Abstract
Feature extraction and selection are very important stages in pattern recognition and computer vision solutions with far-reaching effects on their performance. In computer-aided diagnosis (CADx) systems, efficiency is affected by its subjectivity to the accuracy of the region of interest (ROI) extraction technique, which is largely dependent on the features extracted. Optimization algorithms are often used to improve the selection of discriminative features which thereby leads to improve accuracy of the CADx systems. This work considers the effects of optimizing selected features in the performance of breast tissue characterization in mammograms. It uses Whale Optimization Algorithm (WOA) to optimize Otsu fitness function of Gray Level Co-occurrence Matrix (GLCM) in extracting the region of interest (ROI). The extracted features were classified into BIRADS scales 1, 2 and 5 using Multiclass Support Vector Machine (MSVM). The performance of the developed algorithm was evaluated using specificity, sensitivity as well as accuracy and compared with other techniques namely Texture Signature (TS), Pixel-Based Morphological (PBM), Natural Language Processing (NLP), and Interactive Data Language (IDL). The result of the developed WOA-Otsu-GLCM-MSVM CADx algorithm for specificity, sensitivity, and accuracy are 96%, 92% and 94%, respectively. The developed algorithm gave an accuracy of 94.4% as against 81.0%, 85.7%, 93.0% and 82.5% for TS, PBM, NLP and IDL methods, respectively. The characterization of the breast tumour using the developed CADx algorithm performed better compared with the conventional methods.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mammogram
- Breast tumour
- Fatty tissue
- Whale optimization algorithm
- Dense tissue
- Gray level co-occurrence matrix
- Multi-support vector machine
- Pectoral muscle
1 Introduction
Pattern recognition (PR) refers to techniques used in computer vision (CV) to recognize distinct patterns in images for classification or grouping. PR is used to determine where an image belong to in a group, it finds use in tasks such as recognition, clustering and classification. It has been employed for applications such as; voice, object, tumour and face detection and recognition among others [1]. Feature extraction and selection are very important stages in PR solutions with far-reaching effects on performance. This implies that the performance of a PR system depends mainly on how well the features are extracted, selected and analyzed to distinguish one class from another.
In biomedical imaging, images of internal organs are captured using several modalities for diseases detection, diagnosis and study. Biomedical imaging types are magnetic resonance imaging (MRI), single-photon emission computed tomography (SPECT), computed tomography (CT), X-ray and Ultrasound among others [2]. Mammograms employ low dose X-rays to scan and capture breast tissues for screening and diagnosis purposes. Computed aided diagnosis (CADx) is a system that analysis biomedical images through feature extraction and pattern recognition technique(s) for diagnostic purposes [3, 4].
CADx usually plays a supporting role rather than substituting doctors or radiologists. The aim of CADx systems is the detection of early abnormality signs in patients which human professionals might miss, such as mammogram architectural distortion. Early detection of diseases, tumours, and other medical conditions can be the difference between life and death, hence, the importance of CADx cannot be overemphasized [5].
Breast tumour is a mass that occurs in the breast region, it turns to cancer when the tumour becomes malignant. Breast tumours have been confirmed to occur mostly in the fibroglandular region of the breast, it is depicted as white spots or patches on the mammogram [3]. Tumours are classified as either benign (unharmful) or malignant/cancerous (harmful). The malignant one is a rapidly growing type which can spread to neighbouring tissues and can sometimes result into metastasis (consequential malignant growth far from the origin cancer). Hence, the malignant type needs urgent care to guard against its spread through metastasis [6]. Cancer results from uncontrolled growth of cluster cells in the human body, this cluster can develop into tumour (a noticeable mass). Malignant breast tumour (cancer) is responsible for a high rate of death among women, efficient CADx system with the help of mammogram can result in early detection which consequently helps reduce this rate [7]. Symptoms of breast cancer may include nipple inversion, rapid change in breast skin or colour, dimpling of the skin and spontaneous discharge of one breast among others [8]. The development of breast tumour detection and classification systems remains a topical issue in the CADx research world [9,10,11].
In the area of medicine, the pink ribbon serves as a symbol to raise public awareness of breast cancer. One of the primary reasons of women’s high mortality rates is breast cancer [12]. BRCA1, BRCA2 (two genes), obesity, birth control pill inhalation, irregular menstrual cycles, higher radiation therapy exposure, and estrogen hormone are all high-risk factors for breast cancer [10, 13, 14]. These factors are to blame for causing cell mutations, which resulted in unrestrained cell growth. Breast soreness is the most common sign of breast cancer, which can be fatal if not detected early. Other symptoms include skin irritation, redness, discomfort, and swelling, which turn ominous with the erosion of nipples or sudden watery discharge from the nipples [10, 14].
Radiologists are saddled with the task of reading mammograms for the identification of possible irregularities. In computer-aided diagnosis (CADx) systems, efficiency is affected by its subjectivity to the accuracy of the region of interest (ROI) extraction technique, which is largely dependent on the features extracted. Earlier researches on breast tumour detection CADx systems adopt manually chosen thresholds for ROI extraction and many are not Breast Imaging Reporting and Data System (BIRADS) based. Optimization algorithms are often used to improve the extraction and selections of discriminative features and consequently result in improved outcomes [11, 15, 16].
This work considers the effects of optimizing Otsu algorithm for ROI extraction to enhance features selection for breast tissue characterization using mammograms. It uses Whale Optimization Algorithm (WOA) to optimize Otsu fitness function producing WOA-Otsu, an automatic ROI extraction technique. GLCM is employed to extract discriminative features from the ROI images to form feature vectors which are then passed on to MSVM for classification. The developed CADx system is WOA-Otsu-GLCM-MSVM algorithm for optimal feature selection of breast tissue for tumour detection and classification.
2 Literature Review
The breast consists of dense tissue known as fibro-glandular tissue (which is made up of glandular with connective tissue) and fatty tissue. The dense tissue appears bright, while fatty tissue appears dark on a mammogram [17]. Tumours do originate from the breast tissue (lobules: milk-producing glands) and ducts that connect it to the nipple. Breast is prevalent in women and the malignant type often leads to death. Tumour (malignant or Benign) occurs as a result of uncontrolled cell mutation in the breast which is felt as a lump or visible on X-ray. Timely detection can lead to effective solutions that can curb mastectomy (surgical breast tissue removal) and consequently decrease re-occurrence probability and mortality rate [18].Breast image is a discrete bi-dimensional function, m (a, b), where m, a and b are amplitude and spatial 2D coordinates, respectively. The function represents the intensities at the corresponding points on the image. The breast image is captured using different imaging modalities as aforementioned.
MRI uses a strong magnetic flux to rearrange the hydrogen atoms of the water in the body to produce hundreds of images representing slices or cross-sections in three-dimensional space. Breast MRI is a potential alternative to mammograms, but the cost is higher than other imaging methods and not widely available as ultrasound and mammography [19]. MRI is the technique of choice in breast tumour diagnosis for women with established cases of breast cancer [20]. MRI images depict the true nature of the established breast tumour, but they come in slices which make the analysis challenging. MRI is not suitable for screening examination by itself because it misses some tumours that mammography (that is screening mammography) can depict [18].
Breast ultrasound is primarily useful in differentiating a solid mass from a cyst and discovering an unnoticeable palpable abnormality on mammogram. It provides a better view of a lesion that is not visible on mammograms. Despite this advantage, ultrasound is not accurate imaging modality in detecting breast microcalcifications. In most inconclusive cases ultrasound serves as a follow up after mammography [21].
Mammograms are X-rays of the breast which displays high intensities region as potential tumours. It has the ability to depict subtle fine scale signs due to its high spatial resolution from low dose X-rays [22]. Full Field Digital Mammography (FFDM) is a type of mammography that uses an electronic device in the place of X-ray film to produce better quality mammograms with lower radiation doses. This enables better picture quality with a lower radiation dose. It is an advanced and reliable technique that could lead to better treatment through early detection of breast tumours [23].
Breast tumours are mostly uncovered after symptoms are noticed, however, many females with the ailment shows no symptoms [22]. Therefore, constant breast tumour check-up is so important. The ability of the radiologist to easily detect breast tumours on a mammogram highly depends on how dense the breast is. Several researches have been done in the area of CADx system development, examples of such are given herein.
Three pre-trained networks (VGG16, VGG19, and ResNet50) behavior were analyzed in [24] for magnification independent breast cancer classification. The result for transfer learning was compared to the fully-trained network on the histopathological imaging modalities. At the same time, the impact of training–testing data size on the performance of the considered networks was investigated. A fine-tuned pre-trained VGG16 with logistic regression classifier produced the best results, with 92.60% accuracy.
A method that employs preprocessing, data augmentation, deep neural network architecture (VGG-16), and gradient boosted trees classifier was presented by Ref. [25]. The system returns 87.2% accuracy on 4-class classification task. For a 2-class classification task on carcinomas detection, it gave 93.8% accuracy, 96.5 sensitivity and 88.0% specificity at the high-sensitivity operating point.
The use of Convolutional Neural Networks (CNNs) to classify hematoxylin and eosin-stained breast biopsy images was proposed by Ref. [26]. Images were categorized into four categories: normal tissue, benign lesion, in situ carcinoma, and invasive carcinoma, as well as two categories: carcinoma and non-carcinoma. The network’s architecture is intended to retrieve information at various scales, including nuclei and overall tissue organization. This design enables the proposed system to be extended to whole-slide histology images.A Support Vector Machine classifier was trained using the features extracted by the CNN. The accuracy for four classes is 77.8%, and for carcinoma/non-carcinoma is 83.3%. The approach has a sensitivity of 95.6% for cancer cases.
Using a Faster region convolutional neural network (Faster R-CNN) and deep CNNs, [27] provide a multistage mitotic cell identification approach. In our research, two available datasets of breast cancer histology (international conference on pattern recognition (ICPR) 2012 and ICPR 2014 (MITOS-ATYPIA-14)) were employed. The method delivers state-of-the-art values of 0.876 precision, 0.841 recall, and 0.858 F1-measure for the ICPR 2012 dataset, and 0.848 precision, 0.583 recall, and 0.691 F1-measure for the ICPR 2014 dataset, which are higher than some earlier methods.
A multiclass CADx system capable of diagnosing breast cancer into four categories was proposed by [28]. The method involves normalizing the hematoxylin and eosin stains to enhance colour separation and contrast. Then, two types of novel features—deep and shallow features—are extracted using two deep structure networks based on DenseNet and Xception. Finally, a multi-classifier method based on the maximum value is utilized to achieve the best performance. The proposed method is evaluated using the BreakHis histopathology data set, and the results in terms of diagnostic accuracy is 92%.
2.1 ROI Extraction Techniques
The Breast Imaging Reporting and Data System (BIRADS) categorises findings of expert radiologists about tumours into classes numbered 0 via 6. This is the standard system used by medical doctors or oncologists to report mammograms results or findings. A recommendation was put forward by [29] that each breast should be examined separately and diagnosed in accordance to BIRADS. The summary of BIRADS categorization is described in Tables 3.1 and 3.2.
The precise ROI extraction or segmentation of the breast regions in mammograms is a vital processing phase in computer analysis of mammograms. This allows for accurate recognition of irregularities as the undue influence of the mammograms background or irrelevant regions would have been removed. It also facilitates enhancements of methods like comparative analysis, that is the automatic comparison of mammograms pairs. The borderline of the breast gives substantial information concerning distortion in pairs of mammograms and this serves as the basis for linking nipple position relative to the skin surface [31].
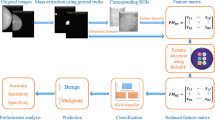
The exactness of ROI extraction technique (otherwise known as segmentation) determines how accurate and efficient a CADx system is. Pectoral muscle presence makes ROI extraction challenging; hence it must be removed from the mammogram for accurate segmentation [32]. The main approaches or procedures of research exploration embedded in the components of the CADx system comprises image acquisition, preprocessing, ROI extraction, features extraction and classification stages as shown in Fig. 3.1.
Mammographic images (mammograms) are readily available in databases of some organizations such as; Mammography Image Analysis Society (MIAS), Medical Information Data Analysis System (MIDAS) database and Digital Database for Screening Mammogram (DDSM) among others.
The segmentation process separates the image pixels into different groups according to their similarities. It is concerned with the demarcation of image portions to communicate and mathematically interpret hidden information. Mammography segmentation is dependent on the motion, shape, colour, spatial configuration and texture of the breast image or its components [33, 34]. Detection of images or their constituents is difficult to attain in many real-world settings. Segmentation of mammograms is made up of duo stages: breast contour outline and pectoral region removal. The pectoral area denotes a high-intensity region in most mammograms and can influence the output image processing. Two preprocessing algorithms are often incorporated, one for breast outline extraction and the other for pectoral area removal [35].
2.2 Optimization Algorithms
Optimization algorithms are techniques that can be used to maximize the best threshold value(s) to split the image into object and background. They are used to optimize Otsu thresholding techniques in this work, to improve the outcome to attain optimal thresholding value. The optimal thresholds are found through between-class variance maximization or within-class variance minimization of the regions which are labelled as thresholds [36]. Optimization algorithms available in the literature are particle swamp optimization (PSO), artificial bee colony (ABC), cat swarm optimization (CSO), whale optimization algorithm (WOA) and genetic algorithm (GA) among others.
WOA is a meta-heuristic algorithm that perform optimization based on the humpback whales bubble net hunting strategy [37]. Validation of WOA was performed using 29 optimization problem mathematical benchmarks with its performance evaluated via comparison with conventional techniques such as PSO, Gravitational Search Algorithm, Fast Evolutionary Programming and Differential Evolution. WOA is shown to outperform most of the compared popular meta-heuristic techniques.
A liver segmentation in MRI images using WOA was proposed in [38]. The technique used WOA for image cluster extraction to aid the segmentation approach. The system was tested with a dataset of 70 radiologists’ approved MRI images. The segmented images were validated using Similarity Index (SI) and Structural Similarity Index Measure (SSIM) among others. The experimental result gave an accuracy of 97.5% using SI and 96.75% using SSIM.
A feature selection technique based on WOA was proposed by [37]. The technique was validated using 18 typical benchmark data sets gotten from UCI respiratory and was compared to three wrapper feature selecting techniques namely GA, PSO and ALO. WOA was found to be better in terms of accuracy and average selection size.
An improved Otsu thresholding based pre- and post-processing technique for soft tissue sacromas (STS) segmentation on MRI images with malignant tumours was reported in [39]. The result of WOA in optimizing Otsu algorithm was evaluated with other techniques such as Differential evaluation, PSO and Grasshopper Optimization. Though the results are close to each other, WOA happen to give better result than others with high robust performance.
WOA was used for clustering by [15] and compared with PSO, ABC, GA, DE and K-means clustering. The proposed method was evaluated with seven standard UCI repository benchmark and one artificial dataset. The results show that WOA based algorithm was the most effective, most robust and easiest to implement of the compared approaches.
WOA was used for feature selection in conjunction with Fuzzy based Relevance Vector Machine (FRVM) classifier by Ref. [16]. This process was compared with PSO-based and other techniques for disease diagnosis. WOA-based technique was shown to converge quicker than the compared techniques to produce a better solution.
A new Image segmentation technique based on multilevel thresholding was proposed by Ref. [1]. WOA was used to optimize thresholding values used for ROI extraction. The obtained results showed that the proposed method performed better in solving multilevel thresholding problems for ROI extraction and produces faster convergence. It is evident from previous studies that WOA outperforms most of the metaheuristic optimization algorithm as it is robust, give faster convergence and higher performance. Hence, this proves promising if implemented on realistic datasets such as MIDAS, MIAS or DDSM.
Mirjalili and Lewis developed a contemporary meta-heuristic optimization algorithm known as WOA [15, 16]. The algorithm comprises of three mathematical models, namely, prey search, prey encircling and bubble-net attacking. These models mimic humpback whales’ traits to perform optimization.
2.3 Feature Extraction
Several techniques have been used to analyse, detect or extract features from mammogram images. Feature extraction translates pixel information into a higher degree depiction of motion, colour, shape, spatial configuration and texture of the breast image or its components. The mined is used for succeeding expression characterization. Feature extraction generally reduces the dimensionality of the input by representing raw images in a reduced form to enable efficient detection or classification [40].
Feature extraction and selection are essential for efficient dimensionality reduction, improved data presentation, prediction performance improvement, data storage requirement reduction, computational requirement reduction and by extension, cost reduction. In a situation where the classifier can get optimal accuracy using the extracted features, feature selection might not be required [39]. Table 3.3 shows the texture features expressions.
A co-occurrence signifies the angular spatial and distance relation on an image subsection of a specific size. GLCM is a matrix obtained from a grayscale image, it is how frequent a pixel with intensity value “i” occurs vertically (90°), horizontally (0°) or diagonally (45° or 135°) with respect to neighbouring pixels with intensity value j. Statistical texture features can be of either first, second or higher-order. The GLCM technique is a method used in mining second-order statistical features [41,42,43]. It is a technique developed by Haralick, which is regularly employed in image feature extractions for the detection and categorization of tumours in mammograms [42, 44, 45].
Many features and classification techniques have already been developed to detect and categorize the lesions as malignant or benign, among others are Artificial Neural Networks (ANN), Hybrid Neural Network Classification (HNN), Support Vector Machines (SVM), K-Nearest Neighbours (KNN), Relevant Vector Machine (RVM), and Fuzzy approaches. But the capacity of SVM to outdo several famous developed methods for the broadly studied problem of microcalcification detection suggests that it is a promising method for object characterization in medical imaging applications [46, 47].
2.4 Evaluation of CAD System
CADx system performance depends on disease, image type and organ. The CADx system findings can be categorised as False Negative (FN), True Positive (TP), True Negative (TN) and False Positive (FP) depending on the presence of abnormality or otherwise. The true or false denotes how CADx decision agrees with actual clinical state and positive or negative represents the decision made by the algorithm [48].
CADx performance is evaluated using several objective evaluation parameters such as sensitivity, accuracy and specificity among others. Equations 3.1, 3.2 and 3.3 are the mathematical definition for sensitivity, specificity and accuracy respectively. The parameter score of a CADx system is directly proportional to its performance.
Higher sensitivity indicates low false negative detection.
Higher specificity indicates low false positive detection.
CADx of breast cancer over the years has greatly contributed to the medical diagnosis development as it is constantly being used by radiologists [9]. CADx accuracy can be improved by reducing FP and FN of the system. Despite advancements in CAD detection systems, there is still a need for optimization of the existing algorithms to produce a more reliable system as there is no one-fit all method. This research tends to detect and characterize breast tumors based on BIRADS scheme of ACR using an optimized segmentation approach.
3 Methodology
The developed method consists of the data acquisition stage, the image preprocessing stage, the WOA-Otsu RoI extraction stage, the feature extraction stage, and the classification stage. The block diagram of the proposed CADx system is shown in Fig. 3.2. All the stages involved were carried out in Python using Jupyter notebook of Anaconda distribution.
MIAS database was used to test the developed system. The database consists of 161 pairs (left and right) of mammography images, out of which 115 were abnormal (64 benign and 51 malignant). The images were also classified into Dense-Glandular (112 images), Fatty (106 images) and Fatty-Glandular (104 images) based on the radiologist report in the database.
Image preprocessing was used for the reduction of image noise. The noises in the images were removed for successful segmentation, image artefact removal, and pectoral region removal. In this study region descriptive method [49] was used to remove pectoral muscle, artefact and high-intensity noise. The median filter was used to filter noise and smoothen the mammograms.
The bilateral comparison stage was done using Breast Images Bilateral Comparison (BIBC) derived from the work of [50]. The left breast and right were set in the same orientation and the BIBC values of each image were computed using Eq. (3.8). BIBC was used to detect asymmetric distortion between the left and right breast. The difference between the breast tissues was compared with a choosing value (0.05). The breasts with higher values than 0.05 are suspected to be with abnormalities. BIBC is derived as shown in Eqs. (3.4) to (3.8).
where ImLis the left breast image, ImIR is the flipped right breast image and BIBC, is the breast image bilateral comparison difference value.
The breast density evaluation stage uses Breast Density Distribution Function (BDDF) defined as in Eq. (3.9):
where: i, j are the spatial coordinates of the function pij, pij is the intensity values greater than 172, N is the total pixels in the images.
The preprocessed images were categorized into fatty and dense breasts using Eq. (3.9). Optimum intensity (threshold) value of 172 was chosen to arrive at a reduced false-positive value and better true negative values, which resulted in an improved classification. The mammograms are categorized using BDDF values. Images with BDDF values less than 0.1 are classified as fatty while others are classified as dense breasts.
The denser the breast tissue is, the difficult it is for radiologists to detect breast cancer. Hence CADx systems for automatic breast tumour detection are more efficient on fatty images.Images with BIBC values less than 0.05 were fed as input into the breast density evaluation stage.
The proposed technique flowchart is shown in Fig. 3.3. The method used WOA to optimize Otsu algorithm for automatic selection of best thresholding values. These optimal values were used to segment the mammograms. The fitness functions used Otsu’s maximum class variance criterion of algorithm to enhance the accuracy of the ROI extraction via two-level thresholding. This improves the accuracy of the classification stage. The input to the proposed WOA-Otsu algorithm is the region descriptive preprocessed mammograms.
The feature extraction stage employed statistical features and GLCM. This technique was applied to the segmented images to extract relevant intensity and texture features such as contrast, uniformity, homogeneity, mean, standard deviation of the object among others. The GLCM features (homogeneity, correlation, contrast and energy) were extracted at 8 different distances and angular orientations (0, 45°, 90°, 135°). In addition, statistical features (mean, standard deviation, variance and median) were added to the bank of features. The combined features were fed into the MSVM.
The classification of the extracted feature vectors was done using the MSVM following BIRADS scale system. The relevant feature vectors were subdivided into train and test data sets in the ratio 0.75 to 0.25 respectively. The appropriate MSVM classifier was trained and tested to categorize the imputed images into normal, benign or malignant tissues based on the BIRADS system.
The system algorithm was implemented in Python using Jupyter notebook IDE of Anaconda distribution version 5.0 on Hewlett Packard (Hp) computer system with processor Intel(R) Core (TM) i3-2350M CPU @ 2.30 GHz and 6.00GB RAM.
The computer system has 750G HDD capacity, 64-bit Operating System (OS), and x64-based processor on Windows 10 professional edition.
The performance of the developed computer-aided diagnosis system was evaluated using specificity, sensitivity, and accuracy.
The parameters TP, TN, FN and FP was derived from the contingency table of the developed system. TP is when the system correctly classified a lesion mammogram, TN is when the system correctly classified a non-lesion mammogram, FN is when the system classified a lesion mammogram as non-lesion and FP is when the system classified a non-lesion mammogram as lesion.
4 Results and Discussions
The 322 mini-MIAS database images were preprocessed using region descriptive method [49] and median filter to eradicate high-frequency noise, artefact and pectoral muscle region. The result achieved in this stage is presented in Table 3.4. The processing stage took 202.32 seconds for the whole 322 images in the database to be preprocessed in Python 3.7.3 using Jupyter notebook of Anaconda Distribution version 5.0, which is approximately 0.6283 s processor time per image this faster preprocessing time compare to time took to obtain the same result in MATLAB as indicated in [50]. The stage successfully processed all the mammograms (322 images). The results of the preprocessed mammograms are shown in Fig. 3.4.
The breast images with BIBC values greater than 0.05 were grouped as BIRADS 3 (suspicious breast tissue) to be examined by an expert radiologist for further diagnosis. A total of 161mammograms out of 322 were classified as BIRAD 3.
The breast BDDF values were used to group the fed-in mammograms (161) into fatty (BDDF value <0.1) and dense (BDDF value ≥0.1) breast images. The dense images were labelled BIRADS 0, which implies that there is a need for additional imaging evaluation. Right and Left breast fatty and dense images are shown in Fig. 3.5.
The images with BDDF values <0.1 (51 fatty breast images) were passed as input into the WOA-Otsu algorithm. The algorithm automatically segments input images into object and background (ROI extraction). It was observed that the normal images were almost completely black as the background (in this case), while the abnormal (benign and malignant) were having various shapes of white masses. Figure 3.6 shows the segmented images, with (a) as a normal mammogram, (b) as benign tumour and (c) as malignant tumour respectively. The segmented images were fed as input into the feature extraction stage.
The feature vector obtained from GLCM at various angular orientations and distances were 128 intensity features extracted, 32 features each contrast, homogeneity, energy and uniformity respectively. Additional texture features were extracted from statistical measures, these are mean, median, standard deviation and variance. The total feature vector extracted is 132 combined features per image, these are fed into the MSVM classifier for training and testing.
The feature data set was divided into train and test data in the ratio 0.75 to 0.25. The 51 mammograms (fatty breast tissues), that consist of 23 normal, 16 benign and 12 malignant gave 39 train and 12 test datasets. The training dataset was used to train the MSVM. The trained system was used to classify the test dataset into malignant, benign and normal. The performance metrics are presented in Table 3.6 which is derived from Table 3.5. The GUI of the model build in Python using the tkinker library is shown in Figs. 3.7, 3.8, and 3.9 as normal image, benign and malignant respectively.
The mini-MIAS database mammograms were used in the development of the computer-aided diagnosis (CADx) system. Both subjective and objective methods of performance evaluation were used to evaluate the system.
The preprocessed image(s) were subjected to visual inspection and compared with the consultant radiologist report presented in the readme file of the database. The results were categorized as successful, acceptable and unacceptable as shown in Fig. 3.4. The results obtained are shown in Table 3.4, 299 images out 322 images (92.86% of the total samples) were successful, 13 images (4.04% of the total samples) were acceptable and 10 images (3.10% of the total samples) were unacceptable.
The mammograms in the bilateral comparison stage with higher BIBC values that were classified as BIRADS3, implies that they are considered to be suspicious images, that is, they are architecturally asymmetrical with their breast pair when set in the same orientation and compared. Since their BIBC value is greater than 0.05, the images were suspected for microcalcification and a follow-up is recommended.
The density evaluation stage used BDDF values to categorize the images into fatty and dense breast image(s). Figure 3.5a, b illustrate samples of fatty and extremely dense breast tissue respectively. The mammograms with BDDF greater than 0.1 that were categorized as BIRADS0 were considered incomplete (inconclusive) diagnoses, that is, there is a need for additional imaging evaluation. The reason being that these mammography images are heterogeneously dense and appears brightly coloured as a potential tumour will appear on a mammogram, hence they are grouped for other imaging evaluation as the radiologist will recommend.
The segmentation stages used the optimum threshold value obtain from WOA-Otsu algorithm to automatically segment the images into objects and background as shown in Fig. 3.6. The mammography images without tumours also known as normal mammograms were observed to dark as the background region of the mammogram after segmentation. This shows an absence of tumour as usually appear white on mammogram after segmentation, Fig. 3.6a shows the output of a normal mammogram. The mammography image with benign cyst were observed to be circumscribed in shape as indicated in Fig. 3.6b while the mammograms with malignant tumours were observed to be speculated in shape as depicted in Fig. 3.6c.
The features mined from the GLCM and statistical measure to the obtained relevant intensity and texture features that were fed into the MSVM classifier were considered, as a hybridized feature that combines two or more techniques is adopted to achieve higher truthfulness [23]. The hybridize features technique adopted combines texture-based and intensity-based features and the total extracted feature for each mammogram is 132 features.
The MSVM was used to categorize the mammography images into BIRADS1, that is normal mammogram, BIRADS2, a benign tumour (non-cancerous abnormality) and BIRADS5, a malignant tumour (cancerous abnormality). The confusion matrix obtained from the classification is shown in Table 3.5. The performance of the system was tested using specificity, sensitivity, and accuracy. Table 3.6 shows the performance of the system derived from the confusion matrix. The system returns specificity of 0.96, sensitivity of 0.92 and accuracy of 0.94.
The CADx System develop in this study was compared with others in the literature that used BIRADS scheme and SVM classifier for mammograms classification including Novel Texture Signature (TS) Based approach proposed by [53], Pixel-Based Morphological Technique (Otsu) technique developed by [18], Natural Language Processing (NLP) method proposed by [52], and Interactive Data Language (IDL) method proposed by Ref. [51]. Table 3.7 shows the performance accuracy of the system as compared with other methods in literature. Figure 3.10 shows the bar chart of the comparison and it observed that the system gave better performance compared to others in the literature with an accuracy of 94.44%.
5 Conclusion
A CADx system to detect the existence or absence of breast tumour have been developed. The use of a median filter for noise elimination at the preprocessing stage proved to be successful for the mammograms’ enhancement. In addition, WOA-Otsu algorithm has resulted in successful automatic detection and segmentation of breast tumours. This has led to a better classification of the breast tissues into normal, benign or malignant by the system.
A novel segmentation technique has been developed that can automatically segment mammograms mitigating the problem of overfitting. This work reiterates that conventional methods coupled with optimization can produce results comparable with deep learning techniques. The developed CAD technique can automatically detect and classify tumours in breast images. The developed WOA-Otsu algorithm can be adopted for tumour detection and categorization in other medical image databases such as liver, kidney, thyroid, and brain among others.
The use of other optimization algorithms in conjunction with WOA-Otsu is suggested. Further research work into how the system can be used to detect and categorize tumours on more dense mammography images is recommended.
Conflict of Interest
There is no conflict of interests.
Funding
There is no funding support.
Data Availability
Not applicable.
References
El Aziz, M. A., Ewees, A. A., Hassanien, A. E., Mudhsh, M., & Xiong, S. (2018). Multi-objective whale optimization algorithm for multilevel thresholding segmentation. In Advances in soft computing and machine learning in image processing (Studies in computational intelligence) (Vol. 730, pp. 23–39). Springer.
Hassanpour, H., Samadiani, N., & Salehi, S. M. (2015). Using morphological transforms to enhance the contrast of medical images. The Egyptian Journal of Radiology and Nuclear Medicine, 46(2), 481–489.
Wang, C., Brentnall, A. R., Cuzick, J., Harkness, E. F., Evans, D. G., & Astley, S. (2017). A novel and fully automated mammographic texture analysis for risk prediction: Results from two case-control studies. Breast Cancer Research, 19, 114.
Priyanka, D., & Chinmay, C. (2021). Application of AI on post pandemic situation and lesson learn for future prospects. Journal of Experimental & Theoretical Artificial Intelligence, 1, 1–24.
Chinmay, C. (2019). Performance analysis of compression techniques for chronic wound image transmission under smartphone-enabled tele-wound network. International Journal of E-Health and Medical Communications (IJEHMC), 10(2), 1–15.
Saidin, N., Ngah, K. U., Shuaib, L. I., & Sakim, H. A. M. Segmentation of breast regions in mammogram based on density: A review. Imaging & Computational Intelligence Group (ICI).
Chakravarthi, R., Nandhitha, N. M., Roslin, S. E., & Selvarasu, N. (2016). Tumour extraction from breast mammographs through hough transform and DNN hybrid segmentation technique. Biomedical Research, 27(4), 1188–1193.
Krishnan, M., Chinmay, C., Banerjee, S., Chakraborty, C., & Ray, A. K. (2009). Statistical analysis of mammographic features and its classification using support vector machine. Expert Systems with Applications, 37, 470–478.
Kaur, P., Singh, G., & Kaur, P. (2019). Intellectual detection and validation of automated mammogram breast cancer images by multi-class SVM using deep learning classification. Informatics in Medicine Unlocked, 16, 100151.
Ganesan, K., Acharya, U. R., Chua, C. K., Min, L. C., Abraham, K. T., & Kwan-Hoong, N. (2013). Computer-aided breast cancer detection using mammograms: A review. IEEE Reviews in Biomedical Engineering, 6, 77–98.
Pavol, Z., Peter, K., Karol, K., Zuzana, D., Hubert, P., Tibor, B., Erik, K., Marek, S., Alena, L., Dominika, V., Tatiana, K., Igor, S., Veronica, H., Jan, B., Zuzana, L., Dietrich, B., Mariusz, A., Walther, K., Jan, D., & Olga, G. (2019). Why the gold standard approach by mammography demands extension by multiomics? Application of liquid Biopsis miRNA profiles to breast cancer disease management. International Journal of MolecularSciences, 20(12), 2878.
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barz, A., & Jemal, A. (2017). Colorectal cancer statistics. CA: a Cancer Journal for Clinicians, 67(3), 177–193.
Ford, D., Easton, D. F., Stratton, M., Narod, S., Goldgar, D., Devilee, P., Bishop, D. T., Weber, B., Lenoir, G., Chang-Claude, J., Sobol, H., Teare, M. D., Struewing, J., Arason, A., Scherneck, S., Peto, J., Rebbeck, T. R., Tonin, P., Neuhausen, S., … Eyfjord, J. (1998). Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. American Journal of Human Genetics, 62, 676–689.
Kallenberg, M., Petersen, K., Nielsen, M., Ng, A. Y., Diao, P., Igel, C., Vachon, C. M., Holland, K., Winkel, R. R., Karssemeijer, N., & Lillholm, M. (2016). Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Transactions on Medical Imaging, 35(5), 1322–1331.
Sayed, G. I., Darwish, A., Hassanien, A. E., & Pan, J.-S. (2017). Breast cancer diagnosis approach based on meta-heuristic optimization algorithm inspired by the bubble-net hunting strategy of whales. In Genetic and evolutionary computing (Advances in intelligent systems and computing). Springer.
Nasiri, J., & Khiyaban, F. M. (2018). A whale optimization algorithm (WOA) approach for clustering. Cogent Mathematics & Statistic, 5, 1483565.
Yogapriya, J., Saravanabhavan, C., & Ila, V. (2018). Medical image retrieval system using local binary patterns,whale optimization & relevance vector machine algorithms. Tagajuornal, 14, 3164–3191.
Adepoju, T. M., Ojo, J. A., Omidiora, E. O., & Olabiyisi, O. S. (2015). Pixel-based morphological technique for breast tumour detection. International Journal of Scientific & Engineering Research, 6(6), 1416–1420.
ACS. (2017). Breast cancer early detection and diagnosis.
Jalalian, A., Mashohor, S., Mahmud, R., Karasfi, B., Saripan, M. I. B., & Ramli, A. R. B. (2017). Review article: Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI Journal, 16, 113–137.
Saslow, D., Boetes, C., & Burke, W. (2007). American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA: A Cancer Journal for Clinicians, 57(2), 75–89.
Raj, J. R., Rahman, S. M. K., & Anand, S. (2016). Preliminary evaluation of differentiation of benign and malignant breast tumors using non-invasive diagnostic modalities. Biomedical Research, 27(3), 596–603.
Adeyemo, T. T., Adepujo, T. M., Sobowale, A. A., Oyediran, M. O., Omidiora, E. O., & Olabiyisi, S. O. (2017). Feature extraction techniques for mass detection in digital mammogram (review). Journal of Scientific Research & Reports, 17(1), 1–11.
Shallu, R. M. (2018). Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express, 4, 247–254.
Rakhlin, A., Shvets, A., Iglovikov, V., & Kalinin, A. A. (2018). Deep convolutional neural networks for breast cancer histology image analysis. In Lecture notes in computer science (Vol. 10882, pp. 737–744). Springer.
Arau’jo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., et al. (2017). Classification of breast cancer histology images using convolutional neural networks. PLoS One, 12(6), e0177544.
Tahir, M., Muhammad, A. M. O., Min, B. L., & Kang, R. P. (2020). Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and Deep CNNs. Journal of Clinical Medicine, 9, 749.
Alkassar, S., Jebur, B. A., Abdullah, M. A. M., Al-Khalidy, J. H., & Chambers, J. A. Going deeper: Magnification invariant approach for breast cancer classification using histopathological images. IET Computer Vision, 15, 151–164.
Adepoju, T. M., Adeyemo, T. T., Fagbola, T. M., Omidiora, E. O., & Olabiyisi, S. O. (2016). Histogram normalization technique for preprocessing of digital mammographic images. LAUTECH Journal of Engineering and Technology, 10(1), 82–87.
Corinne, B., Salma, A., Kim, V. N., Daniel, V., Clarisse, D., & Robert, S. (2007). BIRADSTM classification in mammography. European Journey of Radiology, 61, 192–194.
Rampun, P., Morrow, J., Scotney, B. W., & Wang, H. (2020). Breast density classification in mammograms: An investigation of encoding techniques in binary-based local patterns. Computers in Biology and Medicine, 122, 1–18.
Aghdam, H., Puig, D., & Solanas, A. (2013). A probabilistic approach for breast boundary extraction in mammograms. Computational and Mathematical Methods in Medicine, 10, 17–26.
Suguna, S. K., & Ranganathan, R. (2017). A new Evolutionanary – Based optimization algorithm for mammogram image processing. International Journal of Pure and Applied Mathematics, 117(17), 241–247.
Cruz, C. F. (2011). Automatic analysis of mammography images: Enhancement and segmentation techniques. Port University.
Karakoyun, M., Baykan, N. A., & Hacibeyoglu, M. (2017). Multi-level thresholding for image segmentation with swarm optimization algorithms. International Research Journal of Electronics & Computer Engineering, 3(3), 654–658.
Kumar, T. G., Murugan, D., & Manish, T. I. (2018). An analysis on road extraction from satellite image using Otsu method and genetic algorithm techniques. WSEAS Transactions on Computers, 17, 42–51.
Mirjalili, S., & Lewis, A. (2016). The whale optimization algorithm. Advances in Engineering Software, 95, 51–67.
Zidan, M., Hassanien, A. E., Hefny, H. A., & Houseni, M. (2017). Liver segmentation in MRI images based on whale optimization algorithm. Multimedia Tools and Applications, 76, 24931–24954.
Ozturk, Ş., Akdemir, B., Ozkaya, U., & Seyfi, L. (2017). Soft tissue sacromas segmentation using optimized otsu thresholding algorithms. International Journal of Engineering Technology, Management and Applied Sciences, 5, 9.
Mafarja, M. M., & Mirjalili, S. (2017). Hybrid Whale Optimization Algorithm with simulated annealing for feature selection. Neurocomputing, 260, 302–312.
Kowsalya, S., & Priyaa, D. S. (2015). A survey on diagnosis methods of breast cancer using mammography. International Journal of Engineering Technology Science and Research, 2, 100–107.
Guyon, I., & Elisseeff, A. (2003). Special issue on variable and feature selection. Journal of Machine Learning Research, 3, 1157–1182.
Mohanaiah, P., Sathyanarayana, P., & GuruKumar, L. (2013). Image texture feature extraction using GLCM approach. International Journal of Scientific and Research Publications, 3(5), 1–5.
Punithavathi, V., & Devakumari, D. (2020) Detection of breast lesion using improved GLCM feature based extraction in mammogram images.
Jagadesh, B. N., & Kumari, L. K. (2021). A GLCM based feature extraction in mammogram images using machine learning algorithms. International Journal of Current Research and Review, 13(5), 145–149.
Mahdi, F. (2015). Application of GLCM technique on mammograms for early detection of breast cancer. Journal of Babylon University/Pure and Applied Sciences, 23(2), 885–890.
Chinmay, C. (2019). Computational approach for chronic wound tissue characterization. Elsevier: Informatics in Medicine Unlocked, 17, 1–10.
Mryka, H. B. (2017). GLCM texture: A tutorial. University of Calgar.
Ojo, J. E., Adepoju, T. M., Omidiora, E. O., Olabiyisi, O. S., & Bello, O. T. (2014). Pre-processing method for extraction of pectoral muscle and removal of artefacts in mammogram. IOSR Journal of Computer Engineering (IOSR-JCE), 16(3), 06–09.
Adepoju, T. M., Ojo, J. A., Omidiora, E. O., Olabiyisi, O. S., & Bello, O. T. (2015). Detection of tumour based on breast tissue categorization. British Journal of Applied Science & Technology, 11(5), 1–12.
Mohamed, S. E., Wahbi, T. M., & Sayed, M. H. (2018). Automated detection and classification of breast cancer using mammography images. International Journal of Science, Engineering and Technology Research (IJSETR), 7, 4.
S. M. Castro, E. Tseytlin, O. Medvedeva, . K. Mitchell, S. Visweswaran, T. Bekhuis and R. S. Jacobson, "Automated annotation and classification of BI-RADS assessment from radiology reports, Journal of Biomedical Informatics" 67, 177–187, 2017.
He, W., Denton, E. R., & Zwiggelaar, R. (2010). Mammographic image segmentation and risk classification using a novel texture signature based methodology. IWDM, 6136, 526–533.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ojo, J.A., Bello, T.O., Idowu, P.O., Solomon, I.D. (2022). Optimal Feature Selection for Computer-Aided Characterization of Tissues: Case Study of Mammograms. In: Chakraborty, C., Khosravi, M.R. (eds) Intelligent Healthcare. Springer, Singapore. https://doi.org/10.1007/978-981-16-8150-9_3
Download citation
DOI: https://doi.org/10.1007/978-981-16-8150-9_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-8149-3
Online ISBN: 978-981-16-8150-9
eBook Packages: Computer ScienceComputer Science (R0)