Abstract
The process of producing proteins from nucleotides is termed as gene expression. Genetic information present in the DNA, is first rewritten, generating RNA, the process of which, is termed as transcription. During transcription, starting with a DNA template, RNA is essentially synthesized (Fig. 10.1). The three major types of RNA generated during transcription are tRNA, mRNA, and rRNA. Other non-coding RNAs (ncRNAs) are also generated. The term “coding RNA” refers to RNAs that are translated to proteins, whereas “non-coding RNA” refers to RNAs that do not code for proteins. Messenger RNAs (mRNAs) serve as templates for translation, and ncRNAs are involved in many regulatory functions in the cell. Recent emerging trends have suggested that many proteins play dual roles, and these are referred to as bifunctional (bifRNAs). Every single gene is transcribed into several copies of mRNA, and each mRNA molecule is capable of generating identical copies of a single protein. The amount of a protein synthesized from a gene is regulated during transcription and translation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 RNA Transcription: Overview
The process of producing proteins from nucleotides is termed as gene expression. Genetic information present in the DNA, is first rewritten, generating RNA, the process of which, is termed as transcription. During transcription, starting with a DNA template, RNA is essentially synthesized (Fig. 10.1). The three major types of RNA generated during transcription are tRNA, mRNA, and rRNA. Other non-coding RNAs (ncRNAs) are also generated. The term “coding RNA” refers to RNAs that are translated to proteins, whereas “non-coding RNA” refers to RNAs that do not code for proteins. Messenger RNAs (mRNAs) serve as templates for translation, and ncRNAs are involved in many regulatory functions in the cell. Recent emerging trends have suggested that many proteins play dual roles, and these are referred to as bifunctional (bifRNAs). Every single gene is transcribed into several copies of mRNA, and each mRNA molecule is capable of generating identical copies of a single protein. The amount of a protein synthesized from a gene is regulated during transcription and translation.
The first step in gene expression where the enzyme RNA polymerase converts a DNA segment into RNA is called transcription. DNA and RNA both make use of nucleotide base pairing as a complementary language.
The direction of flow of genetic material was established as the central dogma by Francis Crick in 1957. This states that the flow of genetic material is in the direction of DNA to RNA to protein and is fundamental to all organisms like complex multicellular organisms as well as unicellular prokaryotes.
This phenomenon came into the picture after the discovery of reverse transcription in retroviruses, which does not follow central dogma. Exceptions also include prions, where information flow is from protein to protein (Fig. 10.2).
Central dogma. The central dogma is a concept in molecular biology that explains how the residue-by-residue transfer of genetic information happens in living organisms. It states that such information transfer follows a particular order and that it does not move from protein to nucleic acid or protein to protein
2 Types of RNA: Overview
The product of transcription is RNA. Four kinds of nucleotides make up the RNA, and phosphodiester bonds link them linearly. The difference between DNA and RNA lies in their structures, mainly the sugar moiety and the type of nucleotides that make up the respective nucleic acids. DNA primarily contains deoxyribose sugar, whereas RNA contains ribose sugar—with an extra –OH group. These structural differences have contributed to the nomenclature of the biomolecules. Also, thiamine (T) is one base pair that is part of DNA, whereas it is replaced by uracil in RNA. The other three bases—adenine (A), cytosine (C), and guanine (G)—are in both of these nucleic acids.
The substitution of thiamine with uracil in RNA leaves us with very few chemical differences between the two molecules, and the base complementary properties remain the same as DNA except the U base pairing with A. However, the structure differs a lot. This can be attributed to the fact that RNA molecules are single-stranded and they have different confirmations. Upon completion of the human genome project, a major discovery was made where studies suggested that only 2% of the entire human genome coded for proteins. But, the amount of the genome that was being transcribed was exceptionally high (62%) indicating that there were RNA molecules in huge amounts that did not code for proteins. These non-coding RNAs were found to be involved in cell cycle regulation, maintaining structural integrity, and other cellular and molecular functions.
Many different types of RNAs are responsible for various functions. The important ones include (Fig. 10.3):
-
mRNA—They are protein-coding messenger RNAs.
-
rRNA—These are ribosomal RNAs that are the main structural components of ribosomes. They are essential for the catalysis of translation.
-
tRNA—These clover leaf-shaped molecules are adapter molecules that facilitate the transfer of amino acids to the site of translation.
Others include:
-
snRNA—The processing of pre-mRNA is mediated by small nuclear RNAs.
-
snoRNA—Processing and modification of pre-rRNAs are facilitated by small nucleolar RNAs.
-
miRNA—MicroRNAs are responsible for post-transcriptional gene regulation.
-
piRNA—These are known as Piwi-interacting RNAs, and they are known to regulate epigenetic and post-transcriptional silencing of transposons that protect germlines from certain transposable elements.
-
lncRNA—Chromatin remodeling is influenced by these long non-coding RNAs. They also regulate gene expression at the transcriptional and post-transcriptional level.
-
circRNAs—These are circular RNAs that regulate gene expression and alternative splicing.
3 Process of RNA Transcription
3.1 Prokaryotes
The mechanism of transcription has remained the same in all living species. Prokaryotic transcription has been extensively studied in bacteria such as E. coli. The transcription of RNA involves three steps: initiation, chain elongation, and termination.
3.2 Promoter Recognition by RNA Polymerase
RNA polymerase locates the target DNA by recognizing the promoter region. Promoter sequences are usually located in the region preceding the transcription start site (TSS) on DNA. The TSS is usually referred to as +1, and the nucleotides towards the 3′ end of the template strand from this point are said to be downstream of the transcript start site. The nucleotides preceding +1 are referred to as −1, and the bases towards the 5′ direction are upstream. Promoters are characterized by the presence of two hexameric AT-rich signature sequences at −10 and −35 bases and are referred to as −10 and −35 elements, respectively (Fig. 10.4) The −10 element is commonly known as the Pribnow box. The −10 and −35 elements are separated by a nonspecific stretch of 17–19 bases. This region is known as the spacer region. Several studies analyzing promoter regions of a large number of bacteria suggest that there are conserved sequences (TATAAT and TTGACA) at −10 and −35 elements, respectively. The activity of a promoter is highly dependent on its ability to bind strongly to the RNA polymerase, thereby increasing its efficiency of inducing conformational changes in the DNA-polymerase close complex leading to opening the DNA duplex and quick disassociation of the promoter. Heterogeneity and variance in the promoter sequences lead to differential levels of gene expression. In eukaryotes, transcription initiation by Pol II is triggered when the core promoters are recognized through the interactions with DNA, transcriptional (co)activators, and modified histones.
Initiation: The core enzyme is aided by the σ factor to locate the transcription binding site. This activity is mediated by specific nucleotide sequences on the DNA known as the promoter regions. Promoter recognition is crucial for the initiation of transcription. Initially, the holoenzyme is attached to the DNA weakly and rapidly slides across it. Once the σ factor locates the promoter region, the holoenzyme adheres tightly to the DNA and specifically interacts with the outer edges of the bases of the DNA.
The tightly bound holoenzyme then initiates transcription and unwinds around 12–14 bases of the DNA duplex forming a transcription-competent open promoter complex. The σ factor binds to the unpaired bases of any one strand of the transcription complex, and the core enzyme starts assembling the complementary ribonucleotides.
This process is tedious and generates a lot of stress. This is because the RNA polymerase stays attached to the promoter region pulling the upstream DNA region into its active site expanding the transcription bubble. This is known as scrunching (Fig. 10.5). This stress coupled along with the steric clashes between the RNA undergoing elongation and the σ element set up abortive transcription in which the newly synthesized short stretches of the RNA are discarded from the transcription machinery, while the RNA polymerase remains in the same place and starts again. After the abortive initiation rounds, and synthesis of around 17 bases, the initiation transitions into elongation. Due to scrunching pressure, the RNA polymerase breaks free off the promoter region leaving behind the σ factor.
Studies establishing that transcription initially follows a “scrunching” mechanism. RNA polymerase stays on the promoter region of the DNA and directs the downstream DNA past its active center and into itself (Kapanidis et al. 2006)
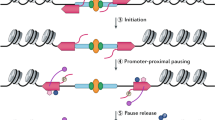
Elongation: After the transcription cycle is set up, the elongation process has to be stabilized. The Pol II machinery equips additional factors to stop the premature dissociation of Pol II. These factors are known as elongation factors, and they associate with Pol II just after initiation. They help the polymerase move along the DNA template on the chromatin network. Pol II goes through something known as “promoter-proximal pausing” after the transcription of 30–50 nucleotides downstream to the TSS (transcription start site). This is depicted in Fig. 10.6. Pol II is released by cyclin-dependent kinase 9 (CDK9) a subunit of positive transcription elongation factor b (P-TEFb). This is facilitated by phosphorylation of several components that make up the transcribing elongation complex (TEC). The P-TEFb works on the core promoters by the COF bromodomain-containing protein 4 (BRD4). A negative elongation factor (NELF) complex is also responsible for promoting Pol II pausing with the help of the DRB sensitivity-inducing factor (DSIF). DSIF directly binds to Pol II, but NELF has a preferential binding activity, and it binds to the assembled DSIF/RNAP II complex.
Pol II-DSIF-NELF: Paused transcription complex (Vos et al. 2018). The negative elongation factor (NELF) as well as the DRB sensitivity-inducing factor (DSIF) protein complexes help stabilize the paused Pol II. NELF binds to the polymerase funnel and forms a bridge between the polymerase units that are mobile, thereafter contacting the trigger loop. This restrains the mobility of Pol II which is necessary for pause release
The NELF complex functionally competes with TFIIF. TFIIF subsequently binds to Pol II and induces conformational changes in the polymerase which are ideal for elongation. Phosphorylation of DSIF, Pol II, and NELF causes dissociation of NELF from Pol II which creates optimum conditions for it to move on to the elongation phase. Certain histone chaperones also help by rapidly disassembling and assembling nucleosomes ahead of the moving Pol II. In eukaryotes, transcription elongation goes hand in hand with RNA processing, and the newly formed RNA molecules are processed in the nucleus, before being exported to the cytoplasm as mature RNAs.
Termination: The terminator signal triggers cascades that cause the core enzyme to dissociate from the template, which releases the newly synthesized RNA transcript and re-associates with the σ factor so that it can start a new round of transcription. The major two kinds of transcription termination mechanisms studied in bacteria are factor-dependent termination (Rho-dependent) and intrinsic (Rho-independent) termination (Fig. 10.7, Table 10.1).
3.3 Eukaryotes
Transcription is carried out by one of three RNA polymerases. The RNAP is recruited based on what kind of RNA is being transcribed. Eukaryotic transcription also involves three stages: (a) initiation, (b) elongation, and (c) termination.
3.4 Initiation of Transcription in Eukaryotes
Prokaryotic RNA transcription is facilitated by RNA polymerases that is capable of binding to a DNA template autonomously, but eukaryotic RNAPs make use of transcription factors. This promoter region is the binding site for these factors, and then polymerase selection takes place based on the requirement. RNAPs and transcription factors form a completed assembly. This binds to the promoter in order to form the pre-initiation complex (PIC). The TATA box is a promoter element which is basically a short stretch of DNA sequence. This found 25–30 base pairs upstream from the transcription start site is the best characterized and most studied core promoter element in eukaryotes. However, the TATA box is only present in about one in ten mammalian genes. Various other core promoter regions are also known to exist.
TBP binds to its binding site “the TATA box.” The binding of TFIID containing the subunit TBP to the TATA box results in the assembly of five more transcription factors. Around the TATA box (Fig. 10.8), these complexes and the RNA polymerase combine forming the pre-initiation complex. Transcription factor II H (TFIIH) drives the unwinding of the DNA double helix in order to provide a DNA template that is single stranded to the moving RNA polymerase. The pre-initiation complex alone is not entirely responsible for transcription initiation. Other than activators, coactivators, repressors, and corepressor proteins are also responsible for regulating transcription. Activators increase the rate of transcription, and repressors decrease the transcription rate.
3.5 Transcription Through Nucleosomes
The release of the polymerase is triggered by the formation of pre-initiation complex, and elongation begins. This phase is characterized as soon as the RNA is synthesized by the polymerase in the direction of 5′ to 3′. In eukaryotes, DNA is present in the form of chromatin and is packed around charged histone molecules. DNA along with histones makes up the nucleosomes. FACT (facilitates chromatin transcription) protein helps in the removal of histones which is mediated by. The disassembly of nucleosomes is also carried out by FACT ahead of a moving Pol II, by the removal of eight histones (Fig. 10.9). This unwinds the chromatin and gives the DNA template access to RNA polymerase II. The nucleosome reassembles the FACT behind the moving and mobile RNA polymerase II. Pol II elongates the newly synthesized RNA until transcription termination signals are encountered.
3.6 Elongation
Twelve protein subunits make up the dynamic RNA polymerase II. This enzyme is a sliding clamp, and single-stranded DNA-binding protein possesses helicase activity. New RNA strands synthesis does not require extra proteins due to this unique multifunctionality of Pol II when compared to replication by the DNA polymerase. But, RNA Pol II requires several accessory proteins for transcription initiation until it is positioned at the +1 initiation nucleotide. After the elongation process has begun, Pol II leaves behind the initiation proteins by a process called “escaping.” This is shown in Fig. 10.10.
The template DNA guides the RNA polymerase to move in the 3′ to 5′ direction. New nucleotides are added by the RNAPs to the 3′ end of the RNA strand and synthesize new RNA strands in the 5′ to 3′ direction. Ahead of the moving RNA Pol, DNA double helix is unwound and simultaneously rewound behind it. When synthesis is happening, 25 unwound DNA base pairs are known to be attached along with new RNA strands which are about 8 nucleotides long.
3.7 Termination
Transcription termination differs based on which RNA polymerase is recruited. A specific (11 bp long in humans; 18 bp in mice) sequence is transcribed by RNA polymerase I that transcribes ribosomal rRNA which contains the sequence. This sequence is recognized by a termination protein called TTF-1 (transcription termination factor for RNA polymerase I) and begins the termination of transcription. From the template DNA strand, RNA polymerase I dissociates causing the release of the new RNA that has just been synthesized. Before the transcription is complete, RNA Pol II cleaves the transcript and is cleaved at an internal site, releasing the upstream portion of the preliminary transcript. This acts as the initial RNA (or pre-mRNA) before further processing can take place. Upon encountering the cleavage site, the end of the gene is reached. A 5′-exonuclease (Xrn2 in humans) digests the remaining transcript as it is being transcribed by Pol II. After the overhanging RNA is digested by Pol II, the 5′-exonuclease catches up to the polymerase II and helps it dissociate from the DNA template strand and concluding transcription.
Where pre-mRNA synthesis is involved, the end of the gene is determined again by the cleavage site. This site is located between an upstream AAUAAA sequence and a downstream GU-rich sequence separated by about 40–60 nucleotides. After transcription of both of these sequences, the CPSF protein binds to the AAUAAA sequence, and the CstF protein binds to the GU-rich sequence (Fig. 10.11).
4 RNA Polymerase: Mechanism
Bacterial RNA polymerase is made up of a core complex consisting of multiple subunits and an initiation factor called the sigma (σ) factor (Fig. 10.12). The core complex has nonspecific polymerase enzyme activity and can bind to DNA and nicks in a nonspecific manner and is known as the core enzyme (E). The σ factor is essential for the sequence-specific transcription activity. The σ factor along with the core enzymes make up the RNA polymerase holoenzyme (Eσ). Most bacterial RNA polymerases are functionally similar, but there are significant structural differences. The E. coli core complex is made of five subunits −α2, β, β′, and ω that possess different functions. Bacteria express several forms of σ factor that recognize and bind different promoter sequences in response to various signals and environmental triggers. In E. coli, the main σ factor is σ70 or σD. It expresses housekeeping genes.
The active site and the DNA-binding cleft of the bacterial RNA polymerase structurally look like a crab claw that possesses two pincers. The β and β′ subunits occupy more than 80% mass of the core enzyme and form the pincers generating a cleft for the entry of the template DNA into the active site of the enzyme. The pincer formed by the β′ subunit is known as the clamp which changes its orientation by swinging between the open and closed conformations. The β and β′ subunits form two double-psi beta-barrel (DPBB) domains. The DPBB domain interacts with the incoming nucleotides with their basic residues on the surface. The DPBB domains are used by most of the cellular RNA polymerases for RNA synthesis. The ribonucleosides enter into the active site of RNA polymerases through a secondary channel, which is a funnel-shaped opening separate from the main channel. The secondary channel contains a binding cavity for accessory factors that control RNA polymerase activity. Two additional motifs from the β′ subunit play very important roles in the RNA synthesis reaction. One motif is the trigger loop involved in catalysis, and the other is the bridge helix used for the translocation of DNA and RNA during the nucleotide addition cycle.
RNA polymerase carries out transcription which is also enzymatically catalyzed. Transcription happens in three distinct phases: initiation, elongation, and termination, which constitute the transcription cycle. In the initiation phase, RNA polymerase recognizes and binds to the DNA at the promoter region which lies upstream of the DNA template. RNA polymerase unwinds the DNA double helix and creates a transcription bubble and exposes 12–14 bases on each strand. One of the helices of DNA acts as the template strand for transcription for the complementary ribonucleotide bases to align. The template strand is commonly referred to as the non-coding strand. The other strand of the DNA double helix will have the same base sequence as the RNA (except uracil instead of thiamine) and is known as the coding strand. RNA polymerase covalently links the base pairs on the template strand. After this step, nine to ten bases of the newly synthesized RNA and the template DNA remain attached forming a temporary DNA-RNA duplex structure. After the synthesis of ten bases, RNA polymerase proceeds to enter the elongation phase. It also involved in the unwinding of the double helical DNA in front and rewinding it from behind. The RNA polymerase moves in the 3′ to 5′ direction of the template, but the direction of chain elongation is in the 5′ to 3′ direction (Fig. 10.13).
At the molecular level, it is understood that the RNA polymerase works by creating phosphodiester bonds between the incoming ribonucleotide triphosphates and the growing chain of RNA. This is a thermodynamically feasible and irreversible reaction. The RNA polymerase adds around 10–100 bases every second. It does not dissociate from the DNA until the transcript is completely formed, and this characteristic is known as processivity. The movement of the RNA polymerase across the template strand happens in such a way that the enzyme is capable of detecting mismatches and other errors. The fidelity of transcription is extensively taken care of because a misincorporated ribonucleotide base leads to disastrous consequences. In case an error is detected, the enzyme moves back onto the template and excises the misincorporated base at the 3′ end exhibiting proofreading activity and replaces it with the correct base. The binding of the RNA polymerase is lenient and allows it to move on the DNA template at different rates. Transcription is terminated at the site where the RNA polymerase recognizes a terminator sequence. The transcript is then released from the transcription bubble. In eukaryotes, RNA polymerases are also involved in the modification of transcripts, a process known as the post-transcriptional modification in which primary transcripts, the firsthand product of the transcription; undergo certain modifications to become functional.
4.1 The Three Eukaryotic RNA Polymerases (RNAPs)
Compared to prokaryotes, mRNA synthesis in eukaryotes is significantly complicated. Eukaryotic transcription involves three polymerases that are comprised of more than ten subunits.
RNA polymerase I: It is a characteristic nuclear substructure in which ribosomal RNA (rRNA) is transcribed, processed, and subsequently assembled into ribosomes. It is found in the nucleolus region. The rRNA molecules are structural RNAs because they offer structural maintenance and support, but they are not translated into proteins but are essential to carry out translation. Majority of the rRNAs, except the 5S rRNA, are synthesized by RNA Pol I.
RNA polymerase II: Main polymerase involved in the synthesis of protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs are subjected to extensive post-transcriptional after. RNA polymerase II transcribes a majority of eukaryotic genes, which includes protein-encoding genes as well as genes that encode for various regulatory RNAs, like microRNAs (miRNAs) and long non-coding RNAs (lncRNAs).
RNA polymerase III: Transcribes structural RNAs including the 5S pre-rRNA, transfer pre-RNAs (pre-tRNAs), and small nuclear pre-RNAs. Small nuclear RNAs are involved in “splicing” pre-mRNAs and regulating transcription factors.
5 Prokaryotic vs Eukaryotic RNA Transcription
The process of transcription is essentially the same in both prokaryotes and eukaryotes. But more steps are involved in eukaryotic transcription (Fig. 10.14). Bacteria and species belonging to archaea require only one type of RNA polymerase, whereas eukaryotes require at least three main enzymes—RNA polymerases I, II, and III (Pol I, II, III), along with polymerases IV and V which are present in plants that transcribe different subsets of RNA. In all three RNA polymerases, the core enzyme is structurally conserved and comprises ten subunits. Additional subunits are located on the periphery. Out of the three, Pol II is known to transcribe the maximum number of genes.
DNA is replicated, and RNA is translated in the same shared space in prokaryotes because of the absence of a nuclear membrane. In eukaryotes, the nucleus is the site of DNA replication and transcription, whereas protein synthesis occurs in the cytoplasm. RNA is exported across the nuclear membrane before it can undergo translation. Transcription and translation are separated by physical barriers. The primary transcript in eukaryotes, which is also known as “heterogeneous nuclear RNA (hnRNA),” is subjected to post-transcriptional processing in order to make a messenger RNA (mRNA) molecule that can pass through the nuclear membrane.
6 Regulation of RNA Transcription
In regulation, transcription factors are key players. Transcription factors are DNA-binding proteins that work by repressing or activating gene transcription. Preferential activity is displayed by some transcription factors. These bind to each other, cis-acting DNA sequences as well as to both DNA and other transcription factors. In order to promote repression or activation, specific promoters act as binding sites for these transcription factors. Certain transcription factors act exclusively as activators or repressors, and some others function as either activators or repressors. The genome of the bacteria E. coli is comprised of around 300 genes that code for proteins that function as transcription factors that up- or downregulate transcription. The functional properties of most of these proteins are still unknown. For the most part, they are known to regulate a large number of genes. Half of all regulated genes are controlled transcription factors such as CRP, FNR, IHF, Fis, ArcA, NarL, and Lrp. A single promoter is known to be controlled by 60 transcription factors. Data inferred from sequence analysis suggest that bacterial transcription factors can be classified into various families and based on these studies. Among these, 12 groups of families have been extensively analyzed and characterized including the LacI, AraC, LysR, CRP, and OmpR families. Bacterial promoter activity also depends on multiple environmental factors and seldom on one signal. Multiple signals are necessary for promoter response. Various transcription factors mediate these events. Many promoters are controlled by two or more transcription factors, with each factor responding to a particular environmental signal.
6.1 Repression of Transcription Initiation
A repressor protein negatively regulates gene expression, by binding to the DNA in order to inhibit the initiation of transcription. An effector molecule essentially binds to the repressor which decides whether or not the repressor is capable of binding to the DNA. Transcriptional control can be best explained by the lac operon expression in E. coli. In this case, the initiation of transcription is brought about from the lac operon promoter by a repressor protein known as LacI. A site known as operator on the DNA acts as the binding site for LacI. This region shares a stretch of sequences with the promoter. Because of the region of similarity, competitive binding of RNAP and LacI repressor takes place, which makes it a requirement that the repressor should be released from the operator for RNAP to bind to the promoter (Fig. 10.15).
Gene regulation by transcription factors and microRNAs (Hobert 2008)
The repressor is subsequently compounded by the binding galactoside, which causes the binding of the RNAP to the promoter region and the initiation of transcription due to the destabilization of the repressor-operator complex (Fig. 10.16). LacI has two additional binding sites in the lac, an upstream site and a downstream site, located in the first gene of the operon. These sites have a lower affinity for the repressor protein, and, likely, they do not directly inhibit transcription initiation.
6.2 Small RNAs
In bacteria several studies have indicated that a subset of small RNAs has been found to regulate transcription in bacteria. An important example is the 6S RNA that inhibits transcription at 70-dependent promoters by binding to the active site of 70-RNAP and competing for DNA binding. It has been proposed that, in the conserved secondary structure of 6S RNA, a single-stranded central bulge within a highly double-stranded molecule that is essential for 6S RNA function is present from which it can be hypothesized that 6S RNA mimics the open conformation of promoter DNA. 6S RNA blocks access to the promoter DNA, and in some cases, it is also used as a template for RNA synthesis.
6.3 Regulation of Transcription Initiation via Changes in DNA Topology
Negatively supercoiled DNA acts as the template for transcription. DNA melting is necessary for the open transcription complex assembly. The degree of supercoiling influences and affects the efficiency of some of the promoters (Fig. 10.17). They are also stimulated by negative supercoiling. The effect of superhelicity on transcription initiation has been demonstrated in several in vitro studies and in in vivo models by gyrase inhibitors, which introduce negative supercoils. Some promoters are also sensitive to the degree of supercoiling, and some are not; the reason for this lies in the fact that the sequence of some promoters is easier to melt.
Epigenetic regulation of gene regulation. Gene expression is regulated by DNA methylation, histone post-translational modifications (PTMs), and the actions of non-coding RNAs, among other mechanisms. To fit within the nucleus, DNA is wrapped around histone proteins creating a higher-order chromatin structure, which can facilitate or prevent access to gene regulatory machinery through steric mechanisms (Torres-Berrío et al. 2019)
7 RNA Processing: Mechanism
RNA processing can be defined as “any type of alteration performed on the RNA after it has been transcribed from DNA to obtain its complete functionality in the cell.”
What Happens During RNA Processing?
After transcription, the RNA is processed before it is exported to the cytoplasm for translation (Fig. 10.18). The hnRNA which is the product of transcription of DNA consists of introns (non-expressing codons) and exons (expressing codons). The introns are excised off from the primary transcript. This process of excising the introns and joining the exons in eukaryotic mRNA (also tRNA and rRNA) is referred to as splicing, usually mediated by a set of protein complexes known as the spliceosomes.
Overview of mRNA processing (Desterro et al. 2020)
7.1 Processing of mRNA
7.1.1 5′ Capping
Eukaryotic mRNA is not stable at the ends and is susceptible to damage thus requiring modification to protect it from ribonucleases. The pre-mRNA hence undergoes capping at the 5′ end immediately after transcription and is then released by Pol II. GTP condensation with triphosphates at the 5′ end is an event that triggers the capping reaction followed by guanine methylation at N-7. This methylation produces the modified guanine or 7-methylguanosine which is attached to the triphosphates of the first transcribed base. Capping of the nascent mRNA protects it from enzymatic degradation by RNAse and also helps in the identification of mRNA by the eukaryotic factors and initiates translation by ribosomes.
It is already known that a protein called m7G cap-binding protein eIF4E binds to the 5′ cap of mRNA and recruits the 40S ribosome subunit to the 5′ end of the mRNA. The ϒ phosphate is removed from the first nucleotide by the enzyme 5′-triphosphatase. The enzyme guanylyl transferase facilitates the attachment of guanosine nucleotide to the first nucleotide of the pre-mRNA. The beta phosphate of the RNA transcript displaces the pyrophosphate group at the 5′ position of the GTP molecule. The cap formation involves a 5′-5′ linkage between the two substrates.
To the terminal end of the RNA, and in the opposite direction, a G residue is added. 5′ diphosphate RNA acts as a substrate for the specific enzyme guanylyltransferase. However, GMP transfer is not catalyzed to give rise to a 5′ monophosphate RNA. Only 5′ end of the pre-mRNAs contain caps, whereas processed 5′ ends followed by endolytic mRNA cleavage do not contain caps.
In the terminal guanine, seven positions are methylated. This event is the first methylation event. The methyl groups are obtained from S-adenosylmethionine in the presence of the enzyme methyltransferase. A cap known as cap 0 carries a single methyl group. The enzyme usually adds methyl groups to 2′ hydroxyl groups on the ribose sugar of the next two nucleotides in the mRNA adjacent to the cap. The addition of methyl group to 2′OH of ribose of the first nucleotide is known as cap-1. The addition of methyl group to 2′OH of the ribose of the second nucleotide and third nucleotide is called cap-2, cap-3, and so on. The decapping enzyme removes the cap and the cap-binding complex, and the mRNA is subjected to degradation after translation (Fig. 10.19).
The mRNA cap is a methylated modification of the 5′ terminus of mRNA. RNA processing and translation factors are recruited to the mRNA cap. The mRNA cap protects transcripts from degradation and defines mRNA as “self.” The formation of the mRNA cap is regulated by cellular signaling pathways. mRNA cap regulation results in changes in gene expression and cell function (Galloway and Cowling 2019)
7.1.2 3′ Polyadenylation
Polyadenylation is a post-transcriptional mechanism in which the addition of poly(A) tail to the messenger RNA at the 3′ end takes place. The poly(A) tail is around 100–250 residues long. The mechanism takes place by endonucleolytic RNA cleavage coupled with the synthesis of polyadenosine monophosphate on the newly formed 3′ end also known as the polyadenylation site. A poly(A) tail is added to the 3′ UTR of newly synthesized pre-mRNAs by the enzyme poly(A) polymerase, which is in turn followed by the recognition of the poly(A) signal and endonucleolytic cleavage of the pre-mRNA at the poly(A) site. Polyadenylation increases the efficacy of mRNA by protecting the 3′ downstream sequences against several nucleases and also plays important roles in mRNA export to the cytosol, its localization, stability, as well as translation. A set of proteins cleave the 3′ segment of the newly synthesized pre-mRNA and then the poly(A) tail. Another important function of the poly(A) tail is to recruit RNases that cleave the RNA. Almost all eukaryotic mRNAs except animal replication-dependent histone mRNAs are polyadenylated. Other important functions of the poly(A) tail include the export of mature mRNA from the nucleus to the cytoplasm, increasing the stability of mRNA and offering protection from cleavage, and signal recognition for the binding of translational factors (Fig. 10.20).
The process of alternative polyadenylation (Gruber and Zavolan 2019)
A multiprotein complex present in the nucleus of eukaryotes primarily targets precursor mRNA. This multi-protein complex excises the 3′ end and adds polyadenyl groups to the cleaved end. The enzyme CPSF (cleavage/polyadenylation specificity factor) specifically binds to the recognition site with the following sequence—5′AAUAAA3′ also known as the polyadenylation signal (PAS) is recognized by the RNA cleavage complex.
When RNA polymerase II recognizes the termination sequence 5′ TTTATT 3′ on the DNA template, transcription is terminated. The polyadenylation machinery is also linked to the spliceosomes.
Alternative polyadenylation (APA) is yet another mechanism of RNA processing which produces distinct 3′ ends on mRNAs. APA is also a gene regulation mechanism in eukaryotes. It is tissue-specific and is extensively studied to understand proliferation and differentiation in cells. Alternative polyadenylation is sometimes used to reduce the length of the coding region which can lead to the expression of different proteins.
7.1.3 RNA Splicing
RNA splicing is an important post-transcriptional process where the non-coding intron sequences are removed from the transcript and the exons are subjected to processing and rejoining. The splicing complex is similar to restriction enzymes that recognize specific sites within the RNA and cleave and ligate the RNA at the cleaved sites. Splicing of pre-mRNA takes place in the nucleus before export.
There has been a significant amount of progress in computational analyses and sequencing methods, and they have led to the discovery of novel splicing isoforms and non-canonical splicing mechanisms. One such example is co-transcriptional splicing. This allows for the epigenetic and epitranscriptomic fine-tuning of gene expression. Studies have revealed that intrinsically disordered domains of RNA Pol II form local condensates, and several splicing factors are known to optimize splicing reactions (Fig. 10.21).
Emerging evidence highlights that the RNA splicing and export machinery can display regulatory potential. Core spliceosome components can display regulatory potential if their levels become limiting for the function of complexes. These findings have important implications for the contribution of selective mRNA processing and export to the development of human cancers and neurodegenerative disorders (Carey and Wickramasinghe 2018)
Exon junction complexes facilitate recursive splicing and also inhibit cryptic splice sites. Circular RNA splicing efficiency is enhanced by the low-efficiency splicing of the flanking introns. Pre-mRNA splicing is crucial in eukaryotic gene expression. Identification of exact splice sites and the accurate removal of introns are also essential for the generation of mRNA and its isoforms. Splicing regulation is mostly well understood. Emerging studies have also revealed that certain non-canonical splicing mechanisms exist. These are important in the regulation of gene expression.
7.1.4 Alternative Splicing
Alternative splicing is a process that allows a messenger RNA (mRNA) to express different forms of proteins (Fig. 10.22). Alternate splicing occurs by the reorganization of the intron and exon sequences in various combinations. This alters the coding sequence of the mRNA. Alternative splicing of the precursor mRNA increases the complexity of gene expression and plays an important role in cell differentiation and growth. Alternative processing is tightly regulated, and this regulation is supervised by the regulatory elements linking both transcription and splicing.
7.1.5 Sequestration as RNP
After nuclear pre-mRNA undergoes complete processing, it is recognized due to the absence of association with splicing factors, and the RNA associated with the functional spliceosome complex is retained in the nucleus. As splicing begins from the cap site and moves towards the polyadenylation site, the hnRNP A1 protein factor binds to the single-stranded RNA molecules that are already exposed. The final processed and fully mature pre-RNA molecules are devoid of any bound splicing factors. Some of the sequences on the proteins are markers for nuclear export signals (NES) and nuclear localization signals (NLS). hnRNP A1 protein also acts as a carrier molecule for mature pre-mRNA (Fig. 10.23).
7.2 Processing of tRNA
Transfer RNA or the tRNA is the primary molecule that facilitates the process of translation. It consists of a single RNA strand made up of 75–95 nucleotides. tRNA is the smallest of the three types of RNA. The 20 amino acids that make up the primary peptide chain all have a specific tRNA that binds to it and transfers it to the growing polypeptide chain during translation. tRNAs are also called adapter molecules. tRNAs have a cloverleaf structure which is stabilized by strong hydrogen bonds between the nucleotides.
All the tRNA molecules have a 3′ end with a conserved 5′-CCA-3′ sequence. Some tRNAs have unusual and modified bases in their primary structure. These unusual bases are mostly a result of post-transcriptional enzymatic modifications of the normal bases in the polynucleotide chain. Two common modifications include pseudouridine (ψU), a derivative of uridine, in which uridine is modified such that the uracil attaches to the ribose to the carbon at the fifth position instead of the nitrogen in the first position, and dihydrouridine (D), also a derivative of uridine where enzymatic reduction of the double bonds between the fifth and the sixth carbon occurs. Other modified bases include hypoxanthine, thymine, and methylguanine. Studies have suggested that cells that do not have these modified bases have shown retarded growth leading to the conclusion that the modified bases have a role in enhanced and better tRNA function.
7.2.1 Secondary and Three-Dimensional Structure of tRNA
There are regions of complementarity within an RNA molecule that enables RAN to form small stretches of double helical patterns which are subsequently stabilized by base pairing. tRNA molecules have a unique conserved pattern showing both single- and double-stranded regions which are also commonly referred to as the cloverleaf model. The cloverleaf model includes significant structures such as the acceptor stem, the ψU loop, the D loop, and the anticodon loop and a fourth variable loop (Fig. 10.24).
(a, b) Secondary and tertiary structures of tRNA. (c) Crystal structure of tRNA (Liu et al. 2015)
X-ray crystallography studies revealed the tertiary structure which takes the shape of the letter L. This structure enabled us to better understand that the orientation of the acceptor stem and anticodon loop and that they are at opposite ends of the adaptor molecule. The acceptor stem and the pseudouracil loop form an extended continuous helix. The anticodon stem associates with the D loop stem to form an extended second helix. The two helices are perpendicular to each other bringing the D loop and the ψU loop together. Interactions such as base stacking, hydrogen bond formation between the bases, and the interaction between bases and the sugar-phosphate backbone stabilize the three-dimensional L-shaped structure and the final confirmation of the tRNA molecule.
7.3 Processing of rRNA
rRNAs account for around 80% of the total RNA present in cells, and they are the main components of ribosomes. Ribosomes are made up of two subunits, a large subunit (the 50S) and a small subunit (30S). Each subunit is made up of specific rRNA molecules. The rRNAs along with proteins and enzymes combine to form ribosomes, which are sites of protein synthesis. The small and large rRNAs contain around 1500 and 3000 nucleotides in prokaryotes such as bacteria and 1800 and 5000 nucleotides in eukaryotes such as humans. The 16S rRNA is the only rRNA in the small subunit of the ribosome and is also called the small subunit rRNA or ss-rRNA. The 5S and 23S are both components of the large subunit of the ribosome. Ribosomes are denoted by the sedimentation unit “S.” In eukaryotes and archaea, four rRNAs are present: 18S in the small subunit and 5S, 5.8S, and 28S in the large subunit. Mitochondria contain 12S and 16S rRNAs. The processing of rRNA is depicted in Fig. 10.25.
8 RNA Editing: Mechanism
RNA editing involves series of molecular processes where the RNA sequence is altered to allow the mature RNA to show variance from the RNA that is encoded by the genomic DNA. Editing includes processes like deletion, insertion, and substitution of the nucleotides. The variation observed in the messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and microRNAs (miRNA) can be attributed to RNA editing. The process of RNA editing occurs in the time interval between the transcription of DNA into mRNA and the translation of this mRNA to protein.
With the discovery of RNA editing, more light is being shed on novel post-transcriptional modifications. RNA editing is facilitated by adenosine and cytidine deaminases acting on DNA and RNA (Fig. 10.26). Adenosine to inosine (A-to-I) editors are members of the ADAR and ADAT protein families. They are important molecules that are crucial in the regulation of alternative splicing and transcription. Other kinds of editors such as cytidine to uridine (C-to-U) editors are members of the AID/APOBEC family and are key players that mediate innate and adaptive immunity and are also responsible for antibody diversification, antibody generation, and antiviral response. These editors are enzymes, and they are present in the nucleus or the cytoplasm. They play a role in the modification of several RNA molecules, including miRNAs, tRNAs, and most importantly mRNAs. Some editors are also capable of editing DNA. Latest technologies such as next-generation sequencing (NGS) have provided us with a large amount of data regarding these post-transcriptional modifications. RNA editing is often implicated in disorders such as cancer and other neurological diseases concerning the brain and the CNS. RNA editing is directly affected by cancer heterogeneity, carcinogenesis, response to treatment, and drug efficacy. Research on RNA editing will lead to the discovery of novel biomarkers identification and diagnostic techniques.
RNA editors such as cytidine and adenosine deaminases are functionally important in regulating cellular processes. (a) Apolipoprotein B is produced in the gut which is mediated by APOBEC1 editing. Glutamate is transformed to a stop codon by C-to-U editing at residue 2153 of hepatic Apo-B100, and a truncated protein Apo-B48 is produced in intestinal cells. (b) The glutamate receptor 2 (GluR2) mRNA at position 607 is edited by ADAR2 in neurons, resulting in change of adenosine to inosine (Christofi and Zaravinos 2019)
What Is Substitution Editing or Site-Specific Base Modification Editing?
A base substitution that causes a significant change in the coding properties or the structure of RNAs is known as substitution editing. These alterations often arise due to chemical changes in the individual nucleotides. Three common deamination reactions give rise to this substitution editing (Fig. 10.27).
This figure depicts the process of RNA editing, where the original guide RNA undergoes post-transcriptional editing which upon translation gives rise to proteins with different functional properties.
8.1 A-to-I Editing
Conversion of A to I: Adenosine deaminases convert an A to inosine (I), which is translated in the form of G by the ribosomes. The abundantly seen type of RNA editing system is A-to-I editing by double-stranded RNA-specific adenosine deaminase (ADAR) enzymes. Data gathered from transcriptomic studies has revealed several “recoding” sites supposedly at which A-to-I editing results in substitutions of bases in protein-coding sequences (Fig. 10.28). The recoding sites are also conserved within lineages and are subjected to positive selection, and they have functional and evolutionary importance. Mapping studies of the editosome complex in various species of the animal kingdom has suggested that most A-to-I editing sites are present within mobile genetic elements in the non-coding parts of the genome and evidence points to the fact that editing of these non-coding sites might have a critical role to play in protection against innate immunity activation by the self-transcripts. Recoding, as well as non-coding events, has been implicated in genome evolution and their deregulation, which could lead to diseased conditions. ADARs are being extensively studied and being adapted for RNA engineering.
A-to-I editing by double-stranded RNA-specific adenosine deaminase (ADAR) enzymes (Eisenberg and Levanon 2018). The deamination of adenosine and formation of inosine, leading to unstable double-stranded RNA base pairing. This alteration leads to reduced production of siRNA, by the dsRNA, and this disrupts the RNAi pathways
8.2 C-to-U Editing
Conversion of C to U: Cytidine deaminases convert a C base in the RNA to uracil (U) apolipoprotein B gene in humans with Apo B100 being expressed in the liver and apo B48 in the intestines of humans (Fig. 10.29).
C-to-U RNA editing of apolipoprotein B. In this figure a ∼35-nucleotide region of apoB RNA flanking the edited base is shown also highlighting apobec-1 and ACF binding to RNA both 5′ and 3′ of the edited base and depicts the presence of additional proteins that may modulate assembly of the holoenzyme
RNA editing by cytidine deamination, the extent to which editing takes place, its regulation, and enzymatic and molecular basis have not been properly established. Hundreds of gene transcripts are known to undergo site-specific C-to-U RNA editing in macrophages and monocytes during M1 polarization and in response to hypoxia and interferons, respectively. This allows for the altering of the amino acid sequences of proteins, especially those that are involved in the viral disease pathogenesis. In single-stranded DNA, cytidines are deaminated by APOBEC3A and also inhibit retrotransposons and viruses. Amino acid residues of APOBEC3A involved in anti-retrotransposition and DNA deamination were also found to affect its RNA deamination activity. In plants, C-to-U editing is seen in the mitochondrial RNA of flowering plants.
8.3 Editing in Mitochondria
8.3.1 Types of RNA Editing in Mitochondria
Protein-coding transcripts (mRNA) are always edited, but introns, rRNA, and tRNA species are also edited in a few cases. In a mitochondrial system, editing restructures many different mRNA transcripts and generates a translatable reading frame. This structured re-tailoring is usually seen in insertion/deletion editing. Editing also gives rise to translation initiation or termination codons. RNA editing affects internal codons; however, substitution editing specifically occurs at first or second positions of codons. Editing often changes the protein properties and causes the protein to significantly deviate from the predicted gene sequence.
In mitochondria, RNA editing systems are known to be mechanistically distinct, and several RNA editing systems are not related mechanistically (Fig. 10.30). The molecular events that occur during editing are phosphodiester bond cleavage and re-ligation. In some cases, editing happens by direct base conversion methods such as deamination. These unique characteristics are discussed along with the concept of the evolution of RNA editing processes. Mitochondrial editing systems are also said to be phylogenetically isolated.
The phylogenetic distribution of most mitochondrial RNA editing systems is restricted, and evidence suggests that these intrinsic mechanisms in the editing systems have evolved from discrete eukaryotic lineages during evolution. Considering U insertion/deletion editing, it has been observed only in kinetoplastid protozoan (Kinetoplastida), in the early-diverging and free-living bodonids (Bodonida), and the late-diverging and parasitic trypanosomatids (Trypanosomatina). This type of editing has not been observed within diplonemids or euglenids. Mitochondrial mRNA editing that occurs in dinoflagellate mitochondria has not been observed in the apicomplexans or ciliates. A different type of editing has been observed in plant organelles (C-to-U and U-to-C) which are not found in any of the green algal mitochondrial systems.
In this way, the mitochondrial editing systems are narrowly restricted and deflect suspicions away from the hypothesis that the editing systems were present in a common eukaryotic ancestor.
8.4 Editing in Plastid
Mitochondria and plastids in terrestrial plants show evidence for post-transcriptional editing from C to U as well as U to C in several transcript sequences. In plastids such as chloroplasts, around 6–20 C bases were found to be deaminated to U bases. RNA analysis studies in mitochondria reveal that in some plants, around 500–1000 U to C conversations is common. Two important factors—(a) PLS-type pentatricopeptide repeat (PPR) protein and (b) multiple organellar RNA editing factors (MORFs, also known as RNA-editing factor interacting protein (RIP))—are trans-acting factors that are involved in this process (Fig. 10.31). MORF9 binding induces significant compressed conformational changes of (PLS)3PPR, revealing the molecular mechanisms by which MORF9-bound (PLS)3PPR has increased RNA-binding activity. Similarly, increased RNA-binding activity is observed for the natural PLS-type PPR protein, LPA66, in the presence of MORF9.
Yan, J., Zhang, Q., Guan, Z. et al. characterize the interactions between a designer PLS-type PPR protein (PLS)3PPR and MORF9 and strongly suggest that RNA-binding activity of (PLS)3PPR is drastically increased on MORF9 binding. Crystal structures of (PLS)3PPR, MORF9, and the (PLS)3PPR-MORF9 complex are shown in the figure
Most plant species do not support U-to-C conversions, and these edits are rare events, specifically two to three edits in the entire mitochondrial RNA sequence. Sites for RNA editing are usually found in the coding regions of mRNAs and rather less in the introns and non-translated regions. It has been observed that in certain situations, RNA editing in tRNA molecules corrects errors and restores normal base pairings. Here, RNA editing becomes crucial because only proper editing can ensure further maturation, accurate folding, and processing of the precursors of tRNA. Figure 10.32 depicts the molecular interactions that influence both RNA editing and chloroplast signaling, essentially suggesting that RNA editing is crucial for normal functioning. In plant organelles, rRNAs are subjected to minimal RNA editing. The mechanisms underlying the recognition of these editing sites, the enzymatic action, and the molecular pathways involved are yet to be determined.
Proposed interactions between RNA editing and chloroplast signaling. In well-functioning photosynthetic cells (left), chlorophyll accumulates in the thylakoid membranes (green), and chloroplasts perform photosynthesis. The GUN1 protein does not accumulate, and thus the chloroplast-to-nucleus signaling that depends on GUN1 is not active. MORF2 contributes to RNA editing (green arrow) and interacts with OPT81, OPT84, and YS1. Light signaling, tissue-specific signals, and the circadian rhythm (not shown) drive high-level expression of PhANGs (black arrow), which promotes chloroplast function (Vo et al. 2019)
In plants, other nucleotide insertions or edits have not been observed. One hypothesis suggests that one reason for RNA editing is that it exists to trigger the activity of a particular RNA-specific C deaminase. These deaminases, however, do not catalyze reverse U-to-C reactions.
Research points to the idea that RNA sequences are involved in guiding the “editosome” editing complexes to specific sites. Cis- or trans-acting RNA molecules facilitate this guiding function but are not native to the sequence regions of the edit sites, but there are no common sequence motifs that have been identified around the different C-to-U conversion sites. In positions preceding the edited Cs, a low amount of G residues has been found. Downstream nucleotides in both mitochondria and plastids are not involved in the editing site specifications, whereas the upstream sequences play a significant role. However, in both the organelles, the upstream region differs in various editing sites, while some sites require only about 5–20 nucleotides and others require around 200 nucleotides. Sequence duplications are also seen in mitochondria. Here as if enough number of upstream sequences are present, RNA editing is accurately maintained. By experimenting in vivo with transgenic plastids, it was proved through upstream and downstream sequence insertions.
Identification of potential RNA editing intermediates suggests that RNA editing in plant organelles is a post-transcriptional process. Partially edited transcripts contain some C bases that have been deaminated to Us. The Cs encoded by the genome exist, in other potential editing sites. Partially edited RNA molecules do not follow a particular order of editing. This means that the hypothetical “editosome” complex does not linearly scan the RNA molecule and the selection of editing sites or regions is arbitrary. These partially edited mRNAs are found in minimal amounts in the plant mitochondria, and they are translated into a family of different proteins. But this assumption only holds for one type of protein sequence, and it is said to be present in the protein complexes of the respiratory chain. These sequences generally are the polypeptide sequences that are best conserved with their respective homologs in other organisms and are selected by their physiological and biochemical functionality. So, it is likely that polypeptides synthesized from unedited RNA molecules would not function properly and such proteins, for example, would hinder the efficiency of respiration in mitochondria.
8.5 Coediting in Virus
In RNA viruses, during transcription of mRNA, the transcription machinery incorporates additional nucleotides that are not specific to the viral genome. For example, in certain paramyxoviruses such as measles, Sendai, parainfluenza, and mumps, viruses around one to ten G residues are inserted at specific editing sites (Fig. 10.33). In Ebola viruses, it is observed that additional A residues are incorporated during transcription.
Certain viruses like the Sendai viruses encode genes to express multiple proteins. They do this with the help of overlapping open reading frames (ORFs) by RNA editing. In viruses like these, the RNA polymerase is capable of reading the same template base more than once, creating insertions that subsequently lead to different mRNAs and generating different types of proteins
So, it is observed that co-transcriptional editing of RNA in RNA viruses happens by insertion of non-templated nucleotides by a mechanism known as “stuttering of the RNA-dependent RNA polymerase.” Specific sequence motifs are present in the viral RNA that can induce the RNA polymerase complex to stutter and repeat the last transcribed nucleotide of the template RNA before resuming transcription.
The replication of Ebola viruses, paramyxoviruses, and other RNA viruses is facilitated by the transcription of RNA-dependent RNA polymerase that is virus encoded resulting in antisense RNA. Stuttering and pausing mechanisms are mostly seen in these types of polymerases at certain nucleotide base combinations, mostly mono- or oligonucleotide tracts (Fig. 10.34). At the nascent mRNA, 3′ ends up to several hundred As are added by the same polymerase, although they are not templated. These As stabilize the mRNA by using a mechanism similar to polyadenylation. mRNA polymerase (complex) pauses at these positions, while the RNA replicase (complex) synthesizes the replication intermediate RNA from the virion RNA. The same RNA polymerase is influenced in a differential manner by additional cofactors, and replication takes place in the virion, while transcription usually occurs in the cytoplasm.
Negative-strand RNA viruses belonging to paramyxoviridae and ebolaviridae are known to polyadenylate mRNA during transcription through a polymerase stuttering mechanism. The viral polymerase acquires a stuttering behavior upon encountering the stop signal present at the end of each gene comprising a stretch of U bases. After each adenine insertion, the RNA polymerase moves back one nucleotide with the mRNA, copying U hundreds of times at the end of viral mRNA thereby producing a poly(A) tail and releasing the polyAdenylated mRNA to stop transcription or scan to restart on the next gene
RNA-dependent RNA polymerase encoded by viruses also pauses and incorporates non-templated nucleotides by the same “stuttering” mechanism around genomic stop codon of the first open reading frame in the unedited mRNA. The reading frame is shifted by insertion of one to two Gs or A bases upstream of the translational stop codon. Upon translation this results in the generation of different proteins with different carboxy-terminal sequences. Compared to the genomically predicted first open reading frame, the extra amino acid sequences in the mumps viruses are known to be double the size.
Transcription termination in E.coli and sequence-specific RNA polymerase pausing and “stuttering” are comparable. Unstable transcription complexes are also induced similarly by sequences at the template sites. Pyrimidine-rich sequences are present along with long U stretches preceding Cs can cause the viral RNA polymerase to slow down, slip back one nucleotide on the template, and incorporate another G nucleotide opposite the same C again. Due to stuttering progression, a sequence of three Gs is altered as initially predicted by the genomic RNA to four Gs in the edited mRNA. RNA viruses facilitate dissociation and realignment of this polymerase-product complex to the previously transcribed nucleotide on the RNA template which can be explained by the specific rate constants of dissociation and RNA-binding protein.
Box 10.1 Scientific Concept: High-Throughput Detection of RNA Processing in Bacteria: Erin E. Gill et al.
A γ-proteobacterium Pseudomonas aeruginosa is a causative agent of opportunistic infections in hospitalized immunocompromised patients and chronic lung infections in patients suffering from cystic fibrosis. P. aeruginosa has been extensively studied because it has devastatingly contributed to human morbidity and mortality. P. aeruginosa is of considerable medical importance due to its metabolic diversity, motility, quorum sensing, ability to produce biofilm, adaptive responses to evade antibiotic stress, and extreme virulence. The main P. aeruginosa strain that is implicated in opportunistic infections is “PAO1.” However, accurate information regarding the transcription start site (TSS) is unavailable to date. The molecular pathways underlying post-transcriptional modifications of RNA transcripts remain elusive in Pseudomonas as well as other organisms. E. Gill et al. suggest that understanding the biochemical makeup of P. aeruginosa, and obtaining the detailed map of TSS and subsequent RNA processing of transcripts and influences virulence, antimicrobial resistance, and other essential cellular functions, is crucial to figuring out the mystery behind the regulation of pathogenesis and drug resistance and the identification of novel drug targets.
Maintaining an inventory of RNA processing sites Transcription Start Sites is necessary to understand cellular processes. RNA sequence-based analysis helps map the set of post-transcriptional modifications occurring in the transcriptomes of organisms. This is a crucial and challenging objective. The completion of transcription results in the occurrence of a series of tightly regulated secondary modifications ultimately leading to the maturation of the RNA transcript. These processes are fundamental to the functionality of many RNAs and also strongly influence the overall behavior of the RNA molecule.
A terminal 5′ triphosphate is present in the primary transcript contains. The conserved pyrophosphatase RppH (YgdP in P. aeruginosa) selectively removes the 5′ triphosphate and leaves a 5′ monophosphate in bacteria. This 5′ monophosphate causes destabilization of the mRNAs by making them susceptible to degradation (Fig. 10.35). The multi-subunit degradosome is an important complex involved in this process. At its core, it contains the 5′ phosphate-sensitive exonuclease/endonuclease RNase E. 5′ monophosphate is known to significantly increase RNase E’s endonuclease activity. Endonucleases that can cleave RNA and leave behind a 5′ phosphate can also result in the production of stable RNAs and activate RNA degradation via degradosome-mediated pathways required for cellular function.
RNA transcription and processing. (Erin E. Gill et al.) (a) Initiation of RNA transcription from a promoter sequence (indicated in red) within the genome and subsequent polymerization of ribonucleoside triphosphate resulting in a 5′ triphosphate at the 5′ end of the nascent mRNA transcript and a 3′ hydroxyl at its 3′ terminus. (b) mRNAs undergoing cleaving by endonucleases to giving rise to two fragments of RNA or can undergoing degradation by exonucleases from 5′ or 3′ termini. (c) RNA processing events that result in either a 5′ triphosphate (dRNA-Seq) or 5′ monophosphate (pRNA-Seq) and that simultaneously contain a terminal 3′ hydroxyl
9 Summary
-
The process of producing proteins from nucleotides is termed as gene expression. Genetic information present in the DNA, is first rewritten, generating RNA, the process of which, is termed as transcription. During transcription, starting with a DNA template, RNA is essentially synthesized. Every single gene is transcribed into several copies of mRNA, and each mRNA molecule is capable of generating identical copies of a single protein.
-
The first step in gene expression where the enzyme RNA polymerase converts a DNA segment into RNA is called transcription. DNA and RNA both make use of nucleotide base pairing as a complementary language.
-
Many different types of RNAs are responsible for various functions. The important ones include mRNA, tRNA, and rRNA.
-
Prokaryotic transcription has been extensively studied in bacteria such as E. coli. The transcription of RNA involves three steps: initiation, chain elongation, and termination. RNA polymerase locates the target DNA by recognizing the promoter region.
-
The core enzyme is aided by the σ factor to locate the transcription binding site. This activity is mediated by specific nucleotide sequences on the DNA known as the promoter regions. Promoter recognition is crucial for the initiation of transcription.
-
After the transcription cycle is set up, the elongation process has to be stabilized. The Pol II machinery equips additional factors to stop the premature dissociation of Pol II. These factors are known as elongation factors, and they associate with Pol II just after initiation.
-
The terminator signal triggers cascades that cause the core enzyme to dissociate from the template, which releases the newly synthesized RNA transcript and re-associates with the σ factor so that it can start a new round of transcription.
-
Bacterial RNA polymerase is made up of a core complex consisting of multiple subunits and an initiation factor called the sigma (σ) factor. The core complex has nonspecific polymerase enzyme activity and can bind to DNA and nicks in a nonspecific manner and is known as the core enzyme (E).
-
At the molecular level, it is understood that the RNA polymerase works by creating phosphodiester bonds between the incoming ribonucleotide triphosphates and the growing chain of RNA. This is a thermodynamically feasible and irreversible reaction. The RNA polymerase adds around 10–100 bases every second.
-
The process of transcription is essentially the same in both prokaryotes and eukaryotes. But more steps are involved in eukaryotic transcription. Bacteria and species belonging to archaea require only one type of RNA polymerase, whereas eukaryotes require at least three main enzymes—RNA polymerases I, II, and III (Pol I, II, III), along with polymerases IV and V which are present in plants that transcribe different subsets of RNA.
-
Transcription factors are DNA-binding proteins that work by repressing or activating gene transcription. Preferential activity is displayed by some transcription factors. These bind to each other, cis-acting DNA sequences as well as to both DNA and other transcription factors. In order to promote repression or activation, specific promoters act as binding sites for these transcription factors.
-
The degree of supercoiling influences and affects the efficiency of some of the promoters. Some promoters are also sensitive to the degree of supercoiling, and some are not; the reason for this lies in the fact that the sequence of some promoters is easier to melt.
-
After transcription, the RNA is processed before it is exported to the cytoplasm for translation. Eukaryotic mRNA is not stable at the ends and is susceptible to damage thus requiring modification to protect it from ribonucleases. The pre-mRNA hence undergoes capping at the 5′ end immediately after transcription and is then released by Pol II.
-
Polyadenylation is a post-transcriptional mechanism in which the addition of poly(A) tail to the messenger RNA at the 3′ end takes place. The poly(A) tail is around 100–250 residues long.
-
RNA splicing is an important post-transcriptional process where the non-coding intron sequences are removed from the transcript and the exons are subjected to processing and rejoining. Alternative splicing is a process that allows a messenger RNA (mRNA) to express different forms of proteins.
-
Transfer RNA or the tRNA is the primary molecule that facilitates the process of translation. It consists of a single RNA strand made up of 75–95 nucleotides. tRNA is the smallest of the three types of RNA.
-
RNA editing involves series of molecular processes where the RNA sequence is altered to allow the mature RNA to show variance from the RNA that is encoded by the genomic DNA. Editing includes processes like deletion, insertion, and substitution of the nucleotides. Conversion of A to I: Adenosine deaminases convert an A to inosine (I), which is translated in the form of G by the ribosomes. Conversion of C to U: Cytidine deaminases convert a C base in the RNA to uracil (U).
-
In a mitochondrial system, editing restructures many different mRNA transcripts and generates a translatable reading frame. This structured re-tailoring is usually seen in insertion/deletion editing. Mitochondria and plastids in terrestrial plants show evidence for post-transcriptional editing from C to U as well as U to C in several transcript sequences.
-
In RNA viruses, during transcription of mRNA, the transcription machinery incorporates additional nucleotides that are not specific to the viral genome.
References
Carey KT, Wickramasinghe VO (2018) Regulatory potential of the RNA processing machinery: implications for human disease. Trends Genet 34(4):279–290. https://doi.org/10.1016/j.tig.2017.12.012. Elsevier Ltd
Christofi T, Zaravinos A (2019) RNA editing in the forefront of epitranscriptomics and human health. J Transl Med 17(1):319. https://doi.org/10.1186/s12967-019-2071-4. BioMed Central Ltd
Desterro J, Bak-Gordon P, Carmo-Fonseca M (2020) Targeting mRNA processing as an anticancer strategy. Nat Rev Drug Discov 19(2):112–129. https://doi.org/10.1038/s41573-019-0042-3. Epub 2019 Sep 25
Eisenberg E, Levanon EY (2018) A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet 19(8):473–490. https://doi.org/10.1038/s41576-018-0006-1. Nature Publishing Group
Galloway A, Cowling VH (2019) mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta Gene Regul Mech 1862(3):270–279. https://doi.org/10.1016/j.bbagrm.2018.09.011. Elsevier B.V.
Gruber AJ, Zavolan M (2019) Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 20(10):599–614. https://doi.org/10.1038/s41576-019-0145-z. Nature Publishing Group
Hobert O (2008) Gene regulation by transcription factors and MicroRNAs. Science 319(5871):1785–1786. https://doi.org/10.1126/science.1151651. American Association for the Advancement of Science
Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314(5802):1144–1147. https://doi.org/10.1126/science.1131399
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8:689–708. https://doi.org/10.1016/j.molp.2015.03.012
Torres-Berrío A et al (2019) Unraveling the epigenetic landscape of depression: focus on early life stress. Dialogues Clin Neurosci 21(4):341–357. https://doi.org/10.31887/DCNS.2019.21.4/enestler. Les Laboratoires Seriver
Vo TV, Dhakshnamoorthy J, Larkin M, Zofall M, Thillainadesan G, Balachandran V, Holla S, Wheeler D, Grewal SIS (2019) CPF recruitment to non-canonical transcription termination sites triggers heterochromatin assembly and gene silencing. Cell Rep 28(1):267–281. e5. https://doi.org/10.1016/j.celrep.2019.05.107
Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P (2018) Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560(7720):607–612. https://doi.org/10.1038/s41586-018-0440-4. Epub 2018 Aug 22
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Sharma, M.G. (2022). RNA Transcription. In: Kar, D., Sarkar, S. (eds) Genetics Fundamentals Notes. Springer, Singapore. https://doi.org/10.1007/978-981-16-7041-1_10
Download citation
DOI: https://doi.org/10.1007/978-981-16-7041-1_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-7040-4
Online ISBN: 978-981-16-7041-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)