Abstract
Population in the globe is estimated to exceed 9 billion by 2050 putting a great challenge to all crop scientists to meet this growing demand. There should be 60% more agricultural production required in 2050 as it was in 2007. The huge shifts from investment in inputs related to fertilizer and pesticides driven technologies to technology based on genetic modifications increasing yields with fewer inputs are indicated. Food and nutrition security have become burning issues in the international discussions at all levels of government as plans are being made to cope up with a changing global climate and increasing global population. One of the most important environmental challenges faced by the developing world is how to meet current food needs without undermining the ability of future generations to meet their needs. Crop production should be adequate to feed the population now and, in the future, also. The current status of agricultural technologies would not be sufficient to meet up the production challenges in future. Innovative technologies have to be exploited in order to enable sufficient food availability in the future. However, even after years of constant efforts by breeders, there are several unanswered issues using traditional methods. In this regard, biotechnology plays an immense role in agriculture by providing better feed and fuel to the growing world.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 Introduction
Critical issues facing agriculture globally include delivery of human health care, reduction in hunger, and increasing energy supply, all in a sustainable manner with optimum animal welfare and minimal negative impact on the environment. The United Nations (U.N.) predicted the world population will exceed nine billion by 2050, improving the quality and quantity of food production is an inevitable necessity. According to the U.N., this doubled food requirement must come from virtually the same land area as today. The U.N. Food and Agriculture Organization (FAO) further stated that 70% of this additional food must come from the use of new and existing agricultural technologies. Therefore, agricultural production faces an exceptional challenge to feed the increasing global population. The existing food production system is under huge pressure to double their food productivity to meet the demands of ever-increasing global population. The annual yield gain reported for major crops like rice, wheat, maize, and soybean (1.2% average) is still less than what is required (2.4%) to reach the goal of doubling global production by 2050. As the world is experiencing high demands for crop production, by 2050 global agricultural production may require to be increased by 60–100% to meet these burgeoning demands as well as there is need to provide food security to the approx 870 million now chronically undernourished (FAO 2012). Food production also suffers from dramatic changes and rapid climate changes including drought, floods, and other disasters. About 80% of world’s population are poor and lives in rural areas which typically rely on local agriculture for their survival needs (FAO 2019). Global yields of major crops are projected to be reduced on average, according to forecasts. For every degree Celsius rise in global mean temperature, wheat, rice, maize, and soybean yields decrease by 6.0%, 3.2%, 7.4%, and 3.1%, respectively (Zhao et al. 2017). CGIAR system (https://www.cgiar.org/) has initiated a ‘Two Degree Initiative for Food and Agriculture’ with the aim of assisting 200 million small scale food producers across the globe to adapt at the speed and scale needed for the current pace of climate change. A significant increase in food production has to be achieved with finite or even depleting land resources and water systems while meeting the demand for ecosystem preservation.
The dynamics of pest pathogens are likely to be influenced by extreme weather conditions, undermining the plant defense response (Atlin et al. 2017). For decades, traditional plant breeding systems have produced a variety of widely accepted high-yielding crop cultivars all over the world. Longer time spent on variety growth and breeding cycles, on the other hand, is a roadblock to plant breeders’ ability to respond quickly to increasing food production demands (Lenaerts et al. 2019). Improving crop productivity rates by breeding entails making significant improvements to our existing plant breeding activities and decisions (Santantonio et al. 2020). Crop improvement has made a major contribution to food security and breeding climate-smart cultivars is thought to be most sustainable way to boost food production. Though, recent studies have argued that current food production practises are insufficient, and that the food system must be transformed. A fundamental change is required within the conventional breeding structure so as to reply satisfactorily to the growing food demands. Crop improvement for food and nutritional security has become a major global concern, particularly in light of population growth and challenges such as climate change and water scarcity. Plant breeding has been very successful in developing improved varieties using conventional tools and methodologies. The success of plant breeding has relied in the utilization of natural and mutant induced genetic variation and in the efficient selection, by using suitable breeding methods, of the favourable genetic combinations. However, existing crop breeding strategies alone will not deliver a high enough rate of crop improvement to satisfy demand in the short or long term. The combination of conventional breeding techniques with genomic tools and approaches is leading to a new genomics-based plant breeding. Genomics-assisted breeding is considered to have the greatest potential for overcoming these challenges and ensuring a sustainable increase of food production by adapting available crops to biotic and abiotic stresses and breeding novel crop varieties. Plant genomics provides breeders with a new set of tools and techniques that allow the study of the whole genome, and which represents a paradigm shift, by facilitating the direct study of the genotype and its relationship with the phenotype. Recombinant DNA technology can help to design almost any desirable characteristic in plants by controlled targeted gene expression. In this new plant breeding context, genomics will be essential to develop more efficient plant cultivars, which are necessary, according to FAO, for the new ‘greener revolution’ needed to feed the world’s growing population while preserving natural resources. Plant genomic data is being utilized in genetic engineering to ensure that better, and fitter varieties of crops are available to ensure food security to the population. Recent progress in genomics technologies has imparted greater strength to the breeders’ toolbox. Using latest techniques such as genomics, biotechnological interventions, speed breeding, genomic selection and genome editing, limitations of traditional breeding could be overcome.
1.2 Crop Improvement and Plant Genetic Resources
One of the most sustainable methods for conserving valuable genetic resources over time while also increasing agricultural production and food security is to use plant genetic resources (PGR) in crop improvement, followed by adoption, cultivation, and consumption or marketing of the improved cultivars by farmers. A more productive use of plant genetic diversity and utilization of plant genetic resources may be a prerequisite to meeting the challenges of growth, food security, and poverty alleviation, according to the Food and Agricultural Organization of the United Nations (FAO) (FAO 1996).
Sources which are available for the improvement of a cultivated plant species are commonly referred to as genetic resources. In traditional plant breeding, genetic resources are the materials which do not require selection for adaption to the target environment and are not immediately useful to the breeders. According to the gene pool concept, genetic resources are categorized into primary gene pool, secondary gene pool, tertiary gene pool, and isolated genes. The primary gene pool consisted of those crop species either itself or other species which are easily crossable with it while the secondary gene pool is consisted of related species that are difficult to cross with the target crop, meaning crossing is less effective and crossing progenies are partly sterile. The tertiary gene pool consists of those species which can only be utilized by advanced techniques like embryo rescue or protoplast fusion. The fourth form of genetic resource may come from related or unrelated plant species, animals, or microorganisms.
The value of various types of genetic resources for crop enhancement largely depends on the crop species. One of the main reasons for the inadequate use of genetic resources in traditional plant breeding is the lack of environmental adaptation of plant genetic resources. Other barriers to PGR use in crop improvement include large performance differences between PGR and actual breeding materials for complex inherited traits such as lack of inbreeding tolerance and unknown affiliation to heterotic pools, as well as genetic issues such as pleiotropy, linkage between desired and undesired PGR alleles, and gene co-adaptation within both breeding populations and plant genetic resources. Genome science tools may be able to finally unlock the genetic potential of our wild and cultivated germplasm resources for the benefit of humanity. The utility of molecular markers and genome research can help the utilization of PGR for crop development in a better way. The revolutionizing advances in plant genomics has evolved from the enrichment and advances made in conventional genetics and breeding, molecular biology, molecular genetics, molecular breeding, and molecular biotechnology in the land of high-throughput DNA sequencing technologies powering the plant research to sequence and understand the genetic compositions, structures, architectures, and functions of full plant genomes. Recent technological development and challenges faced in the field of agriculture have led to the emergence of various genomic tools that can be used to explore and exploit the plant genomes for crop improvement. Next Generation Sequencing (NGS) technologies are allowing the mass sequencing of genomes and transcriptomes, which is producing a vast array of genomic information.
1.3 Biotechnological Interventions
Advances in genomics have allowed scientists to decode genomes for any crop species, as well as knowledge on genes responsible for essential agronomic traits, in the modern period. Gene information can be used to speed up breeding programmes and develop better, higher-yielding varieties. In some cases, the only crop improvement options are genetically modified (GM) crops. The success of Bt cotton has already been realized in our country, as India has gone from being an importer to a major exporter in just a few years. Biotech crops will help farmers in increasing productivity according to a study from international service for the acquisition of agri-biotech applications, which also claims that biotech crops have traditionally been the fastest-adopted crop technology, with farmers satisfied with their benefits and high adoption rates.
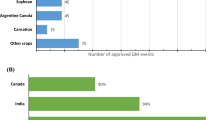
Biotech crop plantings have increased 113-fold since 1996, covering 2.5 billion hectares, demonstrating that biotechnology is the world’s fastest-growing crop technology. Adoption rates of major crops are close to 100% in countries with long histories of high adoption, such as the USA, Brazil, Argentina, Canada, and India, suggesting that farmers prefer this crop technology to traditional varieties. In 2018, more versatile biotech crops with different traits became available on the market to meet the needs of farmers and consumers. Globally, from 1996 to 2018, biotech crops generated economic gains at the farm level worth US$ 167.8 billion. Pesticide use has been decreased by 620 million kilogrammes by utilizing biotech crops. In 2016 alone, fewer insecticides spray reduced carbon dioxide emissions by 26.7 billion kilograms, equivalent to taking 11.9 million cars off the road for a year. Biotech crops have helped 18 million small farmers and their families to overcome the cycle of poverty. According to the Global Status of Commercialized Biotech/GM Crops in 2018, biotech crops were grown and imported in 70 countries in 2018, marking the 23rd year of continuous biotech crop adoption. 26 countries (21 developing and 5 developed) planted 191.7 m ha of biotech crops in 2017, up 1.9 m ha from the previous year’s total. Farmers around the world are continuing to embrace biotech crops, indicating that biotech crops are already helping to address global problems such as poverty, malnutrition, and climate change.
Global agricultural productivity gains are showing a change away from heavy investments in increased fertilizer and pesticide inputs and toward technology-driven changes (including genetic modification) that increased yields with fewer units of input. Increasing population, dwindling agricultural land and water bodies, decreasing productivity, and growing environmental and agricultural issues all contribute to the need for long-term technical interventions in the sector to ensure global food and nutrition stability.
Agricultural biotechnology has been successfully used for decades to increase food production and productivity by creating insect, disease, and herbicide tolerant varieties as well as environmentally sustainable biological products. These products reduce the use of inorganic materials and chemicals in agriculture while increasing productivity and crop nutrition through modern nutrient absorption, their enhancement. Extremely promising novel techniques like Next Generation Sequencing (NGS), Cisgenics, and Genome Editing are being extensively used by the many countries. Some of the ‘high-tech’ novel technologies considered to be very promising in near future in agricultural biotechnology include genome sequencing technologies for crop breeding, RNAi-based gene silencing technologies, new plant breeding techniques (NPBT) including site specific mutagenesis and deploying genes from cross-compatible species through transgenesis (gene transfer), breeding with transgenic inducer line, RNA-dependent DNA methylation (RdDM), reverse breeding, agro-infiltration, grafting techniques, and speed breeding.
1.4 Genomics for Crop Improvement
Genomics is the field of science which correlates genomes at its various levels of structure, function, and evolution. It is aimed towards mapping of genes, their interaction and editing for the betterment of humans. Its function is accomplished with the use of techniques like sequencing and in silico analysis. The emerging field of genomics came into existence during the end of last century with revolutionary vision towards understanding of living forms and it reflects to be the most promising approach in upcoming decades also (Lander 1996; Lander and Weinberg 2000). The advantage towards the approach was founded with the discovery of nucleic acids sequencing by Sanger and coworkers in UK and by Maxam and Gilbert in USA in 1977. The discerning technique led to unlock the whole plan of the improvement of an organism by deciphering the sequence of bases of DNA. The revolution on genomics research was commenced with the crucial phase of Human Genome Project and now has become vital for human welfare in relation to pharmaceutical industry as well as agriculture. The completion of genome and expressed sequence tag (EST) sequencing and gene discovery projects for several crop species like rice, Arabidopsis, sorghum, maize, and soybean based on first-generation Sanger sequencing methods have generated a wealth of genomic and genic sequence information including fully characterized known and candidate genes, transcription factors, and regulatory sequences. With the combination of traditional and high-throughput sequencing platforms, there has been a tremendous increase in genomic resources available, including expressed sequence tags (ESTs), BAC end sequence, genetic sequence polymorphisms, gene expression profiling, whole genome (re)sequencing, and genome wide association studies.
Plant genomics, the study of whole plant genome, their organization and evolutionary patterns along with the functional analysis has become the heart of crop improvement programme. The invention of DNA chip technology is graceful as it allows studying genome wide gene expression patterns with the ease as it simultaneously checks thousands of genes. The gene expression and their regulation with reference to the growth, development, and defense of plants are now the area to be focused with use of translational genomics. The accumulation of huge genomic data with the progress of sequencing technologies can fulfill the need of manipulation in gene expression in order to develop or stimulate responses towards various biotic and abiotic stresses.
Environmental influences, such as extremes of drought, salinity, and temperature, which impose water deficit stress, place significant restrictions on plant productivity (Boyer 1982). More stress tolerant crops need grow to overcome these constraints and increase production quality in the face of a budding world population. Traditional breeding strategies that intended to use genetic variation resulting from varietal germplasm, interspecific or intergeneric hybridization, induced mutations, and somaclonal variation in cell and tissue cultures have only had limited success; very few new plant introductions have resulted in increased stress tolerance under field conditions (Flowers and Yeo 1995). The complexity of stress tolerance characteristics, low genetic variation of yield components under stress conditions, and the absence of successful selection techniques restrict conventional approaches. In addition, quantitative trait loci (QTLs) associated with tolerance at one stage in development can vary from those associated with tolerance at other stages. Desirable QTLs along with the introgressed tolerance trait may require extensive breeding to restore desirable characteristics. Nonetheless, as the resolution of the genetic and physical chromosome maps of major crops strengthens, marker assisted selection of definite secondary yield-related characteristics (e.g. anthesis-silking interval, osmotic adjustment or alteration, membrane stability, or physiological tolerance indices) will prove to be very useful. This strategy could be used in conjunction with pyramiding strategies or sequential selection for accumulation of physiological yield-component traits.
Previous attempts to strengthen drought, high salinity, and low temperature tolerance by breeding and genetic modification have had limited success due to the genetic complexity of stress responses. Progress is now expected through comparative genomics studies of an evolutionarily diverse range of model species, and by the utilization of techniques including expressed sequence tag high-throughput analysis, large-scale parallel gene expression analysis, targeted or random mutagenesis, and gain of function or complementation of mutants.
Agricultural challenges and recent technological advances led to the introduction of high-throughput instruments to explore and manipulate plant genomes for crop improvement. The goal of these genomics-based approaches is to decode the entire genome, including genetic and intergenic regions, in order to gain insights into plant molecular responses, which in turn will provide specific crop improvement strategy. Genomics approaches for crop improvement against stresses are functional, structural, and comparative genomics. Advances in genomics technologies have provided a more thorough analysis of crop genomes and a deeper understanding of stress tolerance mechanisms dynamics. Apart from stress tolerance mechanism, research has also centred on molecular mechanisms regulating stress mediated signalling in plants and the underlying regulatory network of interacting proteins. In order to generate stress resistant crops, functional elucidation of genes involved in these regulatory pathways is intended. Next generation sequencing (NGS) technologies allow mass sequencing of genomes and transcriptomes, creating a broad range of genomic knowledge. Through the advancement of bioinformatics, the study of NGS data has made it possible to discover new genes and regulatory sequences regulating important traits. Many genomic regions associated with significant traits linked to abiotic stress tolerance have also been established with the generation of countless numbers of markers and their use in genome-wide association studies. The discovery of new genes, the determination of their patterns of expression in response to abiotic stress, and an enhanced understanding of their functions in adaptation to stress (acquired by the use of functional genomics) would provide the foundation for successful engineering strategies that lead to greater tolerance to stress.
1.4.1 Whole Genome Sequencing
With the fact of growing global population, changing climate, and environmental pressure, there is an urgent need to accelerate breeding novel crops with higher production, drought or heat tolerance, and less pesticide usage. Advances in genomics offer the potential to speed up the process of developing crops with promising agronomic traits. The recent advent of high-throughput next-generation whole genome and transcriptome sequencing, array-based genotyping, and modern bioinformatics approaches have enabled to produce huge genomic and transcriptomic resources globally on a genome-wide scale in diverse crop genotypes. Moreover, the integration of structural, functional, and comparative genomics including epigenomics with marker-assisted breeding (MAB)/genomics-assisted breeding has been implicated to be an effective approach for identification of genes/QTLs and expressed QTLs (eQTLs) and their regulatory sequences involved in expression of an individual trait in crop plants. The integration of available traditional and modern -omics resources/approaches comprehensively with genomics assisted breeding will certainly decode the molecular and/or gene regulatory networks for identification of functionally relevant novel gene-associated targets and alleles controlling the complex quantitative yield and stress tolerance traits in crop plants.
Whole genome sequencing (WGS) is a laboratory technique which determines the whole DNA sequence of an organism’s genome at once. DNA sequencing methods and computer technique assemble the tremendous biological sequence data that uncovers the order of bases in a whole genome of an organism (Saraswathy and Ramalingam 2011). In 1979, whole genome shotgun sequencing was used for small genomes which range from 4000 to 7000 base pairs (Staden 1979). The first genome of Haemophilus influenzae was sequenced in 1995 (Fleischmann et al. 1995). Further in 2000, the sequencing of almost an entire human genome was completed (Lander et al. 2001). These previously used techniques for WGS were slow, labour-intensive, and costly (Kwong et al. 2015). Wandering-spot analysis was a method reported by Gilbert and Maxam that sequence 24 bp and it was time consuming, and labour required (Gilbert and Maxam 1973). But Sanger sequencing changed the whole scenario when it came into the play as this is technical advances automated, dramatically speed up, also termed as the chain-termination or dideoxy method.
The advancement of WGS helped in identification of the disease associated variants such as complex genomic regions, inaccurate variant calling, detection of SNP, and the phase of the locus, etc. In the process of genomic annotation and analysis, important point is the causation between genomic variants and disease association. It helps to obtain explicit understanding on variation effects of individuals (Albert and Kruglyak 2015). Quantitative assay of biological interactions, downstream effectors such as transcription factors are now feasible with the help of next generation sequencing for genome wide sequencing. Due to advancement in computational and experimental science provided a better understanding of transposable elements sequences in genomic assays and a renewed idea for the importance of TE biology (O'Neill et al. 2020). As a result, plant breeders can use NGS data to discover regulatory sequences and their relative positions as well as can establish molecular markers for marker assisted selection (MAS).
The decoding of entire genomes for a number of plant species has become possible thanks to with the advances in DNA sequencing technology. The sequencing of multiple genomes opens up new possibilities for pan-genomic studies aimed at identifying essential and core genes in crop species. Genomic technologies make effective use of germplasm stored in global repositories for their characterization and utilization.
For crop improvement programmes genetic diversity plays a major role and genetic variations from landraces in crop breeding have been successfully exploited in crops like rice for dwarfing genes, wheat, and barley for mlo alleles (Mascher et al. 2019). Domestication and modern breeding lead to the narrow genetic variations of current crop breeding programmes. Genome-scale studies of large germplasm panels have emerged as a valuable resource for understanding genomic variation dynamics during domestication and selective breeding in recent years (Zhao et al. 2015). For example, recent sequencing of multiple accessions in various crop species in conjunction with genome-wide association studies (GWAS) has aided in the identification of key genomic regions linked to crop domestication and improvement (Varshney et al. 2017). The availability of a reference genome sequence has prompted the sequencing of several accessions of a plant species in order to conduct genome-scale research.
1.4.2 Marker Assisted Selection and QTL Mapping
Crop improvement for various traits depends upon the identification of desirable genes and genotypes harbouring such genes. Identification of such genes and genotypes is facilitated by mapping QTL, finding the tightly linked marker with the QTL and eventually utilizing that markers/QTL in marker assisted selection. Marker-assisted selection is an indirect selection method in which a trait of interest is chosen based on a trait-linked marker. A good MAS necessitates the mapping and close association of a gene to a marker otherwise, it is difficult to analyse or evaluate using traditional methods.
Molecular markers have been generally used to facilitate target gene introgression using the backcross scheme (marker-assisted backcrossing, MABC). MABC also facilitates the recovery of recurrent parent genotype and the elimination of donor parent genome flanking the target gene for minimizing linkage drag. MABC is well suited for introgression of oligogenes and large effect QTLs for defect correction of an otherwise superior variety that is used as the recurrent parent. The identification of diverse strains or hybridization with elite cultivars is needed, to expand the genetic base of core breeding material. Numerous studies have been conducted on the evaluation of genetic diversity in breeding material for all crops. DNA markers have proven to be an invaluable tool for describing genetic tools and providing breeders with more accurate knowledge to aid in parent selection. MAS has been employed for the improvement of many traits in different crops. It has been extensively used for improved access and utilization of germplasm resources, QTL mapping, gene pyramiding, and backcross breeding.
1.4.3 High Throughput Phenotyping
Despite recent advances in genomics, a lack of appropriate phenotyping data (phenomics data) has resulted in weak gene/QTL discovery, which has hampered progress in genomics-assisted crop improvement programmes. As a consequence, high-throughput, reliable, and comprehensive trait data are needed to understand the genetic contribution to phenotype variation. Sustaining and rising crop yields with the benefits of modern genetics methods now depend on phenomics rapid advancement. In a single day, modern phenomics tools intend to record data on characters such as plant production, architecture, growth, biomass, photosynthesis, and so on for hundreds to thousands of plants. As a result of automation, remote control, and data analysis pipelines acquiescent to high throughput phenotyping platforms permitted screening of huge plant populations, germplasm collections, breeding content, and mapping populations with improved precision and accuracy in phenotypic trait acquisition while reducing labour input.
The environment plays a critical role in plant phenomics because most of the essential traits in plants are quantitative in nature and heavily influenced by environmental factors. Plant breeders also want to create crop varieties that have good buffering and stability and can perform well in a variety of environments. As a result, any crop phenomics strategy needs accurate documentation of the experimental environmental conditions (e.g., rainfall, temperature, photoperiod, and soil characteristics).
Phenotyping strategies are categorized into forward phenomics and reverse phenomics. Forward phenomics helps in selecting and identifying superior genotypes while reverse phenomics dissects the best genotypes to discover why they are superior. The forward phenomics offers immediate candidate germplasm for breeding, while the reverse phenomics is a long-term strategy for developing improved crop ideotypes. Reverse phenomics entails applying a variety of new methods to a small set of germplasm in order to uncover common techniques that are responsible for stress tolerance or yield capacity. The phenomics data has been used to find genes/QTL via QTL mapping, association mapping, and genome-wide association studies (GWAS) for crop improvement using genomics-assisted breeding (GAB).
1.4.4 Bioinformatics for Next-Generation Plant Breeding in Plant Genomics
Numerous bioinformatics-based analytical methods are well known in many areas of plant genomic science including comparative genomic analysis, phylogenomics and evolutionary analysis and genome-wide association research. Based on NGS technologies, many autonomous and ultra-high-throughput platforms have recently been developed by big companies such as Roche, Illumina, Applied Biosystems, and so on. All of them are well-fitted for the broad sequence requirements of the present and even future. When entire genomes have been sequenced, it is a vital method to identify and explain the gene and non-coding material in these sequences. For this reason, comparative genomic analysis of plants has emerged as a new area of modern biotechnology, because its main purpose is to predict functions for many unknown genes through the study of significant differences and similarities between organisms. However, in the available datasets of orthologs formed from the same ancestor, these genes are expected to appear. Phylogenomics is known as molecular phylogenetic analysis, in which genomic database sets are used to predict gene function and investigate the evolutionary relationships between organisms. GWAS has a powerful plant species application to classify phenotypic variability in loci correlated with characteristics, as well as allelic variation in candidate genes that resolve quantitative and complex characteristics.
Multiple techniques, including microarrays, expressed cDNA sequence tag (EST) sequencing, serial gene expression analysis (SAGE) tag sequencing, massively parallel signature sequencing (MPSS), RNA-Seq, also known as ‘Whole Transcriptome Shotgun Sequencing’, can determine the expression of many genes by measuring mRNA levels (WTSS). Protein microarrays and high throughput (HT) mass spectrometry (MS) may provide a snapshot of the proteins found in a biological specimen. In making sense of the protein microarray and HT MS data, bioinformatics is very involved. Gene regulation is the dynamic phenomenon of events that ultimately lead to an increase or decrease in the activity of one or more proteins by a signal, possibly an extracellular signal such as a hormone. To explore different steps in this method, bioinformatics techniques have been applied. For instance, gene expression can be regulated in the genome by nearby elements. The analysis of promoters includes the identification and review of sequence motifs in the DNA surrounding a gene’s coding region. The degree to which the region is transcribed into mRNA is influenced by these motifs. Enhancer elements which are located far away from the promoter regions can also regulate gene expression via 3D (three dimensional) looping interactions. Such associations can be determined using bioinformatic analysis of chromosome conformation capture experiments.
In short, along with advances in bioinformatics, the recent wealth of plant genomic resources has enabled plant researchers to gain a fundamental and systematic understanding of economically important plant and plant processes that are critical for advancing crop improvement. Despite these exciting achievements, there remains a critical need for effective tools and methodologies to advance plant biotechnology, address issues that are difficult to solve using current approaches, and facilitate the translation of this newly discovered knowledge to improve the productivity of plants in next-generation plant breeding.
1.5 Advances in Genomics
Crop improvement has become easier with the advent of genomics technologies. Plant breeding for sustainable crop improvement has gained new momentum with the availability of next generation sequencing technologies, modern plant breeding approaches like association mapping, genome wide association selection, advanced backcross QTL analysis, modern genotyping technologies like mass spectrometry allowing SNP discrimination and identification of SNPs based on difference in mass to charge ratio of amplified fragments, SNP arrays.
Identification of large number of single nucleotide polymorphisms (SNPs) with the help of high-throughput genotyping is boosting up the execution of genome wide association studies (GWAS), which relate DNA variants to phenotypes of study. GWAS has enabled the mapping of the genomic loci in diverse set of population which are associated with economically important characters including resistance to biotic and abiotic stress, yield, and quality. There are many genotyping methods available, but among them whole genome resequencing (WGR), reduced representation sequencing (RRS), and SNP arrays are the three most commonly used genotyping techniques in GWAS.
WGR and RRS are based on NGS technologies and bioinformatics pipelines to facilitate alignment of reads to a reference genome while SNP arrays are made up of allele specific oligonucleotide (ASO) probes (which include target SNP loci and flanking regions) that are fixed on a solid support and used to cross-examine complementary sequence from DNA samples and deduce genotypes based on the hybridization signal. Choosing the most suitable (cost-effective) genotyping method for crop GWAS generally requires a thorough review of several factors, including the study’s intent and scope, crop genomic features, and technical and economic issues associated with each genotyping method.
1.6 Conclusion and Future Perspective
Biotechnology has the ability to play a part in securing food and nutrition. The completion of whole genome sequencing of crop plants resulted in a plethora of molecular markers, some of which have the potential to speed up the plant breeding process while also solving genetic purity and adulteration issues. Biotechnology tools deserve to be embraced in the fight against food and nutrition insecurity. This can be made available to small-scale farmers with little or no risk to human health or the environment if reasonable biosafety legislation and policies are in place. As a result, in a world where inaction leads to the deaths of thousands of children, we must not overlook any aspect of a potential solution, like agricultural biotechnology.
Recent advances in genomics research have provided geneticists, biologists, and breeders with a range of cutting-edge tools and technologies that help breeding programmes be more precise and effective. As reference genome assemblies become more widely accessible, gene discovery and trait modulation methods have changed dramatically. In addition to advance genomic researches, gene editing methods in plants for elucidating candidate genes and genetic interactions can be used.
According to Varshney et al. (2019) breeding strategies such as marker-assisted back crossing (MABC) are better suited to removing defects from mega-varieties; however, increasing genetic gains per unit time necessitates rapid population improvement driven by genome-wide predictions and associations. The growing availability of multi-omics data and high-dimensional phenotypic data is exposing the potential challenges associated with data handling and interpretation. Plant breeders must be properly trained, and this will play a key role in adopting more advanced methods for crop improvement, such as systems biology-driven breeding (Lavarenne et al. 2018). To provide solutions for sustainable agriculture, such concerted initiatives involving several disciplines will be critical.
References
Albert FW, Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16(4):197–212
Atlin GN, Cairns JE, Das B (2017) Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob Food Secur 12:31–37
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
FAO (1996) Global plan of action for the conservation and sustainable utilization of plant genetic resources and the Leipzig declaration, adopted by the international technical conference on plant genetic resources. Food and Agricultural Organization of the United Nations, Leipzig, Germany, Rome
FAO WFP, IFAD (2012) The state of food insecurity in the world 2012. Economic growth is necessary but not sufficient to accelerate reduction of hunger and malnutrition. FAO, Rome
FAO (2019) Agriculture and climate change – challenges and opportunities at the global and local level – collaboration on climate-smart agriculture. Food and Agriculture Organization (FAO) of the United Nations, Rome. 52 pp.
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Merrick JM (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269(5223):496–512
Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next? Aust J Plant Physiol 22:875–884
Gilbert W, Maxam A (1973) The nucleotide sequence of the lac operator. Proc Natl Acad Sci 70(12):3581–3584
Kwong JC, McCallum N, Sintchenko V, Howden BP (2015) Whole genome sequencing in clinical and public health microbiology. Pathology 47(3):199–210
Lander ES (1996) The new genomics: global views of biology. Science 274(5287):536–539
Lander ES, Weinberg RA (2000) Journey to the center of biology. Science 287(5459):1777–1782
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Proctor MJ (2001) Initial sequencing and analysis of the human genome
Lavarenne J, Guyomarc’h S, Sallaud C, Gantet P, Lucas M (2018) The spring of systems biology-driven breeding. Trends Plant Sci 23:706–720
Lenaerts B, Collard BCY, Demont M (2019) Improving global food security through accelerated plant breeding. Plant Sci 287:110–207
Mascher M, Schreiber M, Scholz U, Graner A, Reif JC, Stein N (2019) Gene bank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat Genet 51:1076–1081
O'Neill K, Brocks D, Hammell MG (2020) Mobile genomics: tools and techniques for tackling transposons. Philos Trans R Soc B 375(1795):20190345
Santantonio N, Atanda SA, Beyene Y, Varshney RK, Olsen M, Jones E et al (2020) Strategies for effective use of genomic information in crop breeding programs serving Africa and South Asia. Front Plant Sci 11:353
Saraswathy N, Ramalingam P (2011) Concepts and techniques in genomics and proteomics. Elsevier
Staden R (1979) A strategy of DNA sequencing employing computer programs. Nucleic Acids Res 6(7):2601–2610
Varshney RK, Saxena RK, Upadhyaya HD, Khan AW, Yu Y, Kim C et al (2017) Whole-genome resequencing of 292 pigeon pea accessions identifies genomic regions associated with domestication and agronomic traits. Nat Genet 49:1082–1088
Varshney RK, Thudi M, Roorkiwal M, He W, Upadhyaya HD, Yang W, Bajaj P et al (2019) Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat Genet 51:857–864
Zhao Y, Li Z, Liu G, Jiang Y, Maurer HP, Wurschum T, Mock HP et al (2015) Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proc Natl Acad Sci U S A 112:15624–15629
Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P et al (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci U S A 114:9326–9331
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Parihar, A., Shiwani, Mondal, S., Singh, P.K., Singh, R.L. (2022). Introduction, Scope, and Applications of Biotechnology and Genomics for Sustainable Agricultural Production. In: Singh, R.L., Mondal, S., Parihar, A., Singh, P.K. (eds) Plant Genomics for Sustainable Agriculture. Springer, Singapore. https://doi.org/10.1007/978-981-16-6974-3_1
Download citation
DOI: https://doi.org/10.1007/978-981-16-6974-3_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-6973-6
Online ISBN: 978-981-16-6974-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)