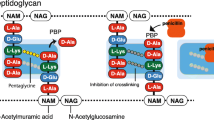
Abstract
Among various β-lactam hydrolyzing enzymes, classified from group A to D, the most genetically as well as biochemically diverse is the class D β-lactamases (DBL), few of which can incapacitate the complete spectrum of β-lactamases. DBLs, like class A and C, are active serine site enzymes, differing from them in amino acid structure. The DBLs form an enzyme substrate complex with β-lactam antibiotics in the periplasmic space leading to their hydrolysis with Ser70 serving as the active site. DBLs can be acquired and natural. Acquired DBLs are classified into narrow spectrum, extended spectrum, and carbapenem-hydrolyzing β-lactamases (CHDLs). Detection of class D β-lactamases is crucial yet challenging due to the lack of appropriate and standardized phenotypic assays. However, currently, molecular detection of the DBL genes is the only standardized method of identification of class D β-lactamases. Intensive research is required for developing rapid and easy detection tools for DBLs and for the discovery of class D specific inhibitors.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
β lactam antibiotics have had been the vanguard of antimicrobial chemotherapy despite their use of over half a century. Of late, majority of these antibiotics are rendered ineffective due to the increasing antimicrobial resistance. Of the various mechanisms of the antimicrobial resistance, presence of bacterial enzymes remains the smartest and traditional mechanism of acquired as well as innate resistance. Among the various β lactam hydrolyzing enzymes, classified from group A to D, the most genetically as well as biochemically diverse is the class D β lactamases (DBLs), few of which can incapacitate the complete spectrum of β lactamases (i.e., penicillins, cephalosporins, and carbapenems) (Leonard et al. 2012; Poirel et al. 2010). Clinically they may be signified into narrow spectrum (effective against earliest generation penicillins and cephalosporins), extended spectrum (hydrolyze later generation cephalosporins), and the most concerning are the DBLs that hydrolyze clinically important carbapenems (e.g., imipenem). They are the largest growing family of β lactamases based on the percentage of new enzymes and their variants (Bush 2013). Their genes may be chromosomal or may be located on plasmids of gram-negative pathogens like Acinetobacter, Shewanella, Pseudomonas, Burkholderia, and few gram-positive microbes also (Sanschagrin et al. 1995; Poirel et al. 2010; Toth et al. 2016). These enzymes are easily transferred between the species due to their association with integrons, insertion sequences, and transposons and are a formidable threat to hospitalized patients (Naas and Nordmann 1999). When associated with other enzymes on the same plasmid they result in synergized phenotypic resistance spectrum narrowing the treatment options (Mendes et al. 2009).
2 General Properties
DBLs, similar to class A and C, are active serine site enzymes differing from them in amino acid structure. In contrast, class B β lactamases have a Zn2+ ion(s) at the active site and are considered as metallo-enzymes (Ambler 1980; Jaurin and Grundstrom 1981; Lamotte-Brasseur et al. 1994). DBLs are highly diverse in sequence and show less than 20% homology with class A and C enzymes (Couture et al. 1992). However, the topological fold is preserved among the three classes and more so within class D (Fisher et al. 2005).
Also known as OXA-type enzymes or oxacillinases, DBLs include more than 400 genetically diverse enzymes described (Bush 2013) predominantly in gram-negative pathogens (e.g., Acinetobacter spp., Pseudomonas aeruginosa, and Enterobacterales), along with the gram-positive pathogens (Walther-Rasmussen and Høiby 2006; Bush and Fisher 2011).
Majority DBLs hydrolyze cloxacillin or oxacillin at a rate of more than 50% higher than that for benzylpenicillin, although this generalization is no longer valid (Fisher et al. 2005; Walther-Rasmussen and Høiby 2006). They are characteristically not inhibited by β-lactamase inhibitors such as clavulanic acid, tazobactam, and sulbactam (Payne et al. 1994; Bush et al. 1995), with a few exceptions, such as OXA-2, OXA-29, and OXA-32 inhibited by tazobactam, and OXA-53, inhibited by clavulanic acid (Franceschini et al. 2001; Mulvey 2004; Naas and Nordmann 1999). However, they are susceptible towards the recently developed inhibitors like avibactum and vaborbactam (Voha et al. 2006; Schneider et al. 2006).
3 Occurrence of DBLs
DBLs genes have been shown to be acquired as well as are naturally present in pathogens as well as environmental microbes (Poirel et al. 2010). The naturally occurring OXA β lactamases are found as cluster in bacterial species (Yoon and Jeong 2020). Few notable innate DBLs include OXA-22 in Ralstonia species (Nordmann et al. 2000; Jiang et al. 2020), OXA-42 like enzymes in Burkholderia pseudomallei (Niumsup and Wuthiekanun 2002), OXA-12 like subfamilies in Aeromonas species (Walsh et al. 1995), OXA-22 subfamilies in Acinetobacter species (Tian et al. 2018; Périchon et al. 2013). Mostly the innate DBLs occur as survival machinery in environmental bacteria (Yoon and Jeong 2020).
Rampant antimicrobial use/misuse/overuse creates a castigatory environment for the clinical bacterial isolates, which thus acquire various resistance mechanisms. The attainment of resistance genes for DBLs is mostly through mobile genetic elements like ISs using transposons or integrons and less commonly by homologous recombination (Yoon and Jeong 2020). The genes for NS and ES DBLs are frequently found as gene cassettes on class 1 integrons or less commonly on class 3 integrons, whereas genes for CHDLs are usually associated with ISs associated with transposons (Yoon and Jeong 2020).
4 Mechanism of Action
The DBLs form an enzyme substrate complex with β-lactam antibiotics in the periplasmic space leading to their hydrolysis. Like other beta-lactamases OXA-β lactamases or DBLs also have Ser70 that serves as the active site. However, DBL has a special hydrophobic active site compared to other β-lactamases.
Lys73 present in the Ser70-X-X-Lys motif in the DBL undergoes N-carboxylation post-translationally to become carbamylated lysine. A strongly hydrophobic active site helps create the conditions that allow the lysine to combine with CO2, and the resulting carbamate is stabilized by a number of hydrogen bonds (Leonard et al. 2012). Trp158 interacts with the carboxylate group of carbamylated Lys73 to form the active site channel. The Ser70 active site in DBL undergoes transient acylation and mimics the penicillin binding proteins (PBPs). The lysine carbamate is essential in acetylation and diacylation step in DBL catalysis, it serves as a general base to activate the serine nucleophile in the acylation reaction and the deacylating water in the second step.
5 Classification
The two classic and most frequently schemes for classification of β lactamases include molecular structure classification using the Ambler method (Ambler 1980) and functional classification using the Bush–Jacoby–Medeiros method (Bush 2013; Bush et al. 1995). β-lactamases are divided into four classes A, B, C, and D in the Ambler classification, by motifs composed of primary sequences constituting the protein molecules. Class A, C, and D β-lactamases use serine at the enzyme active center, whereas class B β-lactamases use metal zinc ions. In functional classification using the Bush–Jacoby–Medeiros method, β-lactamases are classified into groups 1–3 based on the hydrolysis of β-lactam substrates and the effect of the inhibitor.
According to the Bush–Jacoby classification based on substrate hydrolysis, DBLs are classified into group 2d. Those hydrolyzing extended spectrum cephalosporins into 2de, carbapenems into 2df and those hydrolyzing both extended spectrum cephalosporins and carbapenems in group 2def (Bush 2013).
DBLs can be divided into acquired and natural. Acquired DBLs are classified into narrow spectrum, extended spectrum, and carbapenem hydrolyzing β lactamases (CHDLs).
-
1.
Acquired narrow spectrum class D β lactamases (NS-DBL): Important examples include OXA 1, OXA 2, OXA 10 (Poirel et al. 2010). Others acquired narrow spectrum DBLs include OXA 9, 18, 12, 20, LCR 1, NPS 1 (Poirel et al. 2010). OXA 30 and 1 are the same, due to an original sequencing error during identification leading to a mistake (Boyd and Mulvey 2006).
OXA 1 has less than 30% homology with plasmid and chromosomal DBLs (Antunes et al. 2014). Since it is a narrow spectrum DBL it hydrolyses amino and ureidopenicillins and decreases the susceptibility to cephalothin, cefotaxime, and cefepime. However, it has no effect on carbapenems and ceftazidime. OXA 1 and OXA 31, which differ from it by two amino acid sequences, possess the ability to hydrolyze cefepime and cefpirome slightly. These can be considered to be extended spectrum DBLs for bacterial species with high level intrinsic impermeability (e.g. Pseudomonas species) and not on bacterial species with low level intrinsic permeability (e.g. E. coli) (Poirel et al. 2010). The OXA 1 gene is allied with class 1 integrons and is surrounded by the integrase and aminoglycoside aminoacyl transferase gene (Siu et al. 2000; Moura et al. 2012).
OXA 2 shares another cluster with its derivatives OXA 3, OXA 15, OXA 21, OXA 32, OXA 34, OXA 36, and OXA 53 and has 30% homology with OXA 1 (Kratz et al. 1983). OXA 2 has been identified in varying clinical species like Pseudomonas aeruginosa, Salmonella Typhimurium, Morganella morganii, Klebsiella pneumoniae, Bordetella bronchiseptica, Aeromonas hydrophila, and even gram-positive microbes like Corynebacterium amycolatum. OXA 2 is historical and can be tracked back to 1970s, characterized by hydrolysis of oxacillin many times higher than for benzylpenicillin (Suzuki et al. 2015). Although grouped with the narrow spectrum DBLs, studies have shown that OXA 2 is a CHDL (Antunes et al. 2014). Unlike other DBLs, it is inhibited by clavulanic acid and tazobactam.
OXA 10 The OXA 10 DBL (formerly known as PSE-2), originally found in Pseudomonas (Matthew and Sykes 1977), is now detected in a wide variety of gram-negative bacterial pathogens (Fournier et al. 2006; Centron and Roy 2002; Kumar and Thomas 2011). It hydrolyses cephalosporins including cefotaxime, ceftriaxone, aztreonam, but not ceftazidime, cephamycins, and carbapenems (Huovinen et al. 1988). Point mutation derivatives of OXA 10 (OXA 11, 13, 16, 28, 35, and 74) show extended spectrum of activity against cephalosporins (Poirel et al. 2010).
Other narrow spectrum DBLs include LCR-1, NPS-1, OXA 20, and OXA 46.
-
2.
Extended spectrum class D β lactamases (ES-DBL): These are mostly point mutation (clustered around the active site tryptophan) derivatives of the narrow spectrum DBLs and obviously pose a greater clinical challenge as they hydrolyze later generation cephalosporins that contain bulkier side chain constituents (e.g. cefotaxime, ceftazidime, and cefepime) (Leonard et al. 2012). Generally, members of the NS-OXA and CHDL transform their substrate profile to that of ES-DBLs. OXA-2 like and OXA-10 like subfamilies primarily consist of ES-DBLs (Yoon and Jeong 2020). OXA 15, derivative of OXA 2, was the first ES-DBL described (Gly replacing Asp at 150 position in the DBL numbering system) (Danel et al. 1997). OXA 32 is another derivative of OXA 2 (Leu 169 Ile substitution) (Poirel et al. 2002). A number of ES-DBL variants of OXA-10 have been identified, which include OXA 11 (with two substitutions at 146 and 167 BDL numbering system) (Hall et al. 1993), OXA 14 (Gly 167 Asp change) (Danel et al. 1995), OXA 16 (Ala114Thr and Gly167 Asp changes) (Danel et al. 1998), OXA 17 (Asn 76 Ser change) (Danel et al. 1999).
Other ES-DBLs like OXA 18, OXA-45 and OXA- 53 are extended spectrum β lactamases which are not structurally related to narrow spectrum OXAs. OXA 18 displays resistance to high level cephalosporins, but not cephamycins and carbapenems and unlike classic OXA DBLs are inhibited by clavulanic acid (Philippon et al. 1997). OXA 45 and OXA 53, similar to OXA 18 confer resistance to wide range of cephalosporins and are inhibited by clavulanic acid. OXA 18 is chromosomal (Naas et al. 2008), OXA 45 plasmid, while OXA 53 gene is plasmid and integron borne (Mulvey 2004).
-
3.
Acquired carbapenem hydrolyzing class D β lactamases (CHDLs): Of most concern are DBLs with the ability to hydrolyze carbapenems leading to treatment failures. Most of the CHDLs are found in Acinetobacter species. Of note is these carbapenem hydrolyzing CHDLs is the inability or low capacity to hydrolyze expanded spectrum cephalosporins (Poirel et al. 2010). OXA 23 (also known as ARI-1) was the first reported CHDL, detected in Acinetobacter baumannii isolate from Scotland and was found to be plasmid mediated (Dortet et al. 2008).
The CHDLs are divided into four subfamilies (OXA 23 like, OXA 24 like, OXA 48 like, and OXA 58 like) based on their phylogenetic origin, and they cluster according to their source of bacterial genera. These CHDLs are encoded as mobile gene in plasmids as identified in clinical strains, whereas the other CHDLs are generally immobile (Yoon and Jeong 2020). Clinically challenging bacteria possessing CHDLs include OXA 23 producing Acinetobacter baumannii, OXA 24 producing Acinetobacter baumannii, OXA 48 producing Enterobacterales, and OXA 58 producing Acinetobacter species (Yoon and Jeong 2020).
-
a.
OXA 23 like subfamily: This subfamily consists of 41 members, most of which are carbapenemases, with the exception of OXA 105 and OXA 481, which are yet to be described (Yoon and Jeong 2020). OXA 23 was the first CHDL to be identified as mentioned above. The other significant member of the first group of CHDLs is OXA 27, identified from Singapore in Acinetobacter baumannii isolate (Afzal-Shah et al. 2001). OXA 27 has been identified in a single isolate as of yet, whereas OXA 23 is widespread clinically in Acinetobacter isolates and has been reported from different parts of the world (Corvec et al. 2007; Stoeva et al. 2008; Feizabadi et al. 2008; Mugnier et al. 2008; Mansour et al. 2008; Dalla-Costa et al. 2003; Valenzuela et al. 2007). Despite the widespread resistance in Acinetobacter species, the inadequacy of OXA-23 to confer resistance to carbapenems in Enterobacterales may be due to their low turnover and high affinity for carbapenems, resulting in weak hydrolysis (Antunes et al. 2014).
-
b.
OXA 24 like subfamily: The group consists of 18 members, all of which have been identified as CHDLs. Few significant enzymes of the group include, OXA-24 (now OXA 40), OXA 25, OXA 26, OXA 72 (Poirel et al. 2010). An original sequencing error in the index type OXA 24 identified later makes it now OXA 40 (Lopez-Otsoa et al. 2002). In contrast to other subfamilies, the genes for OXA 24 like subfamily are not associated with the ISs or integron associated components but are flanked by inverted repeats homologous to the XerC/XerD binding sites, signifying mobilization of gene by site specific recombination (Merino et al. 2010; D’Andrea et al. 2009). OXA 24 like producing isolates are found to be endemic in Portugal since the mid-1990s (Grosso et al. 2011); however, recently they have been disseminated in other regions of the world leading to clinical concerns (Dortet et al. 2016; Pagano et al. 2017).
-
c.
OXA-58 like subfamily: The third identified group, with a total of seven carbapenem members, also found only in Acinetobacter species has OXA-58 as its prototype and has been often associated with hospital outbreaks. All the seven members are CHDLs. OXA 58 hydrolyses penicillins and carbapenems, but not cefepime, ceftazidime, and cefotaxime, whereas cefpirome hydrolyzed only weakly (Poirel et al. 2005). OXA-58 producing isolates have been isolated from different regions among different bacterial species, with Acinetobacter baumannii global clone 2 being the major host carrying genes for OXA-58 like enzyme (Hamidian and Nigro 2019; Lowe et al. 2018; Taşbent and Özdemir 2015; Higgins et al. 2010).
-
d.
OXA-48 like subfamily: OXA-48 was first identified in plasmid carried gene, in a carbapenem resistant Klebsiella pneumoniae isolate from Istanbul, Turkey in 2001 (Poirel et al. 2004). The OXA-48 subfamily has been merged with OXA-548 subfamily and together comprises 101 enzymes (Yoon and Jeong 2020). OXA-48 is a DBL with highest catalytic activity against imipenem, but is unable to hydrolyze extended spectrum cephalosporins (Zong 2012). OXA-48 occurs primarily in Enterobacterales. Nonetheless, occurrence of chromosomal OXA-48 in Shewanella species is intrinsic (Zong 2012). OXA-48 has been reported in various hospital outbreaks and is reported frequently with NDM-1 producing Enterobacterales (Balm et al. 2013; Avolio et al. 2017).
-
a.
6 Naturally Occurring Class D β Lactamases
Naturally occurring chromosomal class D β lactamase genes have been described in several species, first one being identified in Aeromonas jandaei (Poirel et al. 2010). OXA-12 (inducible) and AmpS are the two DBLs produced by Aeromonas jandaei (Rasmussen et al. 1994; Walsh et al. 1995). Chromosomally located OXA-22 is found in Ralstonia pickettii, leading to intrinsic resistance to penicillins, narrow spectrum cephalosporins, ceftazidime, and aztreonam (Nordmann et al. 2000). OXA-61 is identified in chromosome of Campylobacter jejuni, OXA-62 in Pandoraea species, and OXA-42 in Burkholderia pseudomallei (Walsh et al. 1995; Alfredson and Korolik 2005; Nordmann et al. 2000). A number of other naturally occurring DBLs are reported across several bacterial species and are considered as their survival mechanism against the environment.
7 Class D β lactamases in Gram-Positive Organisms
DBLs are occur frequently in the Bacillaceae family and the environmental isolates of family Clostridiaceae and Eubacteriaceae (Toth et al. 2016). Due to the lack of an arginine residue conserved in all known serine β lactamases, the DBLs in gram-positive organisms engage a unique substitute binding mode. This binding mode differentiates them not only from the DBLs of gram-negative bacteria but also from class A and C enzymes (Toth et al. 2016). DBLs among gram-positive cocci are not yet reported.
8 Detection of Class D β lactamases
Detection of class D β lactamases is crucial yet challenging due to the lack of appropriate and standardized phenotypic assays unlike various rapid and easy tests available for class A, B, and C enzymes. However, few properties of DBLs can be utilized for their early detection.
-
1.
Inhibition of OXA-13 and its variant OXA-19 by imipenem: placing an imipenem disc in proximity to cefsulodin (which is easily hydrolyzed by OXA-13 in the absence of imipenem), decreasing the zone of inhibition of cefsulodin can be used for identification of these DBLs (Mugnier et al. 1998). This feature is also shown by other DBLs like OXA-10 and can be utilized for their identification (Poirel et al. 2010).
-
2.
Synergy tests using clavulanic acid discs: DBLs whose activity is inhibited by clavulanic acid or tazobactam (OXA-12, 18, 45, 46) may be identified by synergy tests using clavulanic acid containing discs (Poirel et al. 2010). Nevertheless, differentiating these from class A ESBL producers is vital.
-
3.
Spectrophotometric analysis: Well-equipped laboratories can utilize crude extracts and UV spectrophotometry to assess the capacity to hydrolyze oxacillin. NaCl inhibition property can be evaluated with a reference substrate like benzyl-penicillin.
However, various drawbacks of this methods are that all DBLs do not hydrolyze oxacillin, in vitro inhibition of OXA enzymes activity is difficult, coproduction of other enzymes which interfere in correct identification.
Currently molecular detection of the DBL genes is the only standardized method of identification of class D β lactamases.
9 Treatment of Class D β-lactamases
In contrast to other class of β-lactamases, no specific inhibitor has had been identified for DBLs. Nevertheless, a number of potential candidates have shown inhibitor activity and can be utilized for degrading these enzymes.
Potential class D β-lactamases inhibitors can be classified into those:
-
1.
Derived from β-lactams includes methylidene penems, penicillin sulfones and
-
2.
Non-β lactams derived include avibactam, phosphonates, boronic acid.
Methylidene penems are potent inhibitors of OXA 1 and given with β-lactams.
Penicillin sulfones (Drawz et al. 2010) are active against OXA1, extended spectrum β-lactamases (OXA10, OXA14, OXA17), and OXA24/40. These inhibitors act by preventing the attack of deacylating water molecule. Their negatively charged sulfinate group mimics C3/C4 carboxylate group of penicillins and interact with a carboxylate recognizing residue on DBL. Studies show that C2 substituted 6-alkylidine penicillins were better than C3 substituted 7-alkylidene cephalosporins sulfones (Pattanaik et al. 2009).
Avibactam has activity against OXA48, given in combination with ceftazidime, ceftaroline, aztreonam (Livermore et al. 2011; Mushtaq et al. 2010) it forms a covalent complex in complex with OXA10 and OXA48 and undergoes ring opening reaction (Docquir et al. 2010).
Phosphonates and boronic acid are novel inhibitors of DBL that do not resemble β-lactams (Antunes and Fisher, 2014). Thiophenyl oxime derived phosphonates and 4,7-dichloro-1-benzothien-2yl-sulfonyl-aminomethyl boronic acid are inhibitors of OXA24/40 (Majumdar et al. 2005; Tan et al. 2011). They work by acylating the enzyme by acting as transition state analogue inhibitors and forming a reversible covalent bond with catalytic serine of enzyme with their phosphorus and boron atom, respectively. Thiophenyl oxime exhibits synergy in combination with imipenem (Tan et al. 2010).
Polycarboxylates are active against OXA46. They work by forming hydrogen bond with active site residues on enzyme, one of the carboxylates also makes ionic interaction with a residue that recognize C3/C4 carboxylate group of β-lactams. Other polycarboxylates, lipophilic aminocitrate, and aminoisocitrate derivatives also inhibit OXA10 (Beck et al. 2009).
Yet, none of the compounds is able to inhibit the entire class D enzymes. This can be attributed to the magnanimous size of the family and the diversity of the members of the group. More research is thus required, to explore inhibitors for the subfamilies existing in the class, if not for the entire family.
10 Conclusion
Class D β lactamases are the largest and most diverse, yet most neglected group of β lactamases. Clinically they should be considered a threat similar to or even greater than other β lactamases, since lack of detection may augment their unseen and rapid spread among the clinical settings. Intensive research is required for developing rapid and easy detection tools for DBLs and also for the discovery of class D specific inhibitors.
Seeing the magnanimous and diverse range of members of this group, and for the unification of the subfamilies, new scheme for their classification should be considered.
References
Afzal-Shah M, Woodford N, Livermore DM (2001) Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 45(2):583–588. https://doi.org/10.1128/aac.45.2.583-588.2001
Alfredson DA, Korolik V (2005) Isolation and expression of a novel molecular class D β-lactamase, OXA-61, from Campylobacter jejuni. Antimicrob Agents Chemother 49:2515–2518
Ambler RP (1980) The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci 289(1036):321–331. https://doi.org/10.1098/rstb.1980.0049
Antunes N, Fisher J (2014) Acquired class D β-lactamases. Antibiotics 3(3):398–434. https://doi.org/10.3390/antibiotics3030398
Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB (2014) Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58(4):2119–2125. https://doi.org/10.1128/aac.02522-13
Avolio M, Vignaroli C, Crapis M, Camporese A (2017) Co-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016. Future Microbiol 12(13):1119–1122. https://doi.org/10.2217/fmb-2017-0041
Balm M, La MV, Krishnan P, Jureen R, Lin R, Teo J (2013) Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect 19(9):E421–E423. https://doi.org/10.1111/1469-0691.12247
Beck J, Vercheval L, Bebrone C, Herteg-Fernea A, Lassaux P, Marchnd-Brynaert J (2009) Discovery of novel lipophilic inhibitors of OXA-10 enzyme (Class D β-lactamase) by screening amino analogs and homologs of citrate and isocitrate. Bioorg Med Chem Lett 19:3593–3597
Boyd DA, Mulvey MR (2006) OXA-1 is OXA-30 is OXA-1. J Antimicrob Chemother 58:224–225
Bush K (2013) Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 1277(1):84–90. https://doi.org/10.1111/nyas.12023
Bush K, Fisher JF (2011) Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-Negative bacteria. Annu Rev Microbiol 65(1):455–478. https://doi.org/10.1146/annurev-micro-090110-102911
Bush K, Jacoby GA, Medeiros AA (1995) A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39(6):1211–1233. https://doi.org/10.1128/aac.39.6.1211
Centron D, Roy PH (2002) Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother 46:1402–1409
Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P (2007) Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob Agents Chemother 51(4):1530–1533. https://doi.org/10.1128/aac.01132-06
Couture F, Lachapelle J, Levesque RC (1992) Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol 6(12):1693–1705. https://doi.org/10.1111/j.1365-2958.1992.tb00894.x
D’Andrea MM, Giani T, D’Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM (2009) Characterization of pABVA01, a plasmid encoding the OXA-24 Carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 53(8):3528–3533. https://doi.org/10.1128/aac.00178-09
Dalla-Costa LM, Coelho JM, Souza HAPHM, Castro MES, Stier CJN, Bragagnolo KL, Rea-Neto A, Penteado-Filho SR, Livermore DM, Woodford N (2003) Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J Clin Microbiol 41(7):3403–3406. https://doi.org/10.1128/jcm.41.7.3403-3406.2003
Danel F, Hall LM, Gur D, Livermore DM (1995) OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) beta- lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 39(8):1881–1884. https://doi.org/10.1128/aac.39.8.1881
Danel F, Hall LM, Gur D, Livermore DM (1997) OXA-15, an extended-spectrum variant of OXA-2 beta-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother 41(4):785–790. https://doi.org/10.1128/aac.41.4.785
Danel F, Hall LMC, Gur D, Livermore DM (1998) OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two Pseudomonas aeruginosa Isolates. Antimicrob Agents Chemother 42(12):3117–3122. https://doi.org/10.1128/aac.42.12.3117
Danel F, Hall LMC, Duke B, Gur D, Livermore DM (1999) OXA-17, a further extended-spectrum variant of OXA-10 β-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother 43(6):1362–1366. https://doi.org/10.1128/aac.43.6.1362
Docquir JD, Benvenuti M, Luca DF, Rossolini GM, Mangini S, Miossec C, Black MT (2010) X ray crystal structure of the Klebsiella pneumoniae OXA-48 class D carbepenemase inhibited by NXL 104. In 50th interscience conference of antimicrobial agents and chemotherapy, abstracts C1-640, Boston, MA
Dortet L, Radu I, Gautier V, Blot F, Chachaty E, Arlet G (2008) Intercontinental travels of patients and dissemination of plasmid-mediated carbapenemase KPC-3 associated with OXA-9 and TEM-1. J Antimicrob Chemother 61(2):455–457. https://doi.org/10.1093/jac/dkm455
Dortet L, Bonnin RA, Bernabeu S, Escaut L, Vittecoq D, Girlich D, Imanci D, Fortineau N, Naas T (2016) First Occurrence of OXA-72-Producing Acinetobacter baumannii in Serbia. Antimicrob Agents Chemother 60(10):5724–5730. https://doi.org/10.1128/aac.01016-16
Drawz SM, Bethel CR, Doppalapudi VR, Sheri A, Pagadala SR, Hujer AM et al (2010) Penicillin sulfone inhibitors of class D β-lactamases. Antimicrob Agents Chemother 54:1414–1424
Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, Soroush S, Mohammadi-Yegane S (2008) Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis 61(4):274–278
Fisher JF, Meroueh SO, Mobashery S (2005) Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev 105(2):395–424. https://doi.org/10.1021/cr030102i
Fournier PE, Vallenet D, Barbe V et al (2006) Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7
Franceschini N, Boschi L, Pollini S, Herman R, Perilli M, Galleni M, Frère JM, Amicosante G, Rossolini GM (2001) Characterization of OXA-29 from Legionella (Fluoribacter) gormanii: molecular class D β-lactamase with unusual properties. Antimicrob Agents Chemother 45(12):3509–3516. https://doi.org/10.1128/aac.45.12.3509-3516.2001
Grosso F, Quinteira S, Peixe L (2011) Understanding the dynamics of imipenem-resistant Acinetobacter baumannii lineages within Portugal. Clin Microbiol Infect 17(8):1275–1279. https://doi.org/10.1111/j.1469-0691.2011.03469.x
Hall LM, Livermore DM, Gur D, Akova M, Akalin HE (1993) OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 37(8):1637–1644. https://doi.org/10.1128/aac.37.8.1637
Hamidian M, Nigro SJ (2019) Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genomics 5(10):e000306. https://doi.org/10.1099/mgen.0.000306
Higgins PG, Dammhayn C, Hackel M, Seifert H (2010) Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65(2):233–238. https://doi.org/10.1093/jac/dkp428
Huovinen P, Huovinen S, Jacoby GA (1988) Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother 32(1):134–136. https://doi.org/10.1128/aac.32.1.134
Jaurin B, Grundstrom T (1981) ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci 78(8):4897–4901. https://doi.org/10.1073/pnas.78.8.4897
Jiang T, Xu J, He F (2020) Genotypic and phylogenetic characterisation of a clinical Ralstonia pickettii strain carrying two novel OXA allelic variants, blaOXA-898 and blaOXA-899, isolated from a bloodstream infection in China. J Glob Antimicrob Resist 21:46–48. https://doi.org/10.1016/j.jgar.2020.02.020
Kratz J, Schmidt F, Wiedemann B (1983) Transposition of a gene encoding OXA-2 -lactamase. J Gen Microbiol 129(9):2951–2957. https://doi.org/10.1099/00221287-129-9-2951
Kumar P, Thomas S (2011) Presence of qnrVC3 gene cassette in SXT and class1 integrons of Vibrio cholerae. Int J Antimicrob Agents 37:280–281
Lamotte-Brasseur J, Knox J, Kelly JA, Charlier P, Fonzé E, Dideberg O, Frère JM (1994) The structures and catalytic mechanisms of active-site serine β-lactamases. Biotechnol Genet Eng Rev 12(1):189–230. https://doi.org/10.1080/02648725.1994.10647912
Leonard DA, Bonomo RA, Powers RA (2012) Class D β-lactamases: a reappraisal after five decades. Acc Chem Res 46(11):2407–2415. https://doi.org/10.1021/ar300327a
Livermore D, Mushtaq S, Warner M et al (2011) Activities of NXL 104 combinations with ceftazidime and aztreonam against carbepenemase producing Enterobacteriaceae. Antimicrob Agents Chemother 55:390–394
Lopez-Otsoa F, Gallego L, Towner KJ, Tysall L, Woodford N, Livermore DM (2002) Endemic carbapenem resistance associated with OXA-40 carbapenemase among Acinetobacter baumannii isolates from a hospital in Northern Spain. J Clin Microbiol 40(12):4741–4743. https://doi.org/10.1128/jcm.40.12.4741-4743.2002
Lowe M, Ehlers MM, Ismail F, Peirano G, Becker PJ, Pitout J, Kock MM (2018) Acinetobacter baumannii: epidemiological and beta-lactamase data from two tertiary academic hospitals in Tshwane, South Africa. Front Microbiol 9:1280. https://doi.org/10.3389/fmicb.2018.01280
Majumdar S, Adediran SA, Nukaga M, Pratt RF (2005) Inhibitors of class D β-lactamases by diacyl phosphates. Biochemistry 44:16121–16129
Mansour W, Poirel L, Bettaieb D, Bouallegue O, Boujaafar N, Nordmann P (2008) Dissemination of OXA-23–producing and carbapenem-resistant Acinetobacter baumannii in a University Hospital in Tunisia. Microb Drug Resist 14(4):289–292. https://doi.org/10.1089/mdr.2008.0838
Matthew M, Sykes RB (1977) Properties of the beta-lactamase specified by the Pseudomonas plasmid RPL11. J Bacteriol 132(1):341–345. https://doi.org/10.1128/jb.132.1.341-345.1977
Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN (2009) Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. In Asia-Pacific nations: report from the SENTRY surveillance program. J Antimicrob Chemother 63(1):55–59. https://doi.org/10.1093/jac/dkn434
Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G (2010) OXA-24 carbapenemase gene flanked by XerC/XerD-Like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob Agents Chemother 54(6):2724–2727. https://doi.org/10.1128/aac.01674-09
Moura A, Pereira C, Henriques I et al (2012) Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res Microbiol 163:92–100
Mugnier P, Casin I, Bouthors A-T, Collatz E (1998) Novel OXA-10-derived extended-spectrum β -lactamases selected in vivo or in vitro. Antimicrob Agents Chemother 42:3113–3116
Mugnier P, Poirel L, Pitout M, Nordmann P (2008) Carbapenem-resistant and OXA-23-producing Acinetobacter baumannii isolates in the United Arab Emirates. Clin Microbiol Infect 14(9):879–882. https://doi.org/10.1111/j.1469-0691.2008.02056.x
Mulvey MR (2004) Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing novel OXA-type -lactamase (blaOXA-53) and 6’-N-aminoglycoside acetyltransferase genes [aac(6’)-I30]. J Antimicrob Chemother 54(2):354–359. https://doi.org/10.1093/jac/dkh347
Mushtaq S, Warner M, Williams G, Critchley I, Livermore D (2010) J Antimicrob Chemother 65:1428–1432
Naas T, Nordmann P (1999) OXA-type beta-lactamases. Curr Pharm Des 5(11):865–879
Naas T, Namdari F, Bogaerts P, Huang TD, Glupczynski Y, Nordmann P (2008) Genetic structure associated with blaOXA-18, encoding a clavulanic Acid-Inhibited Extended-Spectrum oxacillinase. Antimicrob Agents Chemother 52(11):3898–3904. https://doi.org/10.1128/aac.00403-08
Niumsup P, Wuthiekanun V (2002) Cloning of the class D beta-lactamase gene from Burkholderia pseudomallei and studies on its expression in ceftazidime-susceptible and -resistant strains. J Antimicrob Chemother 50(4):445–455. https://doi.org/10.1093/jac/dkf165
Nordmann P, Poirel L, Kubina M, Casetta A, Naas T (2000) Biochemical-genetic characterization and distribution of OXA-22, a chromosomal and inducible class D β -lactamase from Ralstonia (Pseudomonas) pickettii. Antimicrob Agents Chemother 44:2201–2204
Pagano M, Rocha L, Sampaio JL, Martins AF, Barth AL (2017) Emergence of OXA-72-producing Acinetobacter baumannii belonging to high-risk clones (CC15 and CC79) in different Brazilian States. Infect Control Hosp Epidemiol 38(2):252–254. https://doi.org/10.1017/ice.2016.287
Pattanaik P, Bethel CR, Hujer AM, Hujer KM et al (2009) Strategic design of an effective β-lactamase inhibitor. J Biol Chem 284:945–953
Payne DJ, Cramp R, Winstanley DJ, Knowles DJ (1994) Comparative activities of clavulanic acid, sulbactam, and tazobactam against clinically important beta-lactamases. Antimicrob Agents Chemother 38(4):767–772. https://doi.org/10.1128/aac.38.4.767
Périchon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, Feldgarden M, Wortman J, Clermont D, Nemec A, Courvalin P (2013) Identification of 50 class D β-lactamases and 65 acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob Agents Chemother 58(2):936–949. https://doi.org/10.1128/aac.01261-13
Philippon LN, Naas T, Bouthors AT, Barakett V, Nordmann P (1997) OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 41(10):2188–2195. https://doi.org/10.1128/aac.41.10.2188
Poirel L, Gerome P, De Champs C, Stephanazzi J, Naas T, Nordmann P (2002) Integron-located oxa-32 gene cassette encoding an extended-spectrum variant of OXA-2 β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 46(2):566–569. https://doi.org/10.1128/aac.46.2.566-569.2002
Poirel L, Héritier C, Tolün V, Nordmann P (2004) Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48(1):15–22. https://doi.org/10.1128/aac.48.1.15-22.2004
Poirel L, Marqué S, Héritier C, Segonds C, Chabanon G, Nordmann P (2005) OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother 49(1):202–208. https://doi.org/10.1128/aac.49.1.202-208.2005
Poirel L, Naas T, Nordmann P (2010) Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother 54(1):24–38. https://doi.org/10.1128/AAC.01512-08
Rasmussen BA, Keeney D, Yang Y, Bush K (1994) Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother 38(9):2078–2085. https://doi.org/10.1128/aac.38.9.2078
Sanschagrin F, Couture F, Levesque RC (1995) Primary structure of OXA-3 and phylogeny of oxacillin-hydrolyzing class D beta-lactamases. Antimicrob Agents Chemother 39(4):887–893. https://doi.org/10.1128/aac.39.4.887
Schneider I, Queenan AM, Bauernfeind A (2006) Novel carbapenem-hydrolyzing oxacillinase OXA-62 from Pandoraea pnomenusa. Antimicrob Agents Chemother 50(4):1330–1335. https://doi.org/10.1128/aac.50.4.1330-1335.2006
Siu LK, Lo JY, Yuen KY et al (2000) β -Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like b-lactamase, OXA-30. Antimicrob Agents Chemother 44:2034–2038
Stoeva T, Higgins P, Bojkova K, Seifert H (2008) Clonal spread of carbapenem-resistant OXA-23-positive Acinetobacter baumannii in a Bulgarian university hospital. Clin Microbiol Infect 14(7):723–727. https://doi.org/10.1111/j.1469-0691.2008.02018.x
Suzuki M, Nishio H, Asagoe K et al (2015) Genome sequence of a carbapenem resistant strain of Ralstonia mannitolilytica. Genome Announc 3:e00405–e00415
Tan Q, Ogawa AM, Painter RE, Park YW, Young K, DiNinno F, P. (2010) 4,7-Dichloro benzothien-2yl sulfonylaminomethyl boronic acid; first boronic acid derived β-lactamase inhibitor with class A, C and D activity. Bioorg Med Chem Lett 20:2622–2624
Tan Q, Ogawa AM, Raghoobar SL, Wisniewski D et al (2011) Thiophenyl oxime derived phosphonates as nano-molar class β-lactamase inhibitors reducing the MIC of imipenem against Pseudomonas aeruginosa and Acinetobacter baumannii. Bioorg Med Chem Lett 20:2622–2624
Taşbent EF, Özdemir M (2015) The presence of OXA type carbapenemases in Pseudomonas strains: first report from Turkey. Mikrobiyol Bul 49(1):26–34. https://doi.org/10.5578/mb.856
Tian J, Zhang G, Ju Y, Tang N, Li J, Jia R, Feng J (2018) Five novel carbapenem-hydrolysing OXA-type β-lactamase groups are intrinsic in Acinetobacter spp. J Antimicrob Chemother 73:3279–3284. https://doi.org/10.1093/jac/dky359
Toth M, Antunes NT, Stewart NK, Frase H, Bhattacharya M, Smith CA, Vakulenko SB (2016) Class D β-lactamases do exist in Gram-positive bacteria. Nat Chem Biol 12(1):9–14. https://doi.org/10.1038/nchembio.1950
Valenzuela JK, Thomas L, Partridge SR, van der Reijden T, Dijkshoorn L, Iredell J (2007) Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J Clin Microbiol 45(2):453–460. https://doi.org/10.1128/jcm.01971-06
Voha C, Docquier JD, Rossolini GM, Fosse T (2006) Genetic and biochemical characterization of FUS-1 (OXA-85), a narrow-spectrum class D β-lactamase from Fusobacterium nucleatum subsp. polymorphum. Antimicrob Agents Chemother 50(8):2673–2679. https://doi.org/10.1128/aac.00058-06
Walsh TR, Hall L, MacGowan AP, Bennett PM (1995) Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J Antimicrob Chemother 36(1):41–52. https://doi.org/10.1093/jac/36.1.41
Walther-Rasmussen J, Høiby N (2006) OXA-type carbapenemases. J Antimicrob Chemother 57(3):373–383. https://doi.org/10.1093/jac/dki482
Yoon EJ, Jeong SH (2020) Class D β-lactamases. J Antimicrob Chemother 76(4):836–864. https://doi.org/10.1093/jac/dkaa513
Zong Z (2012) Discovery of bla(OXA-199), a chromosome-based bla(OXA-48)-like variant, in Shewanella xiamenensis. PLoS One 7(10):e48280. https://doi.org/10.1371/journal.pone.0048280
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Khan, F., Chaudhary, B., Khan, A.U. (2022). Class D Type Beta-Lactamases. In: Shahid, M., Singh, A., Sami, H. (eds) Beta-Lactam Resistance in Gram-Negative Bacteria. Springer, Singapore. https://doi.org/10.1007/978-981-16-9097-6_7
Download citation
DOI: https://doi.org/10.1007/978-981-16-9097-6_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-9096-9
Online ISBN: 978-981-16-9097-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)