Abstract
The advent of Next Generation Sequencing (NGS) and thereby the access to huge metagenomic data have opened the flood gates of genetic information. Although the field of microbiome has evolved, the field of virome is a relatively new addition. The co-existence of viruses with the host may be localized or systemic in nature. Growing evidence suggests that apart from the gut, organ or tissue specific virus niche does exist. However, the big question that requires attention is the nature of interaction of these viruses with the host. While viral infections and the concurrent co-infections with bacterial pathogens have been established as a factor in disease severity, reports on viral co-infections are significantly limited. Thus a huge conundrum exists on whether the pathogenic trait of viruses is an inclusive property of the virome. Whether a mutualistic relationship exists between viruses and if it is beneficial to the host is still a big question requiring in-depth investigation. In this chapter we aim to provide key details on the basis and the origins of the human virome, distribution and host interaction with a special emphasis on the nature of this association.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
The human virome is a part of one of the most complex ecosystems in the world, namely the microbiome. It represents a repertoire of all the viruses that inhabit the human body which includes the eukaryotic viruses, the virus-derived elements that are inserted into host chromosomes (endogenous retroviruses) and the bacteriophages that are capable of infecting the inhabitant bacteria and archaea (prokaryotic viruses) (Virgin 2014). A large number of viruses have been reported to inhabit humans, while only half of them are pathogenic (Parker 2016). Non-pathogenic viruses are referred to as “commensal” viruses as they survive by either integrating into the host chromosome or by infecting bacteria without causing any clinical outcome. However, pathogenic ones have been shown to affect human health by causing acute, persistent, or latent infections which in many instances are detrimental to the host. The bulk of the healthy human virome comprises of the bacteriophages, which are capable of infecting bacteria present in the intestine and other parts of the human body. A study on the diversity of gut virome in 1-year-old infants has found a strong correlation between diversity and the manner of birth (McCann et al. 2018). Several pathogenic viruses are also transmitted to the newborn during vaginal delivery as well as during breastfeeding. The gut of the healthy neonates is devoid of any viruses at the time of birth. The direct transmission of the virus strains from the mother to the infant was validated by sequencing the fecal sample of the baby and the breast milk of the mother. Human milk viruses are important in shaping the gut virome of the infants and are also important in the overall immune development (Mohandas and Pannaraj 2020).
Over the past decade, several studies have shed light on the link between the microbiome and the various states of human health. Nevertheless, a better understanding of the virome is still lacking when compared to the other players of the microbiome. This lag in knowledge is attributed to the absence of a universal viral sequence similar to the 16S rRNA present in the bacteria. Most often, the virus-enriched preparations do not align with any reference sequences and hence are represented as the viral “dark matter” (Roux et al. 2015). The dark matter hence might comprise several novel and highly divergent viruses, which collectively constitute the “viral assemblage”.
2 Factors Influencing the Distribution and Diversity of Virome
The microbiota of the human body is enriched with distinct microbial flora, which accommodates the growth of a variety of viruses. Several factors are known to affect the distribution and diversity of the human virome (Fig. 10.1). A major constituent of the human virome is bacteriophage, and the type and distribution of the microbial community has a significant impact on the distribution and diversity of the virome. Marked differences exist within the microbiome based on the anatomical site of distribution, and parallels can be drawn likewise with the virome (Abeles et al. 2014; Reyes et al. 2010). The establishment of human virome inside the body is closely associated with the initial colonization. Various viruses, including Zika virus, HIV, rubella virus, herpes simplex virus, and human papillomavirus, are known to adopt a vertical mode of transfer during pregnancy (Leeper and Lutzkanin 3rd 2018; Arora et al. 2017). According to Breitbart et al., neonates lack virome at birth but are gradually colonized after a week of age (Breitbart et al. 2008). This is supported by yet another study which identified a high diversity of gut virome in the neonates after just few weeks of birth (Lim et al. 2015). Breastfeeding is an important factor in shaping the gut viral community in the neonates. An abundance in viral population was observed in infants who were exclusively fed formula than those dependent entirely or partially on breast milk (Liang et al. 2020b). Considering that breast milk is a rich source of maternal antibodies, immune cells, lactoferrin, mucin, and milk oligosaccharides which can restrict the invasion of a wide variety of viruses including influenza virus, rotavirus, enterovirus, norovirus, and SARS-CoV, breast milk while protecting neonates from deadly viruses can aid in the adaptation and colonization of beneficial viruses (Lang et al. 2011; Turin and Ochoa 2014; Simister 2003; Pou et al. 2019; Albrecht and Arck 2020; Berlutti et al. 2011; Wicinski et al. 2020).
The diet, age, and sex of a person can also modulate the diversity of the virome (Fig. 10.1). It has been reported that oral viromes were similar in people with the same diet or oral bacterial population and between people from the same household or family (Robles-Sikisaka et al. 2013). This is compounded by a study that indicated an inter-individual variation of the gut microbiome in response to diet changes (Minot et al. 2011). Within the oral cavity, significant variation in phage communities has been observed in the saliva, dental plaque, and subgingival and supragingival biofilms (Wang et al. 2016a, b). Sex-specific variation inside the oral viral community has been reported, wherein it was identified that the genotype of the oral virome in an individual is highly personalized and gender-specific (Abeles et al. 2014). The age and immune status of an individual are also key factors which can affect the richness of viral communities. Gregory et al. showed that the enrichment of eukaryotic viruses most importantly human Annelloviruses is high during infancy and then decreases with childhood and remains constant and low through the rest of life which corresponds to the fact that the patterns in viral diversity is age-dependent (Gregory et al. 2020).
There is also a direct correlation between the abundance of the viral population and an individual’s immune status. Studies have shown that boosting immunity is an effective strategy in enhancing anti-viral immunity in the gut. Metagenomic analysis of viral population in the gut of an X-linked severe combined immunodeficiency patient revealed a viral population rich in adenovirus and bocaviruses and upon immune reconstitution, the gut microbiota was normalized and the viral infections were cleared (Clarke et al. 2018a). A positive effect of bacteria or bacterial components in restricting viral infection has also been demonstrated wherein it was shown that flagellin exposure activated the immune response and restricted rotavirus infection in mice (Zhang et al. 2014). Alternatively, the administration of recombinant IFNλ could effectively clear persistent norovirus infection (Nice et al. 2015). These observations highlight that effective cross-talk exists between microbiome components and the host which is pivotal in clearing pathogenic viruses which are also part of the virus assemblage.
Host genetics also contributes to the virome composition and diversity. Studies have demonstrated that monozygotic twins have a similar microbiome compared to dizygotic twins (Goodrich et al. 2016; Goodrich et al. 2014). However, few studies have identified that the environment of an individual has a significant role in shaping the microbiome rather than host genetics (Rothschild et al. 2018). Several studies have also claimed a significant variation in human virome associated with geographic location (Holtz et al. 2014). However, it is not always true, for instance, Polyomavirus species collected from individuals of different geographic regions showed very low genetic diversity (Foulongne et al. 2012; Rascovan et al. 2016). Children with diarrhea from two locations within Australia have shown a significant variation of eukaryotic viromes with a differential prevalence of Adenoviridae and Picornaviridae (Holtz et al. 2014). Thus it is clear that viral abundance and diversity is dependent on multiple factors. As more and more systematic studies are carried out, it would emerge that many other factors besides those described above also contribute to the assemblage of viruses in the human virome.
3 System-Wise Distribution of the Human Virome
3.1 Ocular Virome
Ocular surface (OS) microbiome constitutes the microbiota that resides on the surface of the conjunctiva and the cornea (micro-organisms that colonize eyelids are considered as a part of skin microbiota) (Lu and Liu 2016). Investigations into the ocular microbiome are a relatively new and emerging area and most of the studies are designed to investigate the prokaryotic residents as opposed to the larger, more inclusive microbial community including virome and mycobiome (Fig. 10.2). Culture-independent metagenomic studies on OS have revealed that, unlike skin or other mucosal tissues, the healthy OS microbiome is sparsely colonized (~100 times less than that of the facial skin or the buccal mucosa) (Doan et al. 2016). Analysis of metagenomic data of OS microbiome from 90 adult healthy individuals showed that approximately 98% of the microbial reads were of bacterial origin while viral and fungal sequences accounted for <1% each (Wen et al. 2017). Doan T. et al. employed biome representational in silico karyotyping (BRiSK), a deep sequencing technique that achieves unbiased representation of all DNA-based metagenomic constituents and uncovered the presence of viruses such as torque teno virus (TTV), multiple sclerosis-associated retrovirus (MRSV), and human endogenous retrovirus K (HERV-K) in the conjunctiva of healthy volunteers. Although less frequent, sequences pertaining to human papillomavirus (HPV), Merkel Cell Polyomavirus (MCV), and Abelson murine leukemia virus were also retrieved by BRiSK. A noteworthy observation however was the high PCR positivity rate for TTV which was as high as 65% in all the conjunctiva samples tested, suggesting that TTV might be a homeostatic resident on ocular surfaces of healthy humans (Doan et al. 2016). However, in an unrelated study, deep sequencing of vitreous biopsies from patients with presumed culture-negative infectious endophthalmitis could identify TTV in all culture-negative samples, although a direct association or causation was not claimed (Lee et al. 2015). Though metagenomic deep sequencing allows a comprehensive analysis of microbial or host genetic materials in the sample, the choice of the genetic material (RNA vs DNA) can limit the power of the microbiome studies in the context of “virome.” Both the metagenomic studies highlighted above used DNA as their starting material which subsequently restrains or impacts the “virome” analysis as the approach is technically “blind” to the group of viruses with RNA as their genetic material.
3.2 Oral Virome
Microbial communities in the oral cavity are diverse comprising fungi, bacteria, and viruses. Traditionally, oral microbiome sampling was limited to saliva; but recent studies have identified distinct virus communities from other oral microenvironments like dental plaques also. Saliva collected from healthy volunteers, when subjected to SYBR-gold nucleic acid staining post sequential filtration, showed ~108 virus-like particles (VLPs) per milliliter, majorly constituting lysogenic bacteriophages. Among the genomes analyzed, more than 90% accounted for prophages, which largely outnumber bacteriophages. Bacteriophages of the order caudovirales, belonging to the families Siphoviridae and Myoviridae, and Podoviridae are the most abundant. Herpesviridae, Papillomaviridae, Anelloviridae, and Redondoviridae families of eukaryotic viruses are also frequently encountered. Unlike many eukaryotic viruses that are asymptomatic in healthy individuals (Table 10.1) (Wylie et al. 2014), redondoviruses are found to be associated with periodontitis in the oral cavity, and their abundance in the oro-respiratory tract has been implicated with worse prognosis in patients admitted in critical care facilities (Abbas et al. 2019a; Perez-Brocal and Moya 2018). The dental plaque has ~108 VLPs per milligram sample and shares a significant proportion of viral homologs with the salivary virome suggesting the presence of some of the viruses in both the niches (Naidu et al. 2014). The oral virome is influenced by the living environment and its composition is temporally regulated (Pride et al. 2012; Robles-Sikisaka et al. 2013).
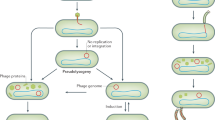
The oral virome is dynamic in nature and it has been suggested that these viruses might serve as a reservoir of pathogenic gene functions, imparting virulence to the resident bacteria of the buccal cavity. The non-exclusive coexistence of both bacteriophages and their hosts in the same ecological niche points toward the existence of both positive and negative interactions between them (Pride et al. 2012). The lysogenic lifestyle of the siphoviruses in the oral cavity, in a dynamic equilibrium with their prokaryotic hosts, makes them excellent vehicles for horizontal gene transfer, potentially imparting antibiotic resistance to the host. On the other hand, myoviruses and podoviruses being predominantly lytic are responsible for the elimination of 20–80% of the oral bacteria. This arms race between phages and bacteria prevent the successful establishment of novel species of bacteria or phages in the oral cavity (Baker et al. 2017).
3.3 Gut Virome
The collective population of both the eukaryotic and prokaryotic viruses colonizing the human gut comprises the human gut virome (Fig. 10.2). Although the total viral loads in the human gut vary from subject to subject, they can range anywhere between 2.2 × 108 and 8.4 × 1010 genome copies per gram of feces (Shkoporov et al. 2019). The human Gut Virome database, a compilation of numerous microbial metagenomic studies across the continents, uncovered 33,242 potentially unique viral population found in the human gut (Gregory et al. 2020). The viruses that infect bacteria (bacteriophages) predominate (>97%) the gut virome and evidences point to a temperate lifestyle exhibited by majority of the phages within the gut ecosystem (Ogilvie and Jones 2015). Most common bacteriophages in the gut virome belonged to the family Siphoviridae, Microviridae, and Myoviridae (Table 10.1). Another family of bacteriophage known as crAssphage and its expansive group of crAss-like phages are the most abundant human-associated virus found in ~50% of the human gut samples, which often comprises 90% of the annotated sequence reads in gut virome-specific metagenome (Shkoporov et al. 2018; Yutin et al. 2018). Though minimal in proportion, several DNA and RNA eukaryotic viruses have also been detected in the feces sample. Most commonly associated RNA viruses included enterovirus, parechovirus, tombamovirus, sapovirus, calciviruses, astroviruses, and picornaviruses (Lim et al. 2015). Members from the family Anelloviridae, Herpesviridae, Adenoviridae, Papillomaviridae, and Polyomaviridae are the main eukaryotic DNA viruses associated with human intestinal virome (Rampelli et al. 2017). Although rare, contigs matching the sequences of megavirome such as Mimivirus and Marseillevirus (Colson et al. 2013), an archaeal virus family (Lipothrixviridae) (Lim et al. 2015), and several plant pathogenic viruses have also been identified in the fecal metagenome (Table 10.1) (Zhang et al. 2006). Similar to their bacterial counterparts in the microbiome, the composition of the gut virome is also dynamic and is mostly shaped during the early years of the development (Lim et al. 2015). In a longitudinal study of the intestinal virome in infants by Lim et al., it was found that the bacteriophage diversity was maximum at the earliest time point tested (month 0) and it eventually decreased with age. They also reported a noticeable shift in the composition of phage community trending toward a relative increase in the abundance of Microviridae family of bacteriophages by the age of 24 months. Interestingly, a parallel analysis of gut bacterial diversity revealed an inverse correlation with the richness of the associated virome in an age-dependent manner indicating dynamic interplay during the early years of life (Lim et al. 2015). Studies with monozygotic twins indicated that, although co-twins and their respective mothers shared similar virome profiles, nevertheless, each subject harbored a distinct and unique individual virome irrespective of the genetic relatedness (Lim et al. 2015; Reyes et al. 2010). Despite high interpersonal variations in fecal virome among the subjects, intrapersonal diversity within individual subjects across time was very low. In a longitudinal study by Reyes et al., it was demonstrated that >95% of the virome was retained over a period of 1 year (Minot et al. 2013). Yet another study suggested that nearly 80% of the virotypes persisted in the stool samples of a subject throughout the study period of 2.5 years (Minot et al. 2011) indicating remarkable long-term genetic stability of the member species. This unusual genomic stability exhibited by the virome despite a hallmark error-prone viral replication system is due to the fact that majority of the phages in the virome exhibit a temperate lifestyle with low mutation rates mainly because the viral genome maintenance involves replication by high fidelity bacterial DNA polymerase. Nevertheless, few members of the viral community (lytic bacteriophages such as members from Microviridae family) had very high substitution rates that propelled the evolution of some long-term virome members over time contributing to interpersonal virome diversity (Minot et al. 2013). It is well established that diet is a key modifier influencing the composition of gut bacterial community. Congruently, Minot et al. showed that a controlled diet can significantly alter gut virome composition where individuals on the same diet converged and showed a tendency toward more similar virome, however not identical (Minot et al. 2011).
3.4 Skin Virome
The human skin is the largest and the most exposed organ in the body which facilitates the inception of a complex ecosystem of cutaneous flora containing bacteria, fungi, and viruses. Investigations into the bacterial and fungal flora of the skin microbiota and their role in health and disease were extensively carried out in the last two decades, however, the studies related to their viral counterpart, the “virobiota,” is still in its infancy. The human skin virobiota is highly diverse. Much like the composition of oral and the gut virome, bacteriophages from the order Caudovirales (mainly Microviridae and Siphoviridae family) largely predominate the niche compared to the viruses that are potent human pathogens (Foulongne et al. 2012). Among the other abundant bacteriophages of skin included Staphylococcus and Propionibacterium phages. Although the relative proportion of phages varied across different anatomical sites and skin microenvironment, >85% of the phages were predicted to exhibit temperate lifestyle (Hannigan et al. 2015). The eukaryotic DNA viruses in the skin include members of the family Papillomaviridae, Polyomaviridae, Circoviridae, and Poxviridae including sequences related to beta and gamma-papillomaviruses (Table 10.1), human polyomavirus 6, 7, and 9, and Merkel cell polyomavirus (MCPyV) (Foulongne et al. 2012; Hannigan et al. 2015). Majority of the reads pertaining to RNA virome from the skin and the nasal swabs from a cohort of patients with primary immunodeficiency could be mapped to DNA viruses of the family Papillomaviridae and Polyomaviridae, suggesting actively replicating DNA viruses in the skin (Tirosh et al. 2018). While most studies focused on the compositional analysis of the whole viral metagenome, a study by Hannigan et al. focused on the variability of the skin virome in the evolutionary context. They identified 106 and 465 hypervariable loci in human papillomavirus (HPV) and Staphylococcus phages respectively which mapped to genes involved in host tropism, immune evasion, virulent gene expression, and utilization of host resources. However, these hypervariable loci expressed low non-synonymous to synonymous ratio thereby suggesting purifying selection with a propensity to maintain the consensus protein sequences (Hannigan et al. 2017). Several factors affect the diversity of the viral communities in the skin, this includes the moisture content and the occlusion status of different anatomic sites. Significant differences in both alpha (within-sample) and beta (between-samples) diversities in the virome and the whole metagenome structure have been reported which strongly depended on the skin microenvironment. The virome structure, but not the whole metagenome, significantly differed at each anatomical site paired over a period of 1 month for each sample, suggesting greater longitudinal stability of whole microbiome when compared to viral communities (Hannigan et al. 2015). Cutaneous microflora is known to influence host immunity in a myriad of ways (reviewed in (Belkaid and Tamoutounour 2016)). However, the immune status of the host also plays a significant role in shaping the ecological constituents of the skin. A remarkable increase in the relative abundance of eukaryotic viruses in the skin of DOC8-deficient patients with primary immunodeficiency compared to healthy controls has been observed (Tirosh et al. 2018). Unbiased high-throughput sequencing has also revealed the association of specific virus with certain diseases, for instance, approximately 25% of the patients with Merkel cell carcinoma (MCC) were identified with a strain of polyomavirus almost identical to human polyomavirus 9 as compared to 0.9% of the healthy controls without MCC (Sauvage et al. 2011). Yet another study reportedly confirmed relatively high levels of MCPyV DNA in the skin samples of patients with MCC as compared to the age-matched healthy controls and other cutaneous cancer patients (Hashida et al. 2016). Metagenomic analysis of skin virome from a single subject with widespread warts could map 30% of all the reads to HPV genomes, particularly HPV2 (Landini et al. 2015). Thus the skin is enriched with viruses that are integral to the microbiome and these viruses can be either symbiotic or pathogenic.
3.5 Respiratory Tract Virome
The human respiratory tract is a dynamic site of interaction of diverse air-borne viruses that cocirculate in the space, forming an ecological niche, facilitating several interspecific interactions (Fig. 10.2). Interactions within this niche are often maintained in a balanced state, failure of which affects the equilibrium and may turn out to be harmful to the host (Young et al. 2015; Willner et al. 2009; Wylie et al. 2012; Clarke et al. 2018b; Abbas et al. 2017, 2019b). Although there are extensive evidences for bacteria-bacteria and bacteria-virus interactions, knowledge on the occurrence of virus-virus interactions is limited (Bosch et al. 2013; Weinberger et al. 2015; McCullers and Rehg 2002). Many viruses have demonstrated selectivity in their attachment and replication in specific regions of the respiratory tract, which largely depends on the type and availability of specific receptors in those regions. Pathogenic viruses that target the respiratory tract have devastated mankind; the most notable ones include the 1918 pandemic flu caused by H1N1 influenza virus and more recent COVID-19 caused by SARS-CoV-2.
The human respiratory virome consists of all the eukaryotic viruses and bacteriophages that are found in the upper respiratory tract and within the lungs. Sampling is the foremost challenge in defining the respiratory tract virome. For example, sampling from the lower respiratory tract requires invasive procedures. Bronchoalveloar lavages are only possible in symptomatic individuals for which we do not have a normal healthy control (Wylie et al. 2012). Studies in patients with cystic fibrosis even showed that there was distinct virus population in different regions of the lung (Willner et al. 2012). Comprehensive studies so far have shown that like the gut virome, many species of phages including those belonging to the order caudovirales and the Inoviridae, and the Microviridae families are present in the lungs. The most pliable source of these phages is either the oral cavity or is derived from the upper respiratory tract. The most notable and widely prevalent families of viruses include those of Anelloviridae, Redondoviridae, Adenoviridae, Papillomaviridae, Herpesviridae, Picornaviridae, and Paramyxoviridae (Table 10.1) (Willner et al. 2009; Young et al. 2015; Wylie et al. 2012; Clarke et al. 2018b; Abbas et al. 2017, 2019b).
4 Other Sites
4.1 The Central and Peripheral Nervous System Virome
The difficulty involved in studying the resident populations of viruses in the nervous system can at least in part be attributed to the difficulty in obtaining samples from normal and healthy individuals. Available literature on the virome of the nervous system is based on the analysis of cerebrospinal fluid. Viruses at times employ multiple mechanisms to breach the blood-brain barrier and can cause acute infection or can integrate into the genome of neuronal cells and be reactivated later. Apart from the pathogenic opportunistic viruses, regions of the nervous system are home to viral communities which included those belonging to Herpesviridae family but mostly predominated by phages. The phages in the CNS were found to be of Siphoviridae, Podoviridae, and Myoviridae families (Ghose et al. 2019).
4.2 Blood Virome
The human blood maybe least expected to harbor viral communities because of the presence of innumerable immune components including an array of immune cells, antibodies, and the complement system. It is known that the human blood is exploited by many pathogenic viruses and viremia, although can serve many times as an indicator of the presence of viruses in the blood stream, is a tool used by these viruses to disseminate to distal sites. Interestingly, certain classes of viruses have been shown to co-habit the blood stream and the well-documented viruses belong to Phycodnaviridae, Picornaviridae, Mimiviridae, Marseilleviridae, and Herpesviridae families (Table 10.1) (Moustafa et al. 2017; Liu et al. 2018). The most compelling evidence was obtained in the case of viruses of Anelloviridae family, which was confirmed by electron microscopy besides the use of other genomic tools (Breitbart and Rohwer 2005). What is the precise role of these viruses in the blood and the nature of their very existence and source are intriguing questions that require systematic investigation.
4.3 Urogenital Tract Virome
The urogenital system of both the male and the female are also found to host a myriad of viruses. The richness of virome in this system is highlighted by the observation that urine samples of healthy individuals contain approximately 1 × 107 virus-like particles per milliliter with phages and papilloma viruses contributing to the overall bulk of viruses (Santiago-Rodriguez et al. 2015; Garretto et al. 2019). Both the male and the female urogenital tracts are home to resident viruses. Analysis of the seminal fluid of healthy men showed that the predominant viral families include Anelloviridae, Herpesviridae, and Papillomaviridae while vaginal swabs from healthy women showed the presence of an abundance of double-stranded DNA phages (Table 10.1) (Jakobsen et al. 2020; Li et al. 2020).
5 Dynamics of Virome–Host Interactions
Viruses are largely believed to be obligate parasites. With technological advancements in the last two decades, we have come to a point where we now know that they are not just parasites that are detrimental to the host, but many of them play dynamic roles in maintaining tissue homeostasis. A “healthy virome” is heterogenous in nature and consists of three components (a) the viruses that systemically enter the human body mainly through food but however do not replicate, (b) viruses that infect prokaryotes and probably the unicellular eukaryotes that comprise the healthy human microbiome, and (c) the viruses that can essentially replicate and persist in humans. Several of them are essential parts of the ecosystem and coexist either temporarily or forever as symbionts. The mode of interaction is governed by multiple factors such as environment, diet, lifestyle, host and virus population structure, and the general health and immunity of the individuals. Due to the influence of the aforesaid factors and due to co-evolution of viruses with the host, the nature of the virus-host relationship does not follow a single pattern, but ranges from aggressive antagonism to mutualism. Therefore, the borderline that dictates the time and the conditions due to which the coexisting viruses may turn to be harmful cannot be well defined. Reports also suggest that the symbiotic association between persistent viruses and the host especially the retroviruses has contributed significantly to the constructive evolution of the host.
Growing evidence suggests that the human virome can act as an immune-modulator and thereby facilitating either the protection from or initiation of diseases. Viruses present inside the human body can interact with both the microbiome and host cells, ultimately resulting in the immune variation inside the host (Shi and Gewirtz 2018). For example, during vertical transmission of mouse mammary tumor virus through maternal milk, the presence of MMTV-bound LPS complex activated TLR-4/MyD88 pathway resulting in the production of the immunosuppressive cytokine IL-10 in the pups, which facilitated the establishment of infection (Kane et al. 2011). Enteric bacteria promote murine norovirus infection of B cells by reducing the efficacy of IFN-λ mediated viral clearance, and depletion of intestinal microbiota by antibiotic treatment reduced mouse norovirus replication in vivo (Jones et al. 2014; Baldridge et al. 2015). Analogous to this, ablation of microbiota negatively affected the initial infectivity of rotavirus and enhanced specific humoral immunity (Uchiyama et al. 2014). Human endogenous retroviruses (HERVs) have been found to be integrated into the human genome and play critical roles in modulating host immunity even in the absence of functional viral proteins (Grandi and Tramontano 2018). The association between human and HERVs represents a typical symbiosis, which can have both beneficial and detrimental effects on the host. HERV-K is a bonafide member of the healthy virome and is found to suppress the spread of invasive melanoma (Singh et al. 2020). Parvovirus B19 (B19) infection in adults is asymptomatic in nature but with a prevalence rate of 25% in human skin biopsies (Bonvicini et al. 2010). However, they are capable of arresting hematopoiesis resulting in anemia and less frequently neutropenia, which may even be life-threatening in immune-compromised patients (Shehi et al. 2020).
Potential beneficial role of viruses in a holobiont is well noted in instances where viruses can even impede further infection or pathogenesis. Classic examples are where the Hepatitis G virus can slow down the progression of HIV infection to acquired immune deficiency syndrome (AIDS) (Tillmann et al. 2001) and that of latent herpes virus having a protective role against Listeria monocytogenes and Yersinia pestis infections (Barton et al. 2007). On the contrary, cytomegalovirus (CMV) exhibits the potential to promote Pneumocystis jiroveci infections (an opportunistic pathogen causing severe pulmonary infections) in immunocompromised patients (Lee et al. 2020). Several trans-kingdom interactions have also been reported. For example., influenza virus by virtue of its neuraminidase activity exposes several bacterial receptors on the cell surface which in turn augments the super infection with Streptococcus pneumoniae or Staphylococcus aureus (Bosch et al. 2013).
Human anelloviruses (AV) represent a group of highly diverse and omnipresent commensal viruses. Their presence in blood, tears, saliva, semen, breast milk, nasal secretion, bile, etc. suggest that these viruses exhibit a broad range of tropism (Kaczorowska and van der Hoek 2020). Children are mostly infected with AVs months after birth, but these viruses have also been detected in children and adults of all ages (Vasilyev et al. 2009; Brassard et al. 2015). Pegiviruses cluster tightly with hepaciviruses which include the hepatitic C virus (HCV), which is a major human pathogen. The incidence rate of pegivirus infection in humans is 5%. Although suspected to be a causative agent of diarrhea, the prototypic pegivirus, formerly known as the hepatitis G virus (HGV), apparently is not linked to any pathology like HCV (Hartlage et al. 2016; Stapleton et al. 2011). They can readily infect and grow in human cell lines and are implicated in fighting the burden of AIDS (Greenhalgh et al. 2019). Another example is that of Picobirnaviruses. Isolation and culturing of these viruses in the lab have proved difficult, and hence the true identity of the host still remains unknown. They have been isolated from the stool samples of individuals with diarrhea of unknown cause (Ganesh et al. 2012; Ganesh et al. 2014) and are suspected to infect bacteria populating the mammalian enteric tract (Krishnamurthy and Wang 2018). Also, infection with human cytomegalovirus can suppress superinfection with HIV-1 (King et al. 2006); and infection with hepatitis A can also suppress hepatitis C virus infection (Deterding et al. 2006). Similarly, hepatitis C can suppress hepatitis B virus replication (Murai et al. 2020). Human papillomaviruses (HPVs) often cause warts which are cleared by the immune system. But a small fraction of the individuals infected with HPVs (HPV-16 and HPV-18) develop cervical cancer. High-risk HPVs, despite being carcinogenic, are also considered to be a symbiont, as the immunity produced against these commensal papillomaviruses offers protection against skin cancers (Strickley et al. 2019).
The human intestine offers a conducive environment for both bacteria and viruses to thrive and this site of residence provides room for cross-talk between bacteria and viruses. The intestine nurtures both symbionts and commensals; however, their biodiversity diversifies during the various stages of development and aging. It also varies along the length of the gut and is influenced by the diet and several other environmental factors. Further, any immunodeficiencies and other host genetic factors affecting the immune regulation in the intestine can tip the balance towards pathological conditions in the intestine (Cadwell et al. 2010; Handley et al. 2016). The bulk of the virome in the intestine consists of bacteriophages followed by DNA viruses such as anelloviruses and herpesvirus and other endogenous retroviruses. However, the benefits and detrimental effects of the resident enteric viruses in healthy individuals and in the diseased need further investigation.
A plethora of diseases has been associated with the dysbiosis in the human gut microbiome (Carding et al. 2015); however, studies that specifically link the gut virome with human diseases have only emerged recently. Inflammatory bowel disease (IBD) is a group of conditions that are characterized by chronic inflammation of the gut. As inflammation is directly related to a heightened immune response of the host, its effect should reflect on the composition of the gut virome in chronic cases of IBD. As expected, alteration in the gut viromes was observed in multiple cohorts of IBD. Patients with IBD had more rich and diverse taxa of bacteriophages, particularly of the order Caudovirales as compared to the controls which were usually accompanied by a significant reduction in the overall bacterial diversity in the IBD fecal microbiome (Norman et al. 2015). Whether the reduction in the diversity of bacterial species in the IBD patients is a causal effect of the increase or abundance of the bacteriophages is unknown. A significant increase in the relative levels of bacteriophages was also observed in patients with type 2 diabetes. Members of the family Podoviridae, Myoviridae, Siphoviridae, and yet unclassified families from the order Caudovirales were significantly abundant in type 2 diabetes when compared to the control group (Ma et al. 2018). Interestingly, a reverse trend was observed in the intestinal virome of type 1 diabetes patients, wherein the overall virome and bacteriophage diversity was less in the patients when compared to the matched control groups. The change in the virome was also associated with the development of autoimmunity which is typically observed in the case with type 1 diabetes (Zhao et al. 2017). The stratification of the gut virome has also been used as a biomarker in the case of hypertension. Han et al. identified a group of 11 and 8 viruses that can act as biomarkers to discriminate the cases with hypertension from the normal control groups respectively. Stratification of hypertension and prehypertension group from the control groups based on the gut virome composition was validated to be superior and more accurate than the gut bacteriome (Han et al. 2018). Dysbiosis of the enteric virome has been reported in colorectal cancer (CRC), wherein the metagenomic samples from CRC patients were associated with significant increase in the diversity of gut bacteriophages primarily exhibiting temperate lifestyle. Overall 22 viral taxa were identified which could be used as biomarkers to discriminate CRC cases from the controls. In this study, it was also suggested that a subgroup of 4 taxonomic markers was found to be strongly associated with poor prognosis and survival outcomes in colorectal cancer (Hannigan et al. 2018; Nakatsu et al. 2018). Although none of the studies linked or confirmed the cause of the condition/disease to the alteration of the gut virome, these incidental studies can be used to predict the status or progression of the disease for better disease management in the future.
6 Translational Prospects of Virome Research
Metagenomic analysis and next-generation sequencing studies have increased the knowledge about human-associated viruses and many studies on the characterization of these viruses revealed their association with several disease phenotypes. For example, a metagenomic analysis in a patient with a respiratory tract infection revealed the presence of gamma papillomavirus; however, it was demonstrated to be unrelated to the disease (Canuti et al. 2014). Large data may or may not provide real-time evidence of the virus populations in a region, and the beneficial or pathogenic nature of viruses thus identified especially in terms of disease may be correlative.
Identification and characterization of novel viruses in niches of microbiota will help us to determine their role in the host and this information can further be used to develop new strategies for disease prevention and therapies. As mentioned earlier the virome as a whole or viral diversity between individuals can be unique. Thus personalized treatment strategies that takes into consideration the virome of an individual can help target the disease effectively and thereby aid in improving the health status of an individual.
It is well documented that the bacteriome of an individual has a direct effect on the abundance of the phage community, and vice versa. Introduction of bacteria derived from the human gut into germ-free mice followed by inoculation of virus-like particles enriched from human feces showed a concomitant decrease of the bacterial population in the host and the levels of bacteria could stabilize only when the phage abundance decreased (Reyes et al. 2013). Phage therapy is widely used to eliminate multi-drug-resistant bacterial population as an alternative to antibiotics (Morozova et al. 2018). One more exciting aspect of phage therapy is that genetically modified phages can control nutrient biosynthesis and degradation especially in obese and dysmetabolic patients (Scarpellini et al. 2015).
The gut microbiome in its entirety is thought to function as an “organ” which coordinates with other bodily functions and has a potentially beneficial effect on human health. Dysbiosis of the gut microbiome composition and function are linked to many pathophysiological conditions and restoration of the same through fecal microbiota transplantation (FMT) has surfaced as a highly effective alternative treatment for patients with recurrent Clostridioides difficile infections (rCDI) and pediatric ulcerative colitis (Fujimoto et al. 2021; Broecker et al. 2016; Nusbaum et al. 2018). The success of the FMT is positively correlated with restructuring of the recipient’s gut bacterial community to levels that are either identical or resemble closely that of the healthy donor (Nusbaum et al. 2018; Fujimoto et al. 2021). Emerging evidence suggests the possible role of the inherent viral community in the outcome of the FMT treatment. An alternative approach to FMT is the treatment with sterile fecal filtrate (residual after removing the bacterial counterparts by a series of centrifugation and filtration steps), which was sufficient to restore normal bowel movements and eliminated all symptoms in patients with rCDI. Interestingly, a subject in the study who was initially treated with FMT with limited to no success, upon treatment with sterile fecal filtrate obtained from the same donor showed better recovery and resolution of the symptoms (Ott et al. 2017). Since serious adverse events including deaths have been reported in FMT (reviewed in Wang et al. 2016a, b), fecal filtrate transfer and thereby the viral assemblage could be a viable alternative to FMT for the treatment of patients with rCDI, particularly in the case of immune-compromised individuals.
The effects that viruses have on human health are highly dependent on their anatomical location inside the host and the interaction with other cells and microbes. All these factors have a direct influence on whether the virus has an advantageous, deleterious, or neutral impact on the host. Studies have shown that human pegivirus (HPgV) infection resulted in the survival of individuals infected with the human immunodeficiency virus (HIV). Greenhalgh et al. showed that HPgV vaccination resulted in reducing morbidity and mortality associated with HIV/AIDS (Greenhalgh et al. 2019). All these studies highlight that there is a pressing need for understanding the interactions of the virome with the host cells and other microbes. This will open up new avenues in targeting disease and improving health.
7 The Human Virome: The Extremes
Viral pathogens are innumerable and diverse in nature and are known to cause potential infections in humans which involve one or many sites. Local transmission results in virus replication and dissemination to distal sites by hematogenous route and results in viremia. Some viruses tap peripheral nerve endings to traverse across and gain access to the CNS. Integration of viral genes in the host genome may be a property of the virus but is also one of the features adopted by the viruses to evade the host immune system and to maintain a steady pool of viral genes which can later be used during virus reactivation. Enrichment of a particular group of virus in the virome has been implicated in disease progression or severity. Such a phenomenon is well documented in Crohn’s disease and ulcerative colitis. Enhanced levels of Caudovirales, Hepadnaviridae, and Hepeviridae were found to be associated with Crohn’s disease or ulcerative colitis or both (Norman et al. 2015; Fernandes et al. 2019; Ungaro et al. 2019; Zuo et al. 2019). A shift in the ratio of Microviridae to Caudovirales has been implicated in the early-onset of inflammatory bowel disease (Liang et al. 2020a). Similarly, a shift in the population dynamics of virulent and temperate phages was observed in Crohn’s disease patients (Clooney et al. 2019). Thus from the available data it is clear that unprecedented increase in specific families of viruses can contribute to disease induction or exacerbation. Besides Crohn’s disease, an increase in enterovirus has been attributed to coeliac disease autoimmunity, while a prevalence of Picobarnoviruses and Tobamoviruses has been reported in pregnant women with type 1 diabetes (Lindfors et al. 2020; Wook Kim et al. 2019). Similarly, the presence of certain phages (Erwinia phage ɸEaH2, Lactococcus phage 1706) in the gut contributed to the development of hypertension (Monaco et al. 2016; Han et al. 2018).
Although outside the context of this chapter, it should be pointed out that viruses are also known to exploit the human microbiome to productively infect the host and also in many instances to evade the host immune system. Bacteria or bacterial components are exploited by many members of the Picornaviridae family including polio virus, coxsackievirus A21, coxsackievirus B5, and echovirus 30 to gain stability in their hostile environment to infect the host (Kuss et al. 2011; Waldman et al. 2017). Enhancement in viral infectivity utilizing the host bacteria has also been reported in the case of rotavirus and reovirus (Uchiyama et al. 2014; Berger et al. 2017). Poliovirus has been shown to exploit the lipopolysaccharide (LPS) and peptidoglycan of the resident microbiome to replicate efficiently in the host (Robinson et al. 2014; Erickson et al. 2018). Such bacterial-virus interaction has been found to enhance poliovirus’s receptor interaction predominantly mediated by LPS and promote poliovirus co-infection (Robinson et al. 2014; Erickson et al. 2018; Kuss et al. 2011). It is therefore quite evident that viruses can utilize the microbiome for their advantage but in some instances they are also known to disrupt the bacterial flora. A comparative analysis of microbiome from children with astrovirus, norovirus, rotavirus, and adenovirus infection showed that a substantial decline in Bifidobacterium and the overall microbiome diversity in the astrovirus-infected cases (Ma et al. 2011).
Reports on the beneficial role of the virome to the host are relatively limited in number, but accumulating evidences from both clinical and animal studies are quite compelling. At least two reports highlight the importance of virome diversity in limiting disease. An overall reduction in viral diversity was identified as one of the contributing factors in type 1 diabetes and acute malnutrition (Zhao et al. 2017; Terho et al. 1983). Especially in the gut which is known to host phages, there exists a constant tussle between the phages and resident bacteria. As described earlier, a shift in the phage property from virulent to temperate supported Crohn’s disease (Clooney et al. 2019). Such a disparity in the virus-bacteria interaction also resulted in exacerbation of ulcerative colitis and growth stunting in children (Khan Mirzaei et al. 2020; Desai et al. 2020). Thus it is clear that the resident phages are important for restricting pathogenic bacteria and supporting the host. Experimental studies on gnotobiotic mice demonstrated that the effects of the lack in bacteria are compensated by murine norovirus that had chronically infected these mice (Kernbauer et al. 2014). Type III interferon induction in the intestine by murine astrovirus could effectively keep the enteric norovirus at bay (Ingle et al. 2019). Thus it can be confidently asserted that the resident human virome can have a positive effect on the overall health status of the host.
8 Conclusions and Future Perspectives
Microbes predominate the host cells in number and were often considered to be associated with human diseases. Technological advancements in the field of genomics have helped us foray into unchartered territories like the human microbiome in which significant progress has been made. It now emerges that the microbiome is an integral part of the host, albeit in a personalized manner. Studies suggest that the role of the human microbiome is multifaceted, playing a major role in the overall physiology. With the emergence of the virus assemblage as a part of the microbiome family, many questions have come to the forefront. Some of the viruses are known to spill over into humans from other species or are transmitted between humans and are either cleared by the host or can have devastating effects on the host. Yet others are known to integrate into the host genome and remain dormant until ambient conditions arise. So the big question is what constitutes the “virus assemblage”? It is now better understood that many viruses reside in the host at specific sites and their relative abundance is dependent on multiple factors. This is quite evident in the case of the neonates where the mother plays an important role in the initial virome enrichment. The biggest conundrum in the field of virome is the source of the viruses in the assemblage as to whether they are resident or are contaminants. The inability to recover viruses, to culture and characterize them, the non-existence of reliable molecular markers and sample contamination, all compound to the problem, adding to the concept of “viral dark matter”. However, of much promise is the evidence, that like the bacterial entities of the microbiome, the virome also plays a pivotal role in contributing to normal host functions and in some cases also adversely affecting the host, resulting in disease conditions. All this points to the fact that the field is in its infancy and serious investigations including setting up of standardization parameters are required to repeal the controversies associated with this field. The cross-talk between organ systems which is highlighted as axes, including the gut-liver axis, gut-brain axis, etc., are noted check points in maintaining normal physiology and homeostasis which is well appreciated. The role of the microbiome in limiting pathogenic bacteria and thereby alleviating major disorders like liver cirrhosis is well appreciated. To what extent the liver virome is involved in these axes remains under-studied and requires in-depth investigation.
The relevance of the resident microbiome in the overall health of an individual is gaining wider recognition. Supplementation of pro-biotics during antibiotic administration and managing of certain metabolic and immune disorders using FMT are emerging as established clinical practices. Clinically, the contribution of virome to the well-being of an individual remains under-appreciated partly due to the limited studies in this area. Alongside the diet and other factors, the existence of cross-talk between the virome and bacteriome is gaining more and more traction. Evidences point to a role of certain viral populations in aggravating disease while others abrogate the symptoms as in the case of Crohn’s disease. Viral pathogens like HIV and H1N1, post-infection generate conditions conducive for secondary infections. One school of thought is the potent immune response initiated by such pathogens can mediate depletion of beneficial bacteria and viruses which indirectly facilitates the survival and spread of the pathogens. More recently, gut bacteriome dysbiosis due to SARS-CoV-2 infection has been attributed as a contributing factor for disease severity and poor recovery. A similar scenario is possible with anti-viral administration, which can deplete the healthy virome besides targeting the pathogen; however, detailed and systematic studies are required to gain further knowledge in this area. Thus the field of virome is riddled with exciting mysteries that require careful unraveling which may have significant implications in understanding the virus niches in the human body and their interaction with the host.
References
Abbas AA, Diamond JM, Chehoud C, Chang B, Kotzin JJ, Young JC et al (2017) The perioperative lung transplant Virome: torque Teno viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant 17(5):1313–1324. https://doi.org/10.1111/ajt.14076
Abbas AA, Taylor LJ, Dothard MI, Leiby JS, Fitzgerald AS, Khatib LA et al (2019a) Redondoviridae, a family of small, circular DNA viruses of the human Oro-respiratory tract associated with periodontitis and critical illness. Cell Host Microbe 25(5):719–729. e714. https://doi.org/10.1016/j.chom.2019.04.001
Abbas AA, Young JC, Clarke EL, Diamond JM, Imai I, Haas AR et al (2019b) Bidirectional transfer of Anelloviridae lineages between graft and host during lung transplantation. Am J Transplant 19(4):1086–1097. https://doi.org/10.1111/ajt.15116
Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK et al (2014) Human oral viruses are personal, persistent and gender-consistent. ISME J 8(9):1753–1767. https://doi.org/10.1038/ismej.2014.31
Albrecht M, Arck PC (2020) Vertically transferred immunity in neonates: mothers, mechanisms and mediators. Front Immunol 11:555. https://doi.org/10.3389/fimmu.2020.00555
Arora N, Sadovsky Y, Dermody TS, Coyne CB (2017) Microbial vertical transmission during human pregnancy. Cell Host Microbe 21(5):561–567. https://doi.org/10.1016/j.chom.2017.04.007
Baker JL, Bor B, Agnello M, Shi W, He X (2017) Ecology of the Oral microbiome: beyond bacteria. Trends Microbiol 25(5):362–374. https://doi.org/10.1016/j.tim.2016.12.012
Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M et al (2015) Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347(6219):266–269. https://doi.org/10.1126/science.1258025
Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS et al (2007) Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447(7142):326–329. https://doi.org/10.1038/nature05762
Belkaid Y, Tamoutounour S (2016) The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol 16(6):353–366. https://doi.org/10.1038/nri.2016.48
Berger AK, Yi H, Kearns DB, Mainou BA (2017) Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog 13(12):e1006768. https://doi.org/10.1371/journal.ppat.1006768
Berlutti F, Pantanella F, Natalizi T, Frioni A, Paesano R, Polimeni A et al (2011) Antiviral properties of lactoferrin–a natural immunity molecule. Molecules 16(8):6992–7018. https://doi.org/10.3390/molecules16086992
Bonvicini F, La Placa M, Manaresi E, Gallinella G, Gentilomi GA, Zerbini M et al (2010) Parvovirus b19 DNA is commonly harboured in human skin. Dermatology 220(2):138–142. https://doi.org/10.1159/000277431
Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D (2013) Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 9(1):e1003057. https://doi.org/10.1371/journal.ppat.1003057
Brassard J, Gagne MJ, Leblanc D, Poitras E, Houde A, Boras VF et al (2015) Association of age and gender with torque Teno virus detection in stools from diarrheic and non-diarrheic people. J Clin Virol 72:55–59. https://doi.org/10.1016/j.jcv.2015.08.020
Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B et al (2008) Viral diversity and dynamics in an infant gut. Res Microbiol 159(5):367–373. https://doi.org/10.1016/j.resmic.2008.04.006
Breitbart M, Rohwer F (2005) Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. BioTechniques 39(5):729–736. https://doi.org/10.2144/000112019
Broecker F, Klumpp J, Schuppler M, Russo G, Biedermann L, Hombach M et al (2016) Long-term changes of bacterial and viral compositions in the intestine of a recovered Clostridium difficile patient after fecal microbiota transplantation. Cold Spring Harb Mol Case Stud 2(1):a000448. https://doi.org/10.1101/mcs.a000448
Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE et al (2010) Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141(7):1135–1145. https://doi.org/10.1016/j.cell.2010.05.009
Canuti M, Deijs M, Jazaeri Farsani SM, Holwerda M, Jebbink MF, de Vries M et al (2014) Metagenomic analysis of a sample from a patient with respiratory tract infection reveals the presence of a gamma-papillomavirus. Front Microbiol 5:347. https://doi.org/10.3389/fmicb.2014.00347
Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ (2015) Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 26:26191. https://doi.org/10.3402/mehd.v26.26191
Clarke EL, Connell AJ, Six E, Kadry NA, Abbas AA, Hwang Y et al (2018a) T cell dynamics and response of the microbiota after gene therapy to treat X-linked severe combined immunodeficiency. Genome Med 10(1):70. https://doi.org/10.1186/s13073-018-0580-z
Clarke EL, Lauder AP, Hofstaedter CE, Hwang Y, Fitzgerald AS, Imai I et al (2018b) Microbial lineages in sarcoidosis. A metagenomic analysis tailored for low-microbial content samples. Am J Respir Crit Care Med 197(2):225–234. https://doi.org/10.1164/rccm.201705-0891OC
Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O'Regan O et al (2019) Whole-Virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26(6):764–778 e765. https://doi.org/10.1016/j.chom.2019.10.009
Colson P, Fancello L, Gimenez G, Armougom F, Desnues C, Fournous G et al (2013) Evidence of the megavirome in humans. J Clin Virol 57(3):191–200. https://doi.org/10.1016/j.jcv.2013.03.018
Desai C, Handley SA, Rodgers R, Rodriguez C, Ordiz MI, Manary MJ et al (2020) Growth velocity in children with environmental enteric dysfunction is associated with specific bacterial and viral taxa of the gastrointestinal tract in Malawian children. PLoS Negl Trop Dis 14(6):e0008387. https://doi.org/10.1371/journal.pntd.0008387
Deterding K, Tegtmeyer B, Cornberg M, Hadem J, Potthoff A, Boker KH et al (2006) Hepatitis a virus infection suppresses hepatitis C virus replication and may lead to clearance of HCV. J Hepatol 45(6):770–778. https://doi.org/10.1016/j.jhep.2006.07.023
Doan T, Akileswaran L, Andersen D, Johnson B, Ko N, Shrestha A et al (2016) Paucibacterial microbiome and resident DNA Virome of the healthy conjunctiva. Invest Ophthalmol Vis Sci 57(13):5116–5126. https://doi.org/10.1167/iovs.16-19803
Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK (2018) Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23(1):77–88 e75. https://doi.org/10.1016/j.chom.2017.11.007
Fernandes MA, Verstraete SG, Phan TG, Deng X, Stekol E, LaMere B et al (2019) Enteric Virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J Pediatr Gastroenterol Nutr 68(1):30–36. https://doi.org/10.1097/MPG.0000000000002140
Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA et al (2012) Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7(6):e38499. https://doi.org/10.1371/journal.pone.0038499
Fujimoto K, Kimura Y, Allegretti JR, Yamamoto M, Zhang YZ, Katayama K et al (2021) Functional restoration of Bacteriomes and Viromes by fecal microbiota transplantation. Gastroenterology 160(6):2089–2102 e2012. https://doi.org/10.1053/j.gastro.2021.02.013
Ganesh B, Banyai K, Martella V, Jakab F, Masachessi G, Kobayashi N (2012) Picobirnavirus infections: viral persistence and zoonotic potential. Rev Med Virol 22(4):245–256. https://doi.org/10.1002/rmv.1707
Ganesh B, Masachessi G, Mladenova Z (2014) Animal picobirnavirus. Virusdisease 25(2):223–238. https://doi.org/10.1007/s13337-014-0207-y
Garretto A, Miller-Ensminger T, Wolfe AJ, Putonti C (2019) Bacteriophages of the lower urinary tract. Nat Rev Urol 16(7):422–432. https://doi.org/10.1038/s41585-019-0192-4
Ghose C, Ly M, Schwanemann LK, Shin JH, Atab K, Barr JJ et al (2019) The Virome of cerebrospinal fluid: viruses where we once thought there were none. Front Microbiol 10:2061. https://doi.org/10.3389/fmicb.2019.02061
Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C et al (2016) Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19(5):731–743. https://doi.org/10.1016/j.chom.2016.04.017
Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R et al (2014) Human genetics shape the gut microbiome. Cell 159(4):789–799. https://doi.org/10.1016/j.cell.2014.09.053
Grandi N, Tramontano E (2018) Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9:2039. https://doi.org/10.3389/fimmu.2018.02039
Greenhalgh S, Schmidt R, Day T (2019) Fighting the public health burden of AIDS with the human pegivirus. Am J Epidemiol 188(9):1586–1594. https://doi.org/10.1093/aje/kwz139
Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB (2020) The gut Virome database reveals age-dependent patterns of Virome diversity in the human gut. Cell Host Microbe 28(5):724–740 e728. https://doi.org/10.1016/j.chom.2020.08.003
Han M, Yang P, Zhong C, Ning K (2018) The human gut Virome in hypertension. Front Microbiol 9:3150. https://doi.org/10.3389/fmicb.2018.03150
Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC et al (2016) SIV infection-mediated changes in gastrointestinal bacterial microbiome and Virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19(3):323–335. https://doi.org/10.1016/j.chom.2016.02.010
Hannigan GD, Duhaime MB, MTT R, Koumpouras CC, Schloss PD (2018) Diagnostic potential and interactive dynamics of the colorectal cancer Virome. mBio 9(6). https://doi.org/10.1128/mBio.02248-18
Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ et al (2015) The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 6(5):e01578–e01515. https://doi.org/10.1128/mBio.01578-15
Hannigan GD, Zheng Q, Meisel JS, Minot SS, Bushman FD, Grice EA (2017) Evolutionary and functional implications of hypervariable loci within the skin virome. PeerJ 5:e2959. https://doi.org/10.7717/peerj.2959
Hartlage AS, Cullen JM, Kapoor A (2016) The strange, expanding world of animal Hepaciviruses. Annu Rev Virol 3(1):53–75. https://doi.org/10.1146/annurev-virology-100114-055104
Hashida Y, Nakajima K, Nakajima H, Shiga T, Tanaka M, Murakami M et al (2016) High load of Merkel cell polyomavirus DNA detected in the normal skin of Japanese patients with Merkel cell carcinoma. J Clin Virol 82:101–107. https://doi.org/10.1016/j.jcv.2016.07.011
Holtz LR, Cao S, Zhao G, Bauer IK, Denno DM, Klein EJ et al (2014) Geographic variation in the eukaryotic virome of human diarrhea. Virology 468-470:556–564. https://doi.org/10.1016/j.virol.2014.09.012
Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, Zhao G et al (2019) Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat Microbiol 4(7):1120–1128. https://doi.org/10.1038/s41564-019-0416-7
Jakobsen RR, Haahr T, Humaidan P, Jensen JS, Kot WP, Castro-Mejia JL et al (2020) Characterization of the vaginal DNA Virome in health and Dysbiosis. Viruses 12(10). https://doi.org/10.3390/v12101143
Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR et al (2014) Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346(6210):755–759. https://doi.org/10.1126/science.1257147
Kaczorowska J, van der Hoek L (2020) Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiol Rev 44(3):305–313. https://doi.org/10.1093/femsre/fuaa007
Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV et al (2011) Successful transmission of a retrovirus depends on the commensal microbiota. Science 334(6053):245–249. https://doi.org/10.1126/science.1210718
Kernbauer E, Ding Y, Cadwell K (2014) An enteric virus can replace the beneficial function of commensal bacteria. Nature 516(7529):94–98. https://doi.org/10.1038/nature13960
Khan Mirzaei M, Khan MAA, Ghosh P, Taranu ZE, Taguer M, Ru J et al (2020) Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27(2):199–212 e195. https://doi.org/10.1016/j.chom.2020.01.004
King CA, Baillie J, Sinclair JH (2006) Human cytomegalovirus modulation of CCR5 expression on myeloid cells affects susceptibility to human immunodeficiency virus type 1 infection. J Gen Virol 87(Pt 8):2171–2180. https://doi.org/10.1099/vir.0.81452-0
Krishnamurthy SR, Wang D (2018) Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology 516:108–114. https://doi.org/10.1016/j.virol.2018.01.006
Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV et al (2011) Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334(6053):249–252. https://doi.org/10.1126/science.1211057
Landini MM, Borgogna C, Peretti A, Doorbar J, Griffin H, Mignone F et al (2015) Identification of the skin virome in a boy with widespread human papillomavirus-2-positive warts that completely regressed after administration of tetravalent human papillomavirus vaccine. Br J Dermatol 173(2):597–600. https://doi.org/10.1111/bjd.13707
Lang J, Yang N, Deng J, Liu K, Yang P, Zhang G et al (2011) Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One 6(8):e23710. https://doi.org/10.1371/journal.pone.0023710
Lee AY, Akileswaran L, Tibbetts MD, Garg SJ, Van Gelder RN (2015) Identification of torque Teno virus in culture-negative endophthalmitis by representational deep DNA sequencing. Ophthalmology 122(3):524–530. https://doi.org/10.1016/j.ophtha.2014.09.001
Lee S, Park Y, Kim SG, Ko EJ, Chung BH, Yang CW (2020) The impact of cytomegalovirus infection on clinical severity and outcomes in kidney transplant recipients with pneumocystis jirovecii pneumonia. Microbiol Immunol 64(5):356–365. https://doi.org/10.1111/1348-0421.12778
Leeper C, Lutzkanin A 3rd (2018) Infections during pregnancy. Prim Care 45(3):567–586. https://doi.org/10.1016/j.pop.2018.05.013
Li Y, Altan E, Pilcher C, Hartogensis W, Hecht FM, Deng X et al (2020) Semen virome of men with HIV on or off antiretroviral treatment. AIDS 34(6):827–832. https://doi.org/10.1097/QAD.0000000000002497
Liang G, Conrad MA, Kelsen JR, Kessler LR, Breton J, Albenberg LG et al (2020a) Dynamics of the stool Virome in very early-onset inflammatory bowel disease. J Crohns Colitis 14(11):1600–1610. https://doi.org/10.1093/ecco-jcc/jjaa094
Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, Bittinger K et al (2020b) The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581(7809):470–474. https://doi.org/10.1038/s41586-020-2192-1
Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM et al (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21(10):1228–1234. https://doi.org/10.1038/nm.3950
Lindfors K, Lin J, Lee HS, Hyoty H, Nykter M, Kurppa K et al (2020) Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: the TEDDY study. Gut 69(8):1416–1422. https://doi.org/10.1136/gutjnl-2019-319809
Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS et al (2018) Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell 175(2):347–359 e314. https://doi.org/10.1016/j.cell.2018.08.016
Lu LJ, Liu J (2016) Human microbiota and ophthalmic disease. Yale J Biol Med 89(3):325–330
Ma C, Wu X, Nawaz M, Li J, Yu P, Moore JE et al (2011) Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr Microbiol 63(3):259–266. https://doi.org/10.1007/s00284-011-9972-7
Ma Y, You X, Mai G, Tokuyasu T, Liu C (2018) A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6(1):24. https://doi.org/10.1186/s40168-018-0410-y
McCann A, Ryan FJ, Stockdale SR, Dalmasso M, Blake T, Ryan CA et al (2018) Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ 6:e4694. https://doi.org/10.7717/peerj.4694
McCullers JA, Rehg JE (2002) Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 186(3):341–350. https://doi.org/10.1086/341462
Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD (2013) Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110(30):12450–12455. https://doi.org/10.1073/pnas.1300833110
Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD et al (2011) The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21(10):1616–1625. https://doi.org/10.1101/gr.122705.111
Mohandas S, Pannaraj PS (2020) Beyond the bacterial microbiome: virome of human Milk and effects on the developing infant. Nestle Nutr Inst Workshop Ser 94:86–93. https://doi.org/10.1159/000504997
Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES et al (2016) Altered Virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19(3):311–322. https://doi.org/10.1016/j.chom.2016.02.011
Morozova VV, Vlassov VV, Tikunova NV (2018) Applications of bacteriophages in the treatment of localized infections in humans. Front Microbiol 9:1696. https://doi.org/10.3389/fmicb.2018.01696
Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y et al (2017) The blood DNA virome in 8,000 humans. PLoS Pathog 13(3):e1006292. https://doi.org/10.1371/journal.ppat.1006292
Murai K, Hikita H, Kai Y, Kondo Y, Fukuoka M, Fukutomi K et al (2020) Hepatitis C virus infection suppresses hepatitis B virus replication via the RIG-I-like helicase pathway. Sci Rep 10(1):941. https://doi.org/10.1038/s41598-020-57603-9
Naidu M, Robles-Sikisaka R, Abeles SR, Boehm TK, Pride DT (2014) Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol 14:175. https://doi.org/10.1186/1471-2180-14-175
Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z et al (2018) Alterations in enteric Virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155(2):529–541 e525. https://doi.org/10.1053/j.gastro.2018.04.018
Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M et al (2015) Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347(6219):269–273. https://doi.org/10.1126/science.1258100
Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC et al (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160(3):447–460. https://doi.org/10.1016/j.cell.2015.01.002
Nusbaum DJ, Sun F, Ren J, Zhu Z, Ramsy N, Pervolarakis N et al (2018) Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol Ecol 94(9). https://doi.org/10.1093/femsec/fiy133
Ogilvie LA, Jones BV (2015) The human gut virome: a multifaceted majority. Front Microbiol 6:918. https://doi.org/10.3389/fmicb.2015.00918
Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA et al (2017) Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152(4):799–811 e797. https://doi.org/10.1053/j.gastro.2016.11.010
Parker MT (2016) An ecological framework of the human Virome provides classification of current knowledge and identifies areas of forthcoming discovery. Yale J Biol Med 89(3):339–351
Perez-Brocal V, Moya A (2018) The analysis of the oral DNA virome reveals which viruses are widespread and rare among healthy young adults in Valencia (Spain). PLoS One 13(2):e0191867. https://doi.org/10.1371/journal.pone.0191867
Pou C, Nkulikiyimfura D, Henckel E, Olin A, Lakshmikanth T, Mikes J et al (2019) The repertoire of maternal anti-viral antibodies in human newborns. Nat Med 25(4):591–596. https://doi.org/10.1038/s41591-019-0392-8
Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA 3rd et al (2012) Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J 6(5):915–926. https://doi.org/10.1038/ismej.2011.169
Rampelli S, Turroni S, Schnorr SL, Soverini M, Quercia S, Barone M et al (2017) Characterization of the human DNA gut virome across populations with different subsistence strategies and geographical origin. Environ Microbiol 19(11):4728–4735. https://doi.org/10.1111/1462-2920.13938
Rascovan N, Monteil Bouchard S, Grob JJ, Collet-Villette AM, Gaudy-Marqueste C, Penicaud M et al (2016) Human Polyomavirus-6 infecting lymph nodes of a patient with an Angiolymphoid hyperplasia with eosinophilia or Kimura disease. Clin Infect Dis 62(11):1419–1421. https://doi.org/10.1093/cid/ciw135
Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F et al (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466(7304):334–338. https://doi.org/10.1038/nature09199
Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI (2013) Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci U S A 110(50):20236–20241. https://doi.org/10.1073/pnas.1319470110
Robinson CM, Jesudhasan PR, Pfeiffer JK (2014) Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15(1):36–46. https://doi.org/10.1016/j.chom.2013.12.004
Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT (2013) Association between living environment and human oral viral ecology. ISME J 7(9):1710–1724. https://doi.org/10.1038/ismej.2013.63
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D et al (2018) Environment dominates over host genetics in shaping human gut microbiota. Nature 555(7695):210–215. https://doi.org/10.1038/nature25973
Roux S, Hallam SJ, Woyke T, Sullivan MB (2015) Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. elife 4. https://doi.org/10.7554/eLife.08490
Santiago-Rodriguez TM, Ly M, Bonilla N, Pride DT (2015) The human urine virome in association with urinary tract infections. Front Microbiol 6:14. https://doi.org/10.3389/fmicb.2015.00014
Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O et al (2011) Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis 17(8):1364–1370. https://doi.org/10.3201/eid1708.110278
Scarpellini E, Ianiro G, Attili F, Bassanelli C, De Santis A, Gasbarrini A (2015) The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis 47(12):1007–1012. https://doi.org/10.1016/j.dld.2015.07.008
Shehi E, Ghazanfar H, Fortuzi K, Gonzalez E, Zeana C (2020) A rare Case of parvovirus B19 infection manifesting as chronic aplastic anemia and neutropenia in a human immunodeficiency virus-infected patient. Cureus 12(12):e12174. https://doi.org/10.7759/cureus.12174
Shi Z, Gewirtz AT (2018) Together forever: bacterial-viral interactions in infection and immunity. Viruses 10(3). https://doi.org/10.3390/v10030122
Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA et al (2019) The human gut Virome is highly diverse, stable, and individual specific. Cell Host Microbe 26(4):527–541 e525. https://doi.org/10.1016/j.chom.2019.09.009
Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, Ross RP et al (2018) PhiCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat Commun 9(1):4781. https://doi.org/10.1038/s41467-018-07225-7
Simister NE (2003) Placental transport of immunoglobulin G. Vaccine 21(24):3365–3369. https://doi.org/10.1016/s0264-410x(03)00334-7
Singh M, Cai H, Bunse M, Feschotte C, Izsvak Z (2020) Human endogenous retrovirus K rec forms a regulatory loop with MITF that opposes the progression of melanoma to an invasive stage. Viruses 12(11). https://doi.org/10.3390/v12111303
Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P (2011) The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92(Pt 2):233–246. https://doi.org/10.1099/vir.0.027490-0
Strickley JD, Messerschmidt JL, Awad ME, Li T, Hasegawa T, Ha DT et al (2019) Immunity to commensal papillomaviruses protects against skin cancer. Nature 575(7783):519–522. https://doi.org/10.1038/s41586-019-1719-9
Terho EO, Lindstrom P, Mantyjarvi R, Tukiainen H, Wager O (1983) Circulating immune complexes and rheumatoid factors in patients with farmer's lung. Allergy 38(5):347–352. https://doi.org/10.1111/j.1398-9995.1983.tb04129.x
Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC et al (2001) Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 345(10):715–724. https://doi.org/10.1056/NEJMoa010398
Tirosh O, Conlan S, Deming C, Lee-Lin SQ, Huang X, Program NCS et al (2018) Expanded skin virome in DOCK8-deficient patients. Nat Med 24(12):1815–1821. https://doi.org/10.1038/s41591-018-0211-7
Turin CG, Ochoa TJ (2014) The role of maternal breast Milk in preventing infantile diarrhea in the developing world. Curr Trop Med Rep 1(2):97–105. https://doi.org/10.1007/s40475-014-0015-x
Uchiyama R, Chassaing B, Zhang B, Gewirtz AT (2014) Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis 210(2):171–182. https://doi.org/10.1093/infdis/jiu037
Ungaro F, Massimino L, Furfaro F, Rimoldi V, Peyrin-Biroulet L, D'Alessio S et al (2019) Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early-diagnosed inflammatory bowel disease. Gut Microbes 10(2):149–158. https://doi.org/10.1080/19490976.2018.1511664
Vasilyev EV, Trofimov DY, Tonevitsky AG, Ilinsky VV, Korostin DO, Rebrikov DV (2009) Torque Teno virus (TTV) distribution in healthy Russian population. Virol J 6:134. https://doi.org/10.1186/1743-422X-6-134
Virgin HW (2014) The virome in mammalian physiology and disease. Cell 157(1):142–150. https://doi.org/10.1016/j.cell.2014.02.032
Waldman P, Meseguer A, Lucas F, Moulin L, Wurtzer S (2017) Interaction of human enteric viruses with microbial compounds: implication for virus persistence and disinfection treatments. Environ Sci Technol 51(23):13633–13640. https://doi.org/10.1021/acs.est.7b03875
Wang J, Gao Y, Zhao F (2016a) Phage-bacteria interaction network in human oral microbiome. Environ Microbiol 18(7):2143–2158. https://doi.org/10.1111/1462-2920.12923
Wang S, Xu M, Wang W, Cao X, Piao M, Khan S et al (2016b) Systematic review: adverse events of fecal microbiota transplantation. PLoS One 11(8):e0161174. https://doi.org/10.1371/journal.pone.0161174
Weinberger DM, Klugman KP, Steiner CA, Simonsen L, Viboud C (2015) Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med 12(1):e1001776. https://doi.org/10.1371/journal.pmed.1001776
Wen X, Miao L, Deng Y, Bible PW, Hu X, Zou Y et al (2017) The influence of age and sex on ocular surface microbiota in healthy adults. Invest Ophthalmol Vis Sci 58(14):6030–6037. https://doi.org/10.1167/iovs.17-22957
Wicinski M, Sawicka E, Gebalski J, Kubiak K, Malinowski B (2020) Human Milk oligosaccharides: health benefits, potential applications in infant formulas, and pharmacology. Nutrients 12(1). https://doi.org/10.3390/nu12010266
Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J et al (2009) Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4(10):e7370. https://doi.org/10.1371/journal.pone.0007370
Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW et al (2012) Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol 46(2):127–131. https://doi.org/10.1165/rcmb.2011-0253OC
Wook Kim K, Allen DW, Briese T, Couper JJ, Barry SC, Colman PG et al (2019) Distinct gut Virome profile of pregnant women with type 1 diabetes in the ENDIA study. Open forum. Infect Dis 6(2):ofz025. https://doi.org/10.1093/ofid/ofz025
Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA (2012) Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7(6):e27735. https://doi.org/10.1371/journal.pone.0027735
Wylie KM, Mihindukulasuriya KA, Zhou Y, Sodergren E, Storch GA, Weinstock GM (2014) Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol 12:71. https://doi.org/10.1186/s12915-014-0071-7
Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E et al (2015) Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant 15(1):200–209. https://doi.org/10.1111/ajt.13031
Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA et al (2018) Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat Microbiol 3(1):38–46. https://doi.org/10.1038/s41564-017-0053-y
Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL et al (2014) Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346(6211):861–865. https://doi.org/10.1126/science.1256999
Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW et al (2006) RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4(1):e3. https://doi.org/10.1371/journal.pbio.0040003
Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW et al (2017) Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A 114(30):E6166–E6175. https://doi.org/10.1073/pnas.1706359114
Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F et al (2019) Gut mucosal virome alterations in ulcerative colitis. Gut 68(7):1169–1179. https://doi.org/10.1136/gutjnl-2018-318131
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Nag, J., Kumar, N.A., Mukesh, R.K., Kunnakkadan, U., Johnson, J.B. (2022). Virome: Sentinels or Marauders in the Microbiome. In: Thomas, S. (eds) Human Microbiome . Springer, Singapore. https://doi.org/10.1007/978-981-16-7672-7_10
Download citation
DOI: https://doi.org/10.1007/978-981-16-7672-7_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-7671-0
Online ISBN: 978-981-16-7672-7
eBook Packages: MedicineMedicine (R0)