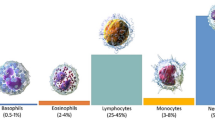
Abstract
White blood cell (WBC) counting is vital for the diagnostic of many diseases. However, the manual counting method operated in hospitals and centers is time-consuming and inefficient. Many automatic processes in classifying WBC types were proposed using deep learning (DL) models. YOLOv4 is an emerging model for detecting objects with promising results in many applications including the medical field. In this paper, YOLOv4’s performance would be tested on classifying four types of WBC: monocyte, lymphocyte, neutrophil, and eosinophil. A mixed database of WBC microscopic images from an online dataset BCCD and a hospital dataset collected from the Vietnamese National Institute of Hematology and Blood Transfusion, comprising of 10,275 images in total after the augmentation, was used for training and testing the model. The results showed that the trained YOLOv4 was able to classify equivalently well all 4 WBC types with the average accuracy of 97.8%, ranking the second-best option when compared to other contemporary studies.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Al-Dulaimi K, Nguyen K, Banks J, Chandran V, Tomeo-Reyes I (2018) Classification of white blood cells using l-moments invariant features of nuclei shape, In: 2018 International conference on image and vision computing New Zealand (IVCNZ), pp. 1–6. New Zealand
Deng J, Xuan X, Li W, Wang Z, Yao H, Wang Z (2020) A review of research on object detection based on deep learning. J Phys Conf Ser (2020):012028
Arakeri MP, Reddy GRM (2015) Computer-aided diagnosis system for tissue characterization of brain tumor on magnetic resonance images. SIViP 9(2):409–425
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. Journal of Big Data 6(1):1–48
Özyurt F (2020) A fused CNN model for WBC detection with MRMR feature selection and extreme learning machine. Soft Comput 24(11):8163–8172
Jiang M, Cheng L, Qin F, Du L, Zhang M (2018) White blood cells classification with deep convolutional neural networks. Int J Pattern Recognit Artif Intell 32(09):1857006
Sahlol AT, Kollmannsberger P, Ewees AA (2020) Efficient classification of white blood cell leukemia with improved swarm optimization of deep features. Sci Rep 10(1):1–11
Sharma M, Bhave A, Janghel RR (2019) White blood cell classification using convolutional neural network. Soft Comput Signal Process 135–143
Toğaçar M, Ergen B, Cömert Z (2020) Classification of white blood cells using deep features obtained from Convolutional Neural Network models based on the combination of feature selection methods. Appl Soft Comput 97:106810
Kutlu H, Avci E, Özyurt F (2020) White blood cells detection and classification based on regional convolutional neural networks. Med Hypotheses 135(97):109472
Yu W, Chang J, Yang C, Zhang L, Shen H, Xia Y, Sha J (2017) Automatic classification of leukocytes using deep neural network. In: 2017 IEEE 12th international conference on ASIC (ASICON), pp 1041–1044
Macawile MJ, Quiñones VV, Ballado A, Cruz JD, Caya MV (2018) White blood cell classification and counting using convolutional neural network. In: 2018 3rd International conference on control and robotics engineering (ICCRE), pp 259–263
Pandian A (2019) Pasumpon: Identification and classification of cancer cells using capsule network with pathological images. J Artif Intell 1(01):37–44
Qin F, Gao N, Peng Y, Wu Z, Shen S, Grudtsin A (2018) Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput Methods Programs Biomed 162:243–252
Habibzadeh M, Jannesari M, Rezaei Z, Baharvand H, Totonchi M (2018) Automatic white blood cell classification using pre-trained deep learning models: ResNet and Inception. In: Tenth international conference on machine vision, p 1069612
Chen J-Z (2021) Design of accurate classification of COVID-19 disease in X-Ray images using deep learning approach. J ISMAC 3(02):132–148
Bochkovskiy A, Wang CY, Liao HYM (2020) Yolov4: optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934
Zou S (2019) A review of object detection techniques. In: 2019 International conference on smart grid and electrical automation (ICSGEA), pp 251–254
Qiao S et al (2020) Automatic detection of cardiac chambers using an attention-based YOLOv4 framework from four-chamber view of fetal echocardiography. arXiv preprint arXiv:2011.13096
Albahli S et al (2020) Melanoma lesion detection and segmentation using YOLOv4-DarkNet and active contour. IEEE Access 8:198403–198414
Punn NS, Sonbhadra SK, Agarwal S, Rai G (2020) Monitoring COVID-19 social distancing with person detection and tracking via fine-tuned YOLO v3 and deepsort techniques. arXiv preprint arXiv:2005.01385
Montalbo FJP (2020) A computer-aided diagnosis of brain tumors using a fine-tuned YOLO-based model with transfer learning. KSII Transa Internet Inf Syst 14(12)
Abdurahman F, Fante KA, Aliy M (2021) Malaria parasite detection in thick blood smear microscopic images using modified YOLOV3 and YOLOV4 models. BMC Bioinformatics 22(1):1–17
Wang X et al (2018) SO-YOLO based WBC detection with Fourier ptychographic microscopy. IEEE Access 6:51566–51576
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Ma N, Zhang X, Zheng HT, Sun J (2018) Shufflenet v2: practical guidelines for efficient CNN architecture design, In: Proceedings of the European conference on computer vision (ECCV), pp 116–131
He K, Zhang X, Ren S, Sun J (2015) Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell 37(9):1904–1916
Liu S, Qi L, Qin H, Shi J, Jia J (2018) Path aggregation network for instance segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 8759–8768
Mish MD (2019) A self regularized non-monotonic neural activation function. arXiv preprint arXiv:1908.08681
Redmon J, Farhadi A (2018) Yolov3: an incremental improvement. arXiv preprint arXiv:1804.02767
Wu D, Lv S, Jiang M, Song H (2020) Using channel pruning-based YOLO v4 deep learning algorithm for the real-time and accurate detection of apple flowers in natural environments. Comput Electron Agric 178:105742
Sajjad M et al (2016) Leukocytes classification and segmentation in microscopic blood smear: a resource-aware healthcare service in smart cities. IEEE Access 5:3475–3489
Ryabchykov O et al (2016) Leukocyte subtypes classification by means of image processing. In: 2016 Federated conference on computer science and information systems (FedCSIS), pp 309–316
Khamael A et al (2018) Classification of white blood cells using l-moments invariant features of nuclei shape. In: 2018 International conference on image and vision computing New Zealand (IVCNZ), New Zealand, pp 1–6
Habibzadeh M, Krzyżak A, Fevens T (2013) Comparative study of shape, intensity and texture features and support vector machine for white blood cell classification. J Theoret Appl Comput Sci 7(1):20–35
Manik S, Saini LM, Vadera N (2016) Counting and classification of white blood cell using artificial neural network (ANN). In: 2016 IEEE 1st international conference on power electronics, intelligent control and energy systems (ICPEICES), pp 1–5
Zhao J, Zhang M, Zhou Z, Chu J, Cao F (2016) Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Comput 55(8):1287–1301
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Trong, L.D., Thanh, T.P., Manh, H.P., Minh, D.N. (2023). Yolov4 in White Blood Cell Classification. In: Jacob, I.J., Kolandapalayam Shanmugam, S., Izonin, I. (eds) Data Intelligence and Cognitive Informatics. Algorithms for Intelligent Systems. Springer, Singapore. https://doi.org/10.1007/978-981-19-6004-8_31
Download citation
DOI: https://doi.org/10.1007/978-981-19-6004-8_31
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-6003-1
Online ISBN: 978-981-19-6004-8
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)