Abstract
Since the advent of molecular genetics, the analysis of polymorphic loci has been used to study diversity between and within species at all levels of biological complexity, including domestic animals. Following technological improvements, different types of molecular markers have been used to characterize population variability, including allozymes, restriction fragment length polymorphisms (RFLPs), microsatellite loci, and single nucleotide polymorphisms. Recently, high-throughput sequencing technologies have made it possible to study molecular diversity at the whole genome level. This chapter summarizes the main aspects of genomic diversity of water buffalo, as revealed by the analysis of different types of molecular markers and with respect to evolutionary history of the two subspecies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 The Water Buffalo Genome
There are two types of Asian water buffalo, the river- and the swamp-type, which are classified into distinct subspecies, Bubalus bubalis bubalis and Bubalus bubalis carabanensis, respectively, which were derived from independent domestication of different ancestral populations of wild Asian buffalo, Bubalus arnee (Cockrill 1981; Kumar et al. 2007a; Yindee et al. 2010; Nagarajan et al. 2015; Wang et al. 2017a; Colli et al. 2018; Sun et al. 2020). Their divergence predated domestication and was likely caused by isolation following the Pleistocenic glaciations (Wang et al. 2017a; Sun et al. 2020). Domestication of river buffaloes occurred in the western Indian subcontinent about 6000 years ago (Kyr BP) (Kumar et al. 2007b; Nagarajan et al. 2015) and in the China-Indo-China border region between 3000 and 7000 years ago for swamp buffaloes (Zhang et al. 2016; Wang et al. 2017a). Following domestication, river buffaloes migrated westwards through southwestern Asia to the Mediterranean basin, while swamp buffaloes spread to south- and north-eastern Asia (Colli et al. 2018; Zhang et al. 2020). There is extensive differentiation between the types in terms of morphology, behavior, and current geographical distribution (Macgregor 1941; Cockrill 1974; Zhang et al. 2020). The two types are genetically differentiated, as revealed by allozymes (Amano et al. 1980; Barker et al. 1997a), microsatellite markers (Barker et al. 1997b; Zhang et al. 2011; Kumar et al. 2006), single-nucleotide polymorphisms (SNPs) (Colli et al. 2018), and whole-genome sequence data (Dutta et al. 2020; Sun et al. 2020). The two types are also differentiated by mitochondrial DNA and Y-chromosome variation (Zhang et al. 2006; Yindee et al. 2010; Zhang et al. 2016; Wang et al. 2017a; Sun et al. 2020).
Both river and swamp buffaloes have five sub-metacentric autosomes, while the remaining autosomes and the sex chromosomes are acrocentric (Iannuzzi and Di Meo 2009). The number of chromosomes (2n) differ between the two types, the river karyotype (2n) being 50 while the swamp karyotype (2n) is 48 (Ulbrich and Fischer 1967; Fischer and Ulbrich 1968). The one-pair difference in chromosome number is the result of a rearrangement creating swamp buffalo chromosome 1 from a tandem fusion translocation between the telomere of river buffalo chromosome BBU4p and the centromere of BBU9 (Fig. 2.1) (Di Berardino and Iannuzzi 1981; Iannuzzi and Di Meo 2009; Luo et al. 2020). Fluorescent in situ hybridization (FISH) and C-banding studies (Di Meo et al. 1995; Tanaka et al. 1999) suggested that this fusion caused the loss of a large portion of heterochromatin and satellite DNA from BBU9, and of the nucleolus organizer region (NOR) present at the telomere of BBU4p (Fig. 2.1). In river buffalo, six nucleolus organizing regions (NOR) are found at the telomeres of chromosomes 3p, 4p, 6, 21, 23, and 24, while only five NORs are found in the swamp buffalo on chromosomes 4p (which corresponds to river buffalo chromosome 3p), 6, 20, 22, and 23 as a result of the tandem fusion translocation (Degrandi et al. 2014). As all chromosome arms pairs are conserved between the two species, river x swamp crosses show 2n = 49 and are fertile, although with a possible reduced fertility due to unbalanced chromosome sorting during meiosis (Iannuzzi and Di Meo 2009).
Tandem fusion translocation between the centromere and the telomere of river buffalo chromosomes 9 and 4p, respectively, originating swamp buffalo chromosome 1. The loss of NORs following the fusion is also indicated. (Modified with BioRender.com from Iannuzzi and Di Meo (2009))
Initial information on the general organization and gene localization on domestic buffalo chromosomes came from the comparison with cattle. Cytogenetic c-banding studies provided the arm-by-arm matching between water buffalo and cattle chromosomes, while Fluorescent In Situ Hybridization (FISH) enabled cattle-derived genes and microsatellite loci to be mapped to water buffalo chromosomes (Amaral et al. 2008; Iannuzzi and Di Meo 2009; Michelizzi et al. 2010). A first-generation whole-genome Radiation Hybrid (RH) map was obtained by mapping 2621 cattle-derived markers on to a river buffalo-hamster panel of hybrid cells (Amaral et al. 2008). This showed that there was good conservation of synteny between cattle and buffalo genomes within linkage groups (Michelizzi et al. 2010).
2 Reference Genome Sequences
A first draft of the domestic buffalo whole-genome sequence Bbu_2.0-alpha was obtained by Illumina paired-end sequencing from a female Murrah river buffalo (Tantia et al. 2011). The genome sequence, which had 17-19X read depth, was assembled by alignment with the bovine genome and was deposited in the NCBI Short Read Archive (SRA) in 2009 under Accession Numbers SRX016621 and SRX015182. The first de novo buffalo genome assembly was created by the International Buffalo Genome Consortium (Williams et al. 2017). A total of 242 Gb raw sequence data derived from Olimpia, a highly homozygous female Mediterranean buffalo, was obtained by combined high-throughput sequencing on the Illumina Genome Analyzer IIx System and Roche 454 FLX titanium platforms, with an approximate 70X genome coverage. This high coverage assembly UMD_CASPUR_WB_2.0 (GenBank Accession Number GCF_000471725.1. Table 2.1) was 2.83 Gb in size, with a 21.94 kb contig N50 and 21,711 protein-coding genes that were annotated based on RNA-seq data from 30 tissues (NCBI SRA project PRJNA207334).
The river buffalo reference genome was further improved using PacBio long read sequence data from the DNA of the same individual used for UMD_CASPUR_WB_2.0 which were combined with Chicago- and Hi-C-based chromatin interaction maps (Cairns et al. 2016) to scaffold a 69X de novo chromosome-level sequence (Low et al. 2019). Indels were corrected using additional Illumina paired-end sequencing. The total length of UOA_WB_1 is 2.66 Gb and contains 509 scaffolds with an N50 of 117.2 Mb (Table 2.1). About 1.53 Gb (58%) of the sequence is haplotype-resolved and has only 383 gaps. The 29 chromosome-level scaffolds are ordered consistently with the buffalo whole-genome radiation hybrid (RH) map and show high conservation of synteny with the homologous chromosomes of Bos taurus genome version UMD3.1 (Fig. 2.2). Annotation of UOA_WB_1 was produced by incorporating information on 3462 buffalo transcripts from GenBank, 1013 buffalo GenBank protein sequences, 50,553 and 13,381 Ref Seq protein from human and cattle, respectively, and RNA sequencing data from more than 50 different buffalo tissue types (Low et al. 2019). This high contiguity assembly constitutes the current domestic buffalo reference genome (Gen Bank Ref Seq GCF_003121395.1), and the annotation can be browsed with the NCBI Genome Data Viewer tool (https://www.ncbi.nlm.nih.gov/genome/gdv/?org=bubalus-bubalis&group=bovidae).
Circular plot of water buffalo chromosomes mapping to B. taurus genome. (From Low et al. (2019))
The trio-binning approach (Koren et al. 2018) in which parental information is used to phase sequence data prior to assembly, has been recently used to create a haplotype-resolved assembly of the Murrah buffalo (Ananthasayanam et al. 2020). PacBio long reads and 10X Genomics linked reads at 166X coverage with an additional 802 Gb of optical mapping data were phased using 274 Gb paired-end data from the parents. Chromosomal level assembly for paternal and maternal genomes with 25 scaffolds and N50 of 117.48 Mb (sire haplotype) and 118.51 Mb (dam haplotype) were achieved; however, this genome is not presently annotated. Recently, a combined PacBio and Hi-C sequencing approaches were used to create the first reference genome sequence for swamp buffalo CUSA_SWP (CNCB-NGDC Accession number GWHAAJZ00000000), estimated to be 2.63 Gb in size (Luo et al. 2020). The chromosome-scale scaffolds of this assembly have an N50 of 117.3 Mb (Table 2.1), representing the 24 swamp buffalo chromosomes and covering 97.5% of the genome.
The availability of high-quality genomic sequences for both river and swamp buffaloes has made it possible to compare the genome arrangement of the two subspecies and to evaluate their relationship in an evolutionary perspective. Analysis of the sequence similarity confirmed that swamp buffalo chromosome 1 arose from the fusion of river buffalo chromosomes 4p and 9. Comparison of the sequences also facilitated the calculation of divergence times based on 6429 single copy orthologs sequences, which showed that the common ancestor of swamp and river buffaloes dates back to 1.1–3.5 million years ago (Luo et al. 2020). This suggests that glacial events known to have occurred at that time may have played a role in separating ancestral buffalo populations on either side of the present-day India-Myanmar border, leading to geographical isolation and genetic divergence. Expanded genes were found in both subspecies, 179 in swamp and 261 in river-type, which may account for the phenotypic differences, particularly in relation to muscle growth and environmental adaptation.
3 Genetic Diversity
The first genetic studies on buffalo were based on variations in protein-coding loci (i.e., allozymes) and microsatellite loci (Barker et al. 1997a, b) in the nuclear genome. Direct sequencing of the control region or of the cytochrome b (Cytb) gene was used to assess maternally inherited mitochondrial DNA (Lau et al. 1998), and sequences from the non-recombining part of Y-chromosome have been used more recently to study the inheritance of the paternal genome (Yindee et al. 2010). High-throughput sequencing and the availability of whole-genome sequence information have made it possible to discover a large number of single-nucleotide polymorphism (SNP) markers and to devise marker panels (Iamartino et al. 2017) for cost-effective genotyping of large numbers of animals to investigate genome diversity and for genome-wide association studies (GWAS).
3.1 Microsatellite Markers
Being highly polymorphic, microsatellite loci that contain Short-Tandem Repeats (STRs) were among the first markers to be widely used for the characterization of the nuclear genomic diversity in both river and swamp buffaloes. Although a panel of 30 microsatellite loci was proposed by ISAG-FAO (FAO 2004), most studies have used different or partially overlapping marker sets. This, together with the difficulties in harmonizing allele scoring of STRs across laboratories, prevented a global scale characterization of water buffalo diversity.
In domestic species, diversity levels are higher close to domestication centers, and decrease as populations move away from it. This pattern of diversity is also seen for both water buffalo subspecies. At the local level, microsatellite-based studies showed that for both buffalo types the highest diversity was found close to the respective likely domestication centers, with a gradual decrease along the dispersal routes. The expected heterozygosity (HE) of river buffalo assessed using STR loci, decreases from HE = 0.71–0.78 for the Murrah, Nili-Ravi, and Kundi group from the Indian subcontinent (Kumar et al. 2006; Vijh et al. 2008) to HE = 0.58–0.68 of the Mediterranean population from Italy (Moioli et al. 2001), which is consistent with the supposed center of domestication in the Indian subcontinent. Similarly, the highest heterozygosity in swamp buffaloes is in Thailand (HE = 0.573), close to the likely domestication center across the China-Indochina border, although overall swamp-type populations display little differentiation from each other all over southeast Asia (Zhang et al. 2011).
A recent meta-analysis made use of model-based clustering of microsatellite marker data from both river and swamp buffaloes (Zhang et al. 2020), which showed a clear difference between the two types and the occurrence of some degree of introgression from the river-type gene pool into swamp buffaloes in Far Southwest China, in Thailand, and to a lesser extent, in the Malaysian peninsula and the Philippines.
3.2 SNP (Single-Nucleotide Polymorphism) Diversity
SNPs represent the most frequent type of mutation occurring genome-wide. Comparison of WGS sequences in mammalian livestock has shown that ca. 30 million SNP loci can be found on average within a species (Alberto et al. 2018; Júnior et al. 2020; Luo et al. 2020). Most SNPs are biallelic, which makes them ideal for the development of array-based genotyping panels and simplifies data analysis.
Cattle SNP arrays have been evaluated for use in buffalo (Michelizzi et al. 2011; Wu et al. 2013; Borquis et al. 2014; Pérez-Pardal et al. 2018). Most loci on the cattle panel can be genotyped successfully in buffalo; however, only a small percentage of cattle SNP are polymorphic in buffalo, e.g., only 926 loci (1.71%) out of the ca. 50K markers included in the Illumina BovineSNP50 Bead Chip and only 16,580 (2.41%) out of the 777K SNP in the Illumina Bovine HD Bead Chip (Michelizzi et al. 2011; Borquis et al. 2014). This shows the low cross-species applicability of SNP arrays, which decreases proportionally to the evolutionary divergence time and that most polymorphisms targeted by SNP panels are generally of recent evolutionary origin.
A buffalo-specific SNP panel including 89,988 SNPs, 5799 probes for quality control and 1784 gender identification probes (Iamartino et al. 2017) is commercially available as the Axiom® Buffalo Genotyping Array (Thermo Fisher). The SNP loci were originally selected to have an even genome-wide distribution using the bovine UMD 3.1 genome to estimate their position but have recently been remapped to the new version of the buffalo reference genome (Low et al. 2019). SNPs were initially identified from whole-genome sequences of Mediterranean, Murrah, Jaffarabadi, and Nili-Ravi buffaloes. As these are all river-type breeds, only ca. 22.74% of the loci are polymorphic in swamp buffaloes (Colli et al. 2018). The 90K panel has also been evaluated in Lowland Anoa (Bubalus depressicornis) and Cape buffalo (Syncerus caffer), resulting in 7652 (7.8%) and 3239 (3.3%) out of 89,988 loci respectively polymorphic in these species (Iamartino et al. 2017).
A multi-species SNP panel has been developed by the European IMAGE consortium to reduce genotyping costs. The IMAGE panel incudes ca. 11K buffalo SNP markers, selected to be evenly distributed genome-wide, to be informative in river and swamp populations by including ancestral polymorphisms, and also to include functional, mtDNA, and Y-chromosome variants (Crooijmans et al. in preparation). Many loci are shared with the Axiom 90K panel to make data integration and meta-analysis possible.
The Axiom Buffalo Genotyping Array has been used to study diversity, linkage disequilibrium, effective population size, genome-wide distribution of runs of homozygosity (ROH), and GWAS have been carried out for milk production, reproduction and disease traits (de Camargo et al. 2015; Colli et al. 2018; Liu et al. 2018; Deng et al. 2019; Du et al. 2019; Guzman et al. 2019). Using genotype data from 20,463 loci that were polymorphic in both river and swamp buffaloes, Colli et al. (2018) evaluated genetic variation, population structure, and gene flow in 30 buffalo populations worldwide. Three distinct gene pools were found, corresponding to individuals of pure river, pure swamp, and admixed river × swamp ancestries. The occurrence of two independent domestication events was confirmed, and several links between populations were identified that were consistent with human migrations, importation, and crossbreeding to improve performance. Association studies have identified candidate genes, that are either trait-specific or with pleiotropic effects, which can be used by breeders to select animals carrying advantageous alleles to improve production traits, particularly milk fat yield and milk protein percentage (Liu et al. 2018). Candidate genes likely to affect milk yield, fat yield, and protein yield have been reported in two genomic regions which respectively harbored the genes MFSD14A, SLC35A3, PALMD and RGS22 and VPS13B (Liu et al. 2018). These regions corresponded to locations on cattle chromosomes BTA3 and BTA14 where QTLs influencing milk performance have been reported in dairy cattle (Harder et al. 2006; Wibowo et al. 2008). The comparison of data from cattle and buffalo suggested that different alleles affect milk traits, for fat production in particular, in the two species (de Camargo et al. 2015; Liu et al. 2018).
Runs of homozygosity (ROHs) are contiguous genomic regions where an individual has inherited the same segment on both chromosomes from a common ancestor (Ceballos et al. 2018). Inbreeding at the genomic level can therefore be estimated from the frequency and length of ROHs. Additionally, ROHs can indicate regions impacted by selective pressure over time. The distribution of ROHs over the genome, their abundance, and length are determined by a number of factors including region-specific recombination rate, GC content, selective pressures, and demographic events (McQuillan et al. 2008; Bosse et al. 2014). The length of the ROH regions is in general inversely proportional to time passed since historic, demographical or selection events, with longer ROHs corresponding to recent inbreeding and shorter ROHs arising from ancient demographic changes as bottlenecks or founder effects (Cardoso et al. 2018). Genomic patterns of runs of homozygosity have been studied in several livestock species to find signatures of natural or human-mediated selection, and to identify advantageous genetic variants that may have become fixed or almost fixed as a consequence (Peripolli et al. 2017). The availability of the Axiom buffalo genotyping array has facilitated studies on the occurrence of ROHs genome-wide in water buffalo populations. Runs of homozygosity have been studied at the local level in Iranian Azeri and Khuzestani, and in Brazilian Murrah river buffaloes (Ghoreishifar et al. 2020; Nascimento et al. 2021), and also in a set of 15 river and 15 swamp buffalo populations worldwide (Macciotta et al. 2021). Particularly long ROHs, up to 10 Mb with mean ROH length per animal of 4.28 ± 1.85 Mb, were found in all Brazilian Murrah individuals, most likely resulting from recent strong selection for milk production traits in this population (Nascimento et al. 2021). Conversely, Iranian breeds showed ROHs about 1 Mb long in all animals, but the total number and total length varied considerably between individuals, suggesting the occurrence of both recent and past inbreeding events which have affected the populations (Ghoreishifar et al. 2020). In both studies, genes and QTLs found within ROH islands bore signatures of selection and were associated with selected traits and functions including body size, muscle and bone development, immune response, milk traits (milk yield, milk fat yield and percentage, milk protein yield and percentage), coat color and pigmentation, reproduction and morphology (Ghoreishifar et al. 2020; Nascimento et al. 2021).
ROH analysis across buffalo types identified >18K homozygous regions overall (Macciotta et al. 2021). Swamp-type buffalo populations possessed more ROHs and had higher genomic inbreeding and number of ROHs per animal. Although differences found between river and swamp populations may have been partly affected by ascertainment bias in the SNP on the array, a convergent signature of selection was found in both river and swamp buffaloes on chromosome 2 where a large ROH island spans genes involved in adaptation to the environment and reproduction (Macciotta et al. 2021).
4 Nuclear Genome Diversity
Huge amounts of whole-genome resequencing data are being produced using high-throughput sequencing platforms. WGS sequence data have been published for water buffaloes from many countries: Bangladesh, Bengal, China, Egypt, India (multiple breeds), Indonesia, Iran, Iraq, Italy, Laos, Myanmar, Nepal, Pakistan, the Philippines, Vietnam, and Thailand (Dutta et al. 2020; El-Khishin et al. 2020; Luo et al. 2020; Mintoo et al. 2020; Sun et al. 2020). Based on 5–10X resequencing data from swamp- and river-type animals from 14 Asian and one European country, ca. 33.5 million SNPs were identified overall for the two subspecies, while 18.7 and 23.7 million SNPs were found to be polymorphic within swamp and river buffalo populations, respectively (Luo et al. 2020). Genetic variation within subspecies has been estimated by calculating nucleotide diversity (θπ), a statistic frequently used in population genetics, which corresponds to the average number of differences per nucleotide site found when the DNA sequences are compared pairwise among all individuals in the sampled set. Overall, swamp buffaloes have lower genetic diversity than river buffalo, with nucleotide diversity values of ca. θπswamp = 0.0018 and θπriver = 0.0027. Among the river-type, the Italian Mediterranean population has a lower-than-average nucleotide diversity, (ca. θπ = 0.002) compared to buffaloes from Pakistan, Iran, and Iraq (ca. θπ = 0.0028) and to Indian breeds (ca. θπ = 0.0025 Luo et al. 2020). This reduction of variability is consistent with the loss of diversity as populations move from their geographic origin and alleles are lost. Gene flow has been seen in the regions of Southeast Asia where the two subspecies hybridize naturally (Luo et al. 2020). Introgression from river buffalo into swamp populations is now used widely to improve productivity, especially in Laos, Myanmar, and the Philippines. Interestingly in Yunnan Province of China, river buffaloes displayed genomic introgression from swamp buffalo (Luo et al. 2020).
Genome-wide analyses have identified hundreds of genes harboring differential selection signatures in swamp and river buffaloes (Dutta et al. 2020; Luo et al. 2020), with a set of 67 genes under selection that are common to both groups (Luo et al. 2020). Pathway and gene ontology analyses of genes under selection in the swamp buffalo identified a significant enrichment in muscle- and cardiac-related functions, nerve development, and behavior, which may account for physical strength and endurance, and docile temperament (Luo et al. 2020). Selective sweeps have also been found in two starch-digestion enzymes genes, which suggests a possible adaptation to reduce rumen acidosis induced by the starch-rich feed traditionally supplied to swamp buffaloes used for traction in several Asian countries (Luo et al. 2020). In the case of river buffaloes, selection signatures have been reported in genes related to body size, fecundity, fetal growth, birth size, and milk production. Selective sweeps common to both subspecies suggest the occurrence of convergent evolution affecting body size, immune response to pathogens, and behavioral changes related to the “domestication syndrome” (Dutta et al. 2020; Luo et al. 2020).
5 Functional Variation
Putative sequence variants affecting a range of phenotypes have been proposed by screening the candidate genes in the water buffalo genome. This strategy identified variations related to coat color, milk production, reproductive performance, and diseases, although the validation of these associations is difficult.
In swamp buffaloes, the most frequent coat color is solid dark gray. Solid white animals are not uncommon with a 10% frequency in some populations. White seems to be dominant over the dark variant (Rife and Buranamanas 1959; Rife 1962), but so far variants that control the white coat have not been identified. Swamp buffalo bulls with a white spotted coat, a phenotype found only in animals from Tana Toraja in Indonesia, are highly prized for ceremonial purposes. Two independent loss of function mutations in the microphthalmia-associated transcription factor (MITF) gene are associated with the white spotted coat of swamp buffalo (Yusnizar et al. 2015). Specific allelic variants have been reported that affect milk production and growth traits. Milk protein percentage in Mediterranean buffalo has been associated with a C > T transition in the signal transducer and activator of transcription 5A (STAT5A) gene and a non-synonymous point mutation in the insulin-like growth factor 2 (IGF2) gene which also has an effect on average daily gain (Abo-Al-Ela et al. 2014; Coizet et al. 2018). SNPs affecting milk yield, fat and protein percentage and yield have been identified within the growth hormone receptor (GHR) gene in Egyptian buffaloes, in which animals carrying the AA homozygous genotype at both missense mutations 380G > A and 836T > A displayed higher productive performance (El-Komy et al. 2020).
The identification of genetic variants related to reproduction and fertility is particularly important for the efficient implementation of breeding plans. In male buffaloes from China variations at two genes, luteinizing hormone beta polypeptide (LHB) and gonadotropin-releasing hormone receptor (GnRHR) were recently found to affect semen quality traits, i.e., volume of the ejaculate and quality of the sperm cells (Cheng et al. 2017; Wang et al. 2017b, 2020). In females, pregnancy rate and susceptibility to anestrus are influenced by variants occurring in the melatonin receptor 1A (MTNR1A) gene and the cytochrome P450 aromatase gene (CYP19A1), respectively (El-Bayomi et al. 2017; Pandey et al. 2019). Susceptibility to disease is a trait that potentially has a significant impact on the viability of livestock production. Susceptibility to tuberculosis and mastitis have been associated with specific mutations. In Mediterranean buffaloes, a G > A transition at nucleotide position 4467 in the 3′-UTR (untranslated region) of the interferon gamma (IFNG) gene disrupts the target site for micro-RNA miR-125b, which seems to be responsible for an increased susceptibility to Mycobacterium bovis (Iannaccone et al. 2018). In Egyptian buffaloes, a C > A transversion in exon 27 of the complement component 3 (C3) gene has been shown to have a significant association with milk somatic cell score, which is correlated with mastitis (El-Halawany et al. 2017).
The heritable defect transverse hemimelia (TH) causes abnormal development of the terminal portion of the limbs, which has varying degrees of severity. A case-control study based on whole-genome sequence data suggests that TH is an oligogenic trait and 13 putative candidate genes have been identified. In particular, variants in wingless-type MMTV integration site family, Member 7a (WNT7A) and SWI/SNF Related, Matrix Associated, Actin Dependent Regulator (SMARCA4) genes in the homozygous state are associated with the extreme forms of the phenotype (Whitacre et al. 2017).
6 Uniparental Genomes Diversity
6.1 Mitochondrial Genome
Mitochondrial DNA (mtDNA) is found at high number of copies per cell and is maternally inherited. The mitochondrial genome is haploid and hence does not recombine, but nevertheless has a high mutation rate because of the absence of repair mechanisms. As a result of these features, mtDNA variation has been used to investigate diversity and phylogenies of the maternal lineage in many animal species (Lenstra et al. 2012). The current water buffalo mtDNA reference sequence (Gen Bank Accession Number NC_006295) is from a swamp-type buffalo belonging to the Haikou breed from China. The swamp buffalo mitochondrial genome has a total size of 16,359 bp, and includes the displacement loop (which is non-coding but contains the origin of replication of the H-strand), two genes coding for 12s and 16s ribosomal RNAs (rRNA), 22 genes for transfer RNAs (tRNA), and 13 genes encoding proteins involved in the mitochondrial electron transport chain and ATP synthesis (Fig. 2.3). Partial copies of varying length of the mtDNA genome have been integrated into the nuclear genome in eukaryotic species in the course of their evolution (Richly and Leister 2004). These nuclear-mitochondrial transpositions (NUMTs) have not been investigated in detail in water buffalo; however, a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for sequences occurring in both mtDNA and nuclear genome reference sequences returns 24 hits, with percentages of identity between 74.89–100% and length 31–5346 bp. The longest uninterrupted hit of 6430 bp is found on chromosome X, and covers positions 9991–16,359 of the mtDNA reference. The water buffalo mtDNA control region, or displacement loop (D-loop) is 909 bp long, similar to other buffalo species, e.g., the tamaraw Bubalus mindorensis (922–925 bp) and the lowland anoa B. depressicornis (927 bp), and also cattle, B. taurus (909 bp) and Bos indicus (912 bp), but is shorter than that of pig (1175 bp), sheep (1179 bp), and goat (1212 bp) due to the lack of repetitive motifs.
Water buffalo mitochondrial DNA organization based on the current B. bubalis mtDNA Reference Sequence (Gen Bank Accession Number NC_006295). Protein, tRNA, and rRNA coding genes are grouped and color coded as follows: purple, tRNAs; red, rRNAs; blue, NADH dehydrogenase; dark green, cytochrome oxidase; light green, ATP synthase; orange, cytochrome b. (The picture was created with Genome VX tool (http://wolfe.ucd.ie/GenomeVx/))
The first mtDNA investigations, based on the analysis of D-loop and Cytb sequence data, highlighted a deep divergence between swamp and river buffalo female lineages and the presence of a hotspot of diversity in animals from southeast Asia, which pointed to domestication in this region (Tanaka et al. 1996; Kikkawa et al. 1997; Lau et al. 1998). The analysis of the entire control region sequence clearly suggested different maternal origin and independent domestication of river- and swamp-type buffaloes (Kumar et al. 2007a; Lei et al. 2007). Large-scale screening of control region variation (Zhang et al. 2016) identified five frequently found swamp buffalo haplogroups, SA1, SA2, SB1, SB2 and SB3, and three rare and highly divergent additional haplogroups, SC, SD, and SE (Figs. 2.4 and 2.5) (Sun et al. 2020). In river buffalo, four haplogroups were found, R1, R2, R3, and R4 (Zhang et al. 2016; Sun et al. 2020). Three are spread across the whole distribution area of river buffalo from the Asian continent to the Mediterranean basin (Figs. 2.4 and 2.5). The R1 haplogroup occurs more frequently (75.4%) compared with R2 (16.4%), and R3 is relatively rare (8.2%) (Fig. 2.4) (Kumar et al. 2007a; Zhang et al. 2016). The R4 haplogroup has only been identified in Italian animals so far (Fig. 2.5. Sun et al. 2020). For both river and swamp buffaloes, mtDNA haplogroup frequencies vary slightly across the distribution areas (Figs. 2.4 and 2.5), following the general trend of a progressive loss of diversity moving away from the domestication centers. The occurrence of rare and highly divergent variants points to the repeated post-domestication introgression of wild buffalo lineages into domestic buffalo (Nagarajan et al. 2015; Wang et al. 2017a), which has also been seen in several livestock species (Bonfiglio et al. 2010; Colli et al. 2015).
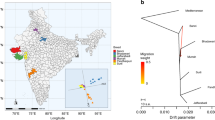
Distribution and frequency of river and swamp buffalo mtDNA haplogroups in different areas of the world based on control region data. The inset shows the phylogenetic relationships between the haplogroups. (From Zhang et al. (2020))
Phylogenetic networks of (a) river and swamp buffalo complete mitochondrial genomes, and (b) Y-chromosome haplotype variation based on 520 SNPs. Gray-shadowed rectangles identify different haplogroups. Edge widths are proportional to the number of mismatches between the joined haplotypes. Geographical provenance of the samples is shown in the top panel. (Modified from Sun et al. (2020))
Using mtDNA data, water buffalo phylogeographic patterns of variation and demographic history have been investigated. MtDNA data show that swamp buffalo populations are strongly partitioned geographically with little gene flow, despite their phenotypic uniformity. In contrast, river buffaloes have a lower haplotypic diversity and a weaker phylogeographic structure (Zhang et al. 2016; Wang et al. 2017a), which may be partly due to the human-mediated effects, including selection for productivity and exchange of animals for the improvement of milk production. Swamp buffaloes in southern China/northern Indochina straddling the Mekong River have the highest diversity, suggesting this region as the likely center of domestication of the swamp type, which is estimated to have occurred ca. 3–7 Kya (Zhang et al. 2016; Wang et al. 2017a). Diversity of river buffaloes is highest in India and gradually decreases westward towards the Mediterranean (Nagarajan et al. 2015), supporting the hypothesis of domestication in the Indian subcontinent. The gradual decline in diversity from the Indian subcontinent suggests that after domestication migration of river buffalo westward towards southwestern Asia and the Mediterranean occurred gradually and without major bottlenecks (Zhang et al. 2016), in agreement with microsatellite- (Moioli et al. 2001; Kumar et al. 2006; Vijh et al. 2008) and SNP-based evidence (Colli et al. 2018).
Whole mitogenomes of swamp buffalo show that Pleistocenic glacial periods, before 11K years ago, played a role in shaping phylogeny and demographic history of the subspecies. Divergence between river- and swamp-type mitochondrial lineages has been estimated to have occurred at the beginning of a glacial period (900–860 Kya). Within swamp buffaloes, the two major macro-lineages diverged at the time of the second glacial event 200–130 Kya (Wang et al. 2017a). Interestingly, most present-day swamp buffalo mitochondrial genome originated from two ancestors dating back to the Last Glacial Maximum 26–19 Kya and further differentiated during the Holocene warm period 11–6 Kya. A substantial bottleneck was observed at 7–3 Kya, corresponding to a reduction in effective population size that occurred at the time of domestication (Wang et al. 2017a).
6.2 Y-Chromosome
The non-recombining part of the Y-chromosome, i.e., the portion lacking a homologous counterpart on the X-chromosome, provides information on the paternal lineage. Low variability of the Y-chromosome has made the identification of Y-chromosome polymorphisms difficult in most livestock species (Hellborg and Ellegren 2004). As a consequence, Y-chromosome variation has been analyzed by sequencing short gene fragments or by typing a few microsatellite loci (Zhang et al. 2006, 2016; Yindee et al. 2010; Wang et al. 2018). Microsatellite-based studies of the buffalo Y-chromosome diversity initially exploited cattle STR loci with limited success. Out of 40 cattle loci tested in Asian swamp buffaloes, only 12 could be satisfactorily amplified and only seven were polymorphic (Wang et al. 2018). Together they described nine haplotypes and four haplogroups, i.e., Y1, Y2, Y3, and Y4. Haplogroup Y1 occurred more frequently (83.4%) across southeastern Asia, while Y2 and Y3 had discontinuous distributions and varying frequencies. The high frequency of Y1 around the Yangtze Valley suggested this as the domestication center. Interestingly, the rare and highly divergent Y4 was found only on Hainan Island, and was interpreted as introgression from an ancient wild Asian buffalo population native to the island (Wang et al. 2018).
Studies of the Y-chromosomes genes of water buffalo have produced sequence data from specific genes, e.g., DEAD-Box Helicase 3 (DBY), Y-linked Zinc finger Y-chromosomal protein (ZFY), and Sex-determining Region Y (SRY) gene segments (Zhang et al. 2006; Yindee et al. 2010; Zhang et al. 2016). Sequencing of SRY identified a single SNP difference between river and swamp buffalo at position 202 of the coding region. PCR genotyping of this SNP was used to reveal male-mediated introgression of river-type buffalo into swamp buffalo in Southern China (Zhang et al. 2006). A 2310-bp Y-chromosome fragment was sequenced in 495 males (450 swamp and 45 river) from 35 populations spanning most of southeastern and central Asia (Zhang et al. 2016). Among swamp buffaloes sequence analysis identified only nine SNPs originating 11 haplotypes, of which four were frequent and seven rare. Only one SNP defining two haplotypes was found in river buffalo (Zhang et al. 2016). Nevertheless, the geographical distribution of the haplogroups and the values of haplotype diversity in swamp buffaloes corroborated the hypothesis of domestication around the border between China and Indochina (Zhang et al. 2016).
Despite the lack of a buffalo Y-chromosome reference assembly, sequencing of whole genomes of about 90 male buffaloes from different geographical areas produced Y-chromosome data (Fig. 2.5) (Sun et al. 2020). A total of 520 Y-chromosome specific SNPs were detected in these data, which identified a clear subdivision of the paternal lineages into swamp- (YS) and river-type clades (YR). Two haplogroups were identified within each clade: swamp-type haplogroups YS1 and YS2 are spread across the whole distribution area of the subspecies, although with varying frequencies, while river-type haplogroups were geographically separated, YR1 and YR2 were detected in southern Asia and Italy, respectively. The YR clade haplotypes have been identified in swamp individuals showing the male-mediated river buffalo introgression into swamp buffalo populations due to crossbreeding plans to improve productivity (Fig. 2.5). Y-chromosome data confirms the clear genetic subdivision between river and swamp paternal lineages, indicating that they originated from different wild ancestral population.
7 Conclusions
Y-chromosome specific loci and mitochondrial genome variation suggest that swamp and river buffaloes were domesticated from different ancestral populations that had already diverged due to prolonged isolation following glacial events during the early Pleistocene period. Likely domestication sites of river and swamp buffaloes were the northwestern Indian subcontinent and the China/Indochina border region, respectively (Kumar et al. 2007b; Yindee et al. 2010; Nagarajan et al. 2015; Zhang et al. 2016). This hypothesis has been confirmed by microsatellite loci (Kumar et al. 2006; Vijh et al. 2008; Zhang et al. 2011, 2020) and genome-wide SNP analysis (Colli et al. 2018). River buffaloes have fewer haplotypes and lower overall diversity compared with swamp buffalo for both mtDNA and Y-chromosome markers, which contrasts with the higher values of genomic diversity estimated in river buffaloes compared to swamp buffalo based on whole-genome sequence data (Luo et al. 2020). These differences may be explained by taking demographic effects into account. Inference drawn from whole-genome sequences showed that around 100 thousand years ago river buffalo ancestors experienced a four times larger population expansion compared to swamp buffalo ancestors (Luo et al. 2020). This expansion of the ancestral river buffalo likely led to increased diversity of which only a small amount was sampled when domestication occurred, resulting in a reduction in uniparental lineages while nuclear genomic diversity was maintained in the background. Another explanation may be that in swamp buffalo domestication captured a proportion of diversity from wild population with a high variability of uniparental markers but a generally lower nuclear genomic diversity. Following domestication swamp buffalo variation increased due to a further introgression from wild populations (Zhang et al. 2016).
References
Abo-Al-Ela HG, El-Magd MA, El-Nahas AF, Mansour AA (2014) Association of a novel SNP in exon 10 of the IGF2 gene with growth traits in Egyptian water buffalo (Bubalus bubalis). Prod Trop Anim Health Prod 46:947–952. https://doi.org/10.1007/s11250-014-0588-3
Alberto FJ, Boyer F, Orozco-terWengel P, Streeter I, Servin B, de Villemereuil P (2018) Convergent genomic signatures of domestication in sheep and goats. Nat Commun 9(1):813. https://doi.org/10.1038/s41467-018-03206-y
Amano T, Namikawa T, Suzuki S (1980) Genetic differences between swamp and river buffaloes in the electrophoretic variations of albumin and transferrin. Proc Jpn Acad Ser B Phys Biol Sci 56:463–468. https://doi.org/10.2183/pjab.56.463
Amaral MEJ, Grant JR, Riggs PK, Stafuzza NB, Filho EAR, Goldammer T et al (2008) A first generation whole genome RH map of the river buffalo with comparison to domestic cattle. BMC Genomics 9:631. https://doi.org/10.1186/1471-2164-9-631
Ananthasayanam S, Kothandaraman H, Nayee N, Saha S, Baghel DS, Gopalakrishnan K et al (2020) First near complete haplotype phased genome assembly of river buffalo (Bubalus bubalis). In: bioRxiv 618785. https://doi.org/10.1101/618785
Barker JSF, Tan SG, Selvaraj OS, Mukherjee TK (1997a) Genetic variation within and relationships among populations of Asian water buffalo (Bubalus bubalis). Anim Genet 28:13
Barker JSF, Moore SS, Hetzel DJS, Evans D, Tan SG, Byrne K (1997b) Genetic diversity of Asian water buffalo (Bubalus bubalis): microsatellite variation and a comparison with protein-coding loci. Anim Genet 28:103–115. https://doi.org/10.1111/j.1365-2052.1997.00085.x
Bonfiglio S, Achilli A, Olivieri A, Negrini R, Colli L, Liotta L et al (2010) The enigmatic origin of bovine mtDNA haplogroup R: sporadic interbreeding or an independent event of Bos primigenius domestication in Italy? PLoS One 5(12):e15760. https://doi.org/10.1371/journal.pone.0015760
Borquis RRA, Baldi F, de Camargo GMF, Cardoso DF, Santos DJA, Lugo NH et al (2014) Water buffalo genome characterization by the Illumina BovineHDBeadChip. Genet Mol Res 13(2):4202–4215. https://doi.org/10.4238/2014.June.9.6
Bosse M, Madsen O, Megens HJ, Frantz LAF, Paudel Y, Crooijmans RPMA et al (2014) Hybrid origin of European commercial pigs examined by an in-depth haplotype analysis on chromosome 1. Front Genet 5:442. https://doi.org/10.3389/fgene.2014.00442
Cairns J, Freire-Pritchett P, Wingett SW, Várnai C, Dimond A, Plagnol V et al (2016) CHiCAGO: robust detection of DNA looping interactions in capture hi-C data. Genome Biol 17(1):127. https://doi.org/10.1186/s13059-016-0992-2
Cardoso TF, Amills M, Bertolini F, Rothschild M, Marras G, Boink G et al (2018) Patterns of homozygosity in insular and continental goat breeds. Genet Sel Evol 50(1):56. https://doi.org/10.1186/s12711-018-0425-7
Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF (2018) Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet 19:220–234. https://doi.org/10.1038/nrg.2017.109
Cheng Y, Gu J, Xue H, Li Q, Liang M, Wang N et al (2017) Identification of four SNPs in LHB gene and their associations with sperm qualities of Chinese buffaloes. Anim Biotechnol 28(3):168–173. https://doi.org/10.1080/10495398.2016.1244068
Cockrill WR (ed) (1974) The husbandry and health of the domestic Buffalo. FAO, Rome
Cockrill WR (1981) The water buffalo: a review. Br Vet J 137(1):8–16
Coizet B, Frattini S, Nicoloso L, Iannuzzi L, Coletta A, Talenti A et al (2018) Polymorphism of the STAT5A, MTNR1A and TNFa genes and their effect on dairy production in Bubalus bubalis. Ital J Anim Sci 17:31–37. https://doi.org/10.1080/1828051X.2017.1335181
Colli L, Lancioni H, Cardinali I, Olivieri A, Capodiferro M, Pellecchia M et al (2015) Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability. BMC Genomics 16(1):1115. https://doi.org/10.1186/s12864-015-2342-2
Colli L, Milanesi M, Vajana E, Iamartino D, Bomba L, Puglisi F et al (2018) New insights on water buffalo genomic diversity and post-domestication migration routes from medium density SNP chip data. Front Genet 9:53. https://doi.org/10.3389/fgene.2018.00053
de Camargo GM, Aspilcueta-Borquis RR, Fortes MR, Porto-Neto R, Cardoso DF, Santos DJA et al (2015) Prospecting major genes in dairy buffaloes. BMC Genomics 16:872. https://doi.org/10.1186/s12864-015-1986-2
Degrandi TM, Pita S, Panzera Y, de Oliveira EHC, Marques JRF, Figueiró MR et al (2014) Karyotypic evolution of ribosomal sites in buffalo subspecies and their crossbreed. Genet Mol Biol 37(2):375–380. https://doi.org/10.1590/s1415-47572014000300009
Deng T, Liang A, Liu J, Hua G, Ye T, Liu S et al (2019) Genome-wide SNP data revealed the extent of linkage disequilibrium, persistence of phase and effective population size in purebred and crossbred buffalo populations. Front Genet 9:688. https://doi.org/10.3389/fgene.2018.00688
Di Berardino D, Iannuzzi L (1981) Chromosome banding homologies in swamp and murrah buffalo. J Hered 72:183–188. https://doi.org/10.1093/oxfordjournals.jhered.a109469
Di Meo GP, Perucatti A, Iannuzzi L, Rangel-Figueiredo MT, Ferrara L (1995) Constitutive heterochromatin polymorphism in river buffalo chromosomes. Caryologia 48:137–145. https://doi.org/10.1080/00087114.1995.10797323
Du C, Deng T, Zhou Y, Ye T, Zhou Z, Zhang S et al (2019) Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus bubalis). Anim Genet 50:207–216. https://doi.org/10.1111/age.12739
Dutta P, Talenti A, Young R, Jayaraman S, Callaby R, Kumar S et al (2020) Whole genome analysis of water buffalo and global cattle breeds highlights convergent signatures of domestication. Nat Commun 11:4739. https://doi.org/10.1038/s41467-020-18550-1
El-Bayomi KM, Saleh AA, Awad A, El-Tarabany MS, El-Qaliouby HS, Afifi M et al (2017) Association of CYP19A1 gene polymorphisms with anoestrus in water buffaloes. Reprod Fertil Dev 30(3):487–497. https://doi.org/10.1071/RD16528
El-Halawany N, Abd-El-Monsif SA, Al-Tohamy Ahmed FM, Hegazy L, Abdel-Shafy H, Abdel-Latif MA et al (2017) Complement component 3: characterization and association with mastitis resistance in Egyptian water buffalo and cattle. J Genet 96(1):65–73. https://doi.org/10.1007/s12041-017-0740-8
El-Khishin D, Ageez A, Saad ME, Ibrahim A, Shokrof M, Hassan LR et al (2020) Sequencing and assembly of the Egyptian buffalo genome. PLoS One 15(8):e0237087. https://doi.org/10.1371/journal.pone.0237087
El-Komy SM, Saleh AA, Abdel-Hamid TM, El-Magd MA (2020) Association of GHR polymorphisms with milk production in buffaloes. Animals 10(7):1203. https://doi.org/10.3390/ani10071203
FAO (2004) Secondary guidelines for development of National Animal Genetic Resources Management Plans. Measurement of Domestic Animal Diversity (MoDAD): Recommended Microsatellite Markers Recommendations of joint ISAG/FAO Standing Committee. http://dad.fao.org/en/refer/library/guidelin/marker.pdf
Fischer H, Ulbrich F (1968) Chromosomes of the Murrah buffalo and its crossbreds with the Asiatic swamp buffalo (Bubalus bubalis). Z Tierzuecht Zuechtungsbiol 84:110–114. https://doi.org/10.1111/j.1439-0388.1967.tb01102.x
Ghoreishifar SM, Moradi-Shahrbabak H, Fallahi MH, Sarghale AJ, Moradi-Shahrbabak M, Abdollahi-Arpanahi R et al (2020) Genomic measures of inbreeding coefficients and genome-wide scan for runs of homozygosity islands in Iranian river buffalo, Bubalus bubalis. BMC Genet 21(1):16. https://doi.org/10.1186/s12863-020-0824-y
Guzman JLG, Lázaro SF, do Nascimento AV, de Abreu Santos DJ, Cardoso DF, Scalez DCB (2019) Genome-wide association study applied to type traits related to milk yield in water buffaloes (Bubalus bubalis). J Dairy Sci 103:1642–1650. https://doi.org/10.3168/jds.2019-16499
Harder B, Bennewitz J, Reinsch N, Thaller G, Thomsen H, Kühn C et al (2006) Mapping of quantitative trait loci for lactation persistency traits in German Holstein dairy cattle. J Anim Breed Genet 123(2):89–96. https://doi.org/10.1111/j.1439-0388.2006.00577.x
Hellborg L, Ellegren H (2004) Low levels of nucleotide diversity in mammalian Y chromosomes. Mol Biol Evol 21(1):158–163. https://doi.org/10.1093/molbev/msh008
Iamartino D, Nicolazzi EL, Van Tassel CP, Reecy JM, Fritz-Waters ER, Koltes JE et al (2017) Design and validation of a 90K SNP genotyping assay for the water buffalo (Bubalus bubalis). PLoS One 12:e0185220. https://doi.org/10.1371/journal.pone.0185220
Iannaccone M, Cosenza G, Pauciullo A, Martino G, Capparelli R (2018) The SNP g.4667G>a at 3′-UTR of IFNG gene is associated with susceptibility to bovine tuberculosis in Mediterranean water buffalo (Bubalus bubalis). Anim Genet 49(5):496–497. https://doi.org/10.1111/age.12698
Iannuzzi L, Di Meo GP (2009) Water buffalo. In: Cockett NE, Kole C (eds) Genome mapping and genomics in domestic animals. Springer Verlag, Berlin, Heidelberg, pp 19–31
Júnior GAF, de Oliveira HN, Carvalheiro R, Cardoso DF, Fonseca LFS, Ventura RV et al (2020) Whole-genome sequencing provides new insights into genetic mechanisms of tropical adaptation in Nellore (Bos primigenius indicus). Sci Rep 10(1):9412. https://doi.org/10.1038/s41598-020-66272-7
Kikkawa Y, Yonekawa H, Suzuki H, Amano T (1997) Analysis of genetic diversity of domestic water buffaloes and anoas based on variations in the mitochondrial gene for cytochrome b. Anim Genet 28:195–201. https://doi.org/10.1111/j.1365-2052.1997.00101.x
Koren S, Rhie A, Walenz BP, Dilthey AT, Bickhart DM, Kingan SB et al (2018) De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol 36:1174–1182. https://doi.org/10.1038/nbt.4277
Kumar S, Gupta J, Kumar N, Dikshit K, Navani N, Jain P et al (2006) Genetic variation and relationships among eight Indian riverine buffalo breeds. Mol Ecol 15:593–600. https://doi.org/10.1111/j.1365-294X.2006.02837.x
Kumar S, Nagarajan M, Sandhu JS, Kumar N, Behl V, Nishanth G (2007a) Mitochondrial DNA analyses of Indian water buffalo support a distinct genetic origin of river and swamp buffalo. Anim Genet 38:227–232. https://doi.org/10.1111/j.1365-2052.2007.01602.x
Kumar S, Nagarajan M, Sandhu JS, Kumar N, Behl V (2007b) Phylogeography and domestication of Indian river buffalo. BMC Evol Biol 7:186–193. https://doi.org/10.1186/1471-2148-7-186
Lau CH, Drinkwater RD, Yusoff K, Tan SG, Hetzel DJS, Barker JSF (1998) Genetic diversity of Asian water buffalo (Bubalus bubalis): mitochondrial DNA D-loop and cytochrome b sequence variation. Anim Genet 29:253–264. https://doi.org/10.1046/j.1365-2052.1998.00309.x
Lei CZ, Zhang W, Chen H, Lu F, Liu RY, Yang XY et al (2007) Independent maternal origin of Chinese swamp buffalo (Bubalus bubalis). Anim Genet 38:97–102. https://doi.org/10.1111/j.1365-2052.2007.01567.x
Lenstra JA, Groeneveld LF, Eding H, Kantanen J, Williams JL, Taberlet P et al (2012) Molecular tools and analytical approaches for the characterization of farm animal genetic diversity. Anim Genet 43:483–502. https://doi.org/10.1111/j.1365-2052.2011.02309.x
Liu JJ, Liang AX, Campanile G, Plastow G, Zhang C, Wang Z et al (2018) Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J Dairy Sci 101(1):433–444. https://doi.org/10.3168/jds.2017-13246
Low WY, Tearle R, Bickhart DM, Rosen BD, Kingan SB, Swale T et al (2019) Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity. Nat Commun 10(1):260. https://doi.org/10.1038/s41467-018-08260-0
Luo X, Zhou Y, Zhang B, Zhang Y, Wang X, Feng T et al (2020) Understanding divergent domestication traits from the whole-genome sequencing of swamp- and river-buffalo populations. Natl Sci Rev 7:686–701. https://doi.org/10.1093/nsr/nwaa024
Macciotta NPP, Colli L, Cesarani A, Ajmone-Marsan P, Low WY, Tearle R, Williams JL (2021) The distribution of runs of homozygosity in the genome of river and swamp buffaloes reveals a history of adaptation, migration and crossbred events. Genet Sel Evol 53:20. https://doi.org/10.1186/s12711-021-00616-3
Macgregor R (1941) The domestic buffalo. Vet Rec 53:443–450
McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L et al (2008) Runs of homozygosity in European populations. Am J Hum Genet 83:359–372. https://doi.org/10.1016/j.ajhg.2008.08.007
Michelizzi VN, Dodson MV, Pan Z, Amaral MEJ, Michal JJ, McLean DJ et al (2010) Water buffalo genome science comes of age. Int J Biol Sci 6:333–349. https://doi.org/10.7150/ijbs.6.333
Michelizzi VN, Wu X, Dodson MV, Michal JJ, Zambrano-Varon J, McLean DJ et al (2011) A global view of 54,001 single nucleotide polymorphisms (SNPs) on the Illumina BovineSNP50 BeadChip and their transferability to water buffalo. Int J Biol Sci 7(1):18–27. https://doi.org/10.7150/ijbs.7.18
Mintoo AA, Zhang H, Chen C, Moniruzzaman M, Deng T, Anam M et al (2020) Draft genome of the river water buffalo. Ecol Evol 9(6):3378–3388. https://doi.org/10.1002/ece3.4965
Moioli B, Georgoudis A, Napolitano F, Catillo G, Lucioli S, Ligda C et al (2001) Genetic diversity between Italian and Greek buffalo populations. Anim Genet Resour 29:31–40. https://doi.org/10.1017/S1014233900001383
Nagarajan M, Nimisha K, Kumar S (2015) Mitochondrial DNA variability of domestic river buffalo (Bubalus bubalis) populations: genetic evidence for domestication of river buffalo in Indian subcontinent. Genome Biol Evol 7(5):1252–1259. https://doi.org/10.1093/gbe/evv067
Nascimento AV, Cardoso DF, Santos DJA, Romero ARS, Scalez DCB, Borquis RRA (2021) Inbreeding coefficients and runs of homozygosity islands in Brazilian water buffalo. J Dairy Sci 104(2):1917–1927. https://doi.org/10.3168/jds.2020-18397
Pandey AK, Gunwant P, Soni N, Kavita KS, Kumar A et al (2019) Genotype of MTNR1A gene regulates the conception rate following melatonin treatment in water buffalo. Theriogenology 128:1–7. https://doi.org/10.1016/j.theriogenology.2019.01.018
Pérez-Pardal L, Chen S, Costa V, Liu X, Carvalheira J, Beja-Pereira A (2018) Genomic differentiation between swamp and river buffalo using a cattle high-density single nucleotide polymorphisms panel. Animal 12(3):464–471. https://doi.org/10.1017/S1751731117001719
Peripolli E, Munari DP, Silva MVGB, Lima ALF, Irgang R, Baldi F (2017) Runs of homozygosity: current knowledge and applications in livestock. Anim Genet 48:255–271. https://doi.org/10.1111/age.12526
Richly E, Leister D (2004) NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21(6):1081–1084. https://doi.org/10.1093/molbev/msh110
Rife DC (1962) Color and horn variations in water buffalo: the inheritance of coat color, eye color and shape of horns. J Hered 53:239–246. https://doi.org/10.1093/oxfordjournals.jhered.a107182
Rife DC, Buranamanas P (1959) Inheritance of white coat color in the water buffalo of Thailand. J Hered 50:269–272
Sun T, Shen J, Achilli A, Chen N, Chen Q, Dang R et al (2020) Genomic analyses reveal distinct genetic architectures and selective pressures in buffaloes. GigaScience 9:1–12. https://doi.org/10.1093/gigascience/giz166
Tanaka K, Solis CD, Masangkay JS, Maeda K, Kawamoto Y, Namikawa T (1996) Phylogenetic relationship among all living species of the genus Bubalus based on DNA sequences of the cytochrome b gene. Biochem Genet 34:443–452. https://doi.org/10.1007/BF00570125
Tanaka K, Matsuda Y, Masangkay JS, Solis CD, Anunciado RV, Namikawa T (1999) Characterization and chromosomal distribution of satellite DNA sequences of the water buffalo (Bubalus bubalis). J Hered 90:418–422. https://doi.org/10.1093/jhered/90.3.418
Tantia MS, Vijh RK, Bhasin V, Sikka P, Vij PK, Kataria RS et al (2011) Whole-genome sequence assembly of the water buffalo (Bubalus bubalis). Indian J Anim Sci 81:38–46
Ulbrich F, Fischer H (1967) The chromosomes of the Asiatic buffalo (Bubalus bubalis) and the African buffalo (Cyncerus caffer). Z Tierzuecht Zuechtungsbiol 83:219–223. https://doi.org/10.1111/j.1439-0388.1966.tb00865.x
Vijh RK, Tantia MS, Mishra B, Kumar STB (2008) Genetic relationship and diversity analysis of Indian water buffalo (Bubalus bubalis). J Anim Sci 86:1495–1502. https://doi.org/10.2527/jas.2007-0321
Wang S, Chen N, Capodiferro MR, Zhang T, Lancioni H, Zhang H et al (2017a) Whole mitogenomes reveal the history of swamp buffalo: initially shaped by glacial periods and eventually modelled by domestication. Sci Rep 7:4708. https://doi.org/10.1038/s41598-017-04830-2
Wang G, Hao L, Cheng Y, Li S, Zhang Y, Lv C et al (2017b) Effects of GnRHR polymorphisms on sperm quality in Chinese water buffalo. Anim Reprod Sci 186:37–43. https://doi.org/10.1016/j.anireprosci.2017.09.001
Wang S, Xia X, Zhang T, Sun T, Dang R, Huang Y et al (2018) Y-chromosome haplotype analysis revealing multiple paternal origins in swamp buffaloes of China and Southeast Asia. J Anim Breed Genet 135:442–449. https://doi.org/10.1111/jbg.12364
Wang S, Zhang Y, Cheng Y, Lu G, Yang R, Geng H et al (2020) Association of SNPs in GnRH gene with sperm quality traits of Chinese water buffalo. Reprod Domest Anim 55:384–392. https://doi.org/10.1111/rda.13634
Whitacre LK, Hoff JL, Schnabel RD, Albarella S, Ciotola F, Peretti V et al (2017) Elucidating the genetic basis of an oligogenic birth defect using whole genome sequence data in a non-model organism, Bubalus bubalis. Sci Rep 7:39719. https://doi.org/10.1038/srep39719
Wibowo TA, Gaskins CT, Newberry RC, Thorgaard GH, Michal JJ, Jiang Z (2008) Genome assembly anchored QTL map of bovine chromosome 14. Int J Biol Sci 4(6):406–414. https://doi.org/10.7150/ijbs.4.406
Williams JL, Iamartino D, Pruitt KD, Sonstegard T, Smith TPL, Low WY et al (2017) Genome assembly and transcriptome resource for river buffalo, Bubalus bubalis (2n = 50). GigaScience 6:1–6. https://doi.org/10.1093/gigascience/gix088
Wu JJ, Song LJ, Wu FJ, Liang XW, Yang BZ, Wathes DC et al (2013) Investigation of transferability of BovineSNP50 BeadChip from cattle to water buffalo for genome wide association study. Mol Biol Rep 40(2):743–750. https://doi.org/10.1007/s11033-012-1932-1
Yindee M, Vlamings BH, Wajjwalku W, Techakumphu M, Lohachit C, Sirivaidyapong S et al (2010) Y-chromosomal variation confirms independent domestications of swamp and river buffalo. Anim Genet 41:433–435. https://doi.org/10.1111/j.1365-2052.2010.02020.x
Yusnizar Y, Wilbe M, Herlino AO, Sumantri C, Noor RR, Boediono A et al (2015) Microphthalmia-associated transcription factor mutations are associated with white-spotted coat color in swamp buffalo. Anim Genet 46:676–682. https://doi.org/10.1111/age.12334
Zhang Y, Sun D, Yu Y, Zhang Y (2006) A Y-linked SNP in SRY gene differentiates Chinese indigenous swamp buffalo and introduced river buffalo. Asian Australas J Anim Sci 19:1240–1244
Zhang Y, Vankan D, Zhang Y, Barker JSF (2011) Genetic differentiation of water buffalo (Bubalus bubalis) populations in China, Nepal and south-East Asia: inferences on the region of domestication of the swamp buffalo. Anim Genet 42:366–377. https://doi.org/10.1111/j.1365-2052.2010.02166.x
Zhang Y, Lu Y, Yindee M, Li K-Y, Kuo H-Y, Ju Y-T et al (2016) Strong and stable geographic differentiation of swamp buffalo maternal and paternal lineages indicates domestication in the China/Indochina border region. Mol Ecol 25:1530–1550. https://doi.org/10.1111/mec.13518
Zhang Y, Colli L, Barker JSF (2020) Asian water buffalo: domestication, history and genetics. Anim Genet 51(2):177–191. https://doi.org/10.1111/age.12911
Acknowledgments
JW was supported by the project LEO: Livestock Environment Open Data, 16.2—PSRN 2014-2020, funded through Fondo Europeo Agricolo per lo Sviluppo Rurale (FEASR).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Colli, L., Williams, J.L., Ajmone-Marsan, P. (2022). Water Buffalo Genomic Diversity. In: Chauhan, M.S., Selokar, N. (eds) Biotechnological Applications in Buffalo Research. Springer, Singapore. https://doi.org/10.1007/978-981-16-7531-7_2
Download citation
DOI: https://doi.org/10.1007/978-981-16-7531-7_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-7530-0
Online ISBN: 978-981-16-7531-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)