
Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM (2004) Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 15:677–687
Monnet V, Gardan R (2015) Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide’. Mol Microbiol 97:181–184
Musk DJ Jr, Hergenrother PJ (2006) Chemical countermeasures for the control of bacterial biofilms: effective compounds and promising targets. Curr Med Chem 13:2163–2177
Ng WL, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222
Novick RP (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449
Novick RP, Geisinger E (2008) Quorum sensing in staphylococci. Annu Rev Genet 42:541–564
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79
Okada M, Sato I, Cho SJ, Iwata H, Nishio T, Dubnau D, Sakagami Y (2005) Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol 1:23–24
Palmer J, Flint S, Brooks J (2007) Bacterial cell attachment, the beginning of a biofilm. J Ind Microbiol Biotechnol 34:577–588
Paraje M (2011) Antimicrobial resistance in biofilms. In: Méndez-Vilas A (ed) Science against microbial pathogens: communicating current research and technological advances. Formatex Research Center, Badajoz, pp 736–744
Parsek MR, Greenberg EP (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33
Parsek MR, Singh PK (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701
Patel R (2005) Biofilms and antimicrobial resistance. Clin Orthop Relat Res 437:41–47
Penesyan A, Gillings M, Paulsen IT (2015) Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20:5286–5298
Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH (1999) Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234
Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO (2015) Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem 7:493–512
Rasmussen TB, Givskov M (2006) Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 296:149–161
Renner LD, Weibel DB (2011) Physicochemical regulation of biofilm formation. MRS Bull 36:347–355
Ruby EG (1996) Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624
Sardesai VM (1995) Role of antioxidants in health maintenance. Nutr Clin Pract 10:19–25
Sauer FG, Remaut H, Hultgren SJ, Waksman G (2004) Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta 1694:259–267
Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A 93:9505–9509
Schauder S, Bassler BL (2001) The languages of bacteria. Genes Dev 15:1468–1480
Sekhar S, Kumar R, Chakraborti A (2009) Role of biofilm formation in the persistent colonization of Haemophilus influenzae in children from northern India. J Med Microbiol 58:1428–1432
Sifri CD (2008) Quorum sensing: bacteria talk sense. Healthcare Epidemiol 47:1070–1076
Simon MI, Crane BR, Crane A (2007) Two-component signaling systems. Academic Press, San Diego
Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764
Solomon JM, Lazazzera BA, Grossman AD (1996) Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev 10:2014–2024
Sordi LD, Muhlschlegal FA (2009) Quorum sensing and fungal-bacterial interactions in Candida albicans: a communication network regulating microbial coexistence and virulence. FEMS Yeast Res 9:990–999
Staley C, Dunny GM, Sadowsky MJ (2014) Environmental and animal associated enterococci. Adv Appl Microbiol 87:147–186
Stevens AM, Dolan KM, Greenberg EP (1994) Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci U S A 91:12619–12623
Sturme MH, Kleerebezem M, Nakayama J, Akkermans AD, Vaugha EE, de Vos WM (2002) Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81:233–243
Sutherland IW (1999) Polysaccharases for microbial exopolysaccharides. Carbohydr Polym 38:319–328
Sutherland IW (2001a) The biofilm matrix – an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227
Sutherland I (2001b) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9
Sutherland IW (2001c) Microbial polysaccharides from Gram-negative bacteria. Int Dairy J 11:663–674
Taga ME, Semmelhack JL, Bassler BL (2001) The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol 42:777–793
van Bodman SB, Willey JM, Diggle SP (2008) Cell-cell communication in bacteria: united we stand. J Bacteriol 190:4377–4391
Van Delden C, Iglewski BH (1998) Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4:551–560
Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR (2005) Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3:383–396
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Watnick PI, Kolter R (1999) Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 34:586–595
West SA, Winzer K, Gardner A, Diggle SP (2012) Quorum sensing and the confusion about diffusion. Trends Microbiol 20:586–594
Williams P, Winzer K, Chan WC, Camara M (2007) Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond Ser B Biol Sci 362:1119–1134
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Subramani, R., Jayaprakashvel, M. (2019). Bacterial Quorum Sensing: Biofilm Formation, Survival Behaviour and Antibiotic Resistance. In: Bramhachari, P. (eds) Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry . Springer, Singapore. https://doi.org/10.1007/978-981-32-9409-7_3
Download citation
DOI: https://doi.org/10.1007/978-981-32-9409-7_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9408-0
Online ISBN: 978-981-32-9409-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)