Abstract
To cope with the growing demand of protein requirement for the world population, aquaculture should be given more attention as the growth of capture fisheries from both sea and freshwater has stagnated in the last few decades. Looking at the data collected by the FAO, it is now clear that aquaculture productivity has surpassed the capture fisheries substantially. Such development has been possible due to factors such as advancement in stock improvement, better health care, optimal food and feed development and better water quality management for species used in commercial aquaculture. Though advancement in all these areas has collectively enhanced the productivity, the increased population pressure has forced the aquaculturists to look for methods to enhance productivity further. Several technologies are now available which may be used to fulfil the requirements of increasing productivity. Transgenic (both auto-transgenic and allo-transgenic) technology has been available for several fish species from the early 1980s, where it has shown that higher growth rate, better health care and tolerance to environmental stress can be modulated for ensuring higher productivity. Problem in implementation of these technologies was the regulatory stumbling block which took more than 20 years to resolve (the first approval was received in 2019). The use of gene editing techniques, especially CRISPR, though developed recently, is increasing at a very rapid pace in many areas of aquaculture that may enhance productivity. The regulatory authorities have taken a proactive decision (at least in one case concerning a Tilapia species altered for higher growth rate) where alteration/deletion of a few bases is not considered as a new genetically modified organism and therefore, could be cultivated at commercial scale without regulatory approvals. Micro RNA technology is also developing at a very fast pace as an advance technique of DNA sequencing and computation in fish species to test and assign the role of new miRNAs. In this chapter, all the aforementioned techniques are described briefly with a few relevant examples of their usage with respect to productivity enhancement of aquaculture species.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 Introduction
Aquaculture is the farming of aquatic organisms including fishes, molluscs, crustaceans and aquatic plants in freshwater or salt water. Farming implies some form of intervention in the rearing process including regular stocking, feeding and protection from predators to enhance productivity. Farming also implies individual or corporate ownership of the stock being cultivated (FAO 2020). The freshwater aquaculture operation may be categorized into extensive, semi-intensive and intensive on the inputs. In an extensive culture system, the organisms survive and grow on the food available in the environment whereas, in the case of semi-intensive culture system, additional food is supplied to enhance the growth and in the intensive culture system, the organisms survive and grow exclusively on the feed supplied from the external source. Productivity is very high in the intensive culture system and so is the expenditure. A modified type of intensive culture system is now in operation viz., the recirculating aquaculture system where all parameters of water quality, feed supply and removal of excretory substances are monitored in real time to ensure maximum productivity. This type of system is used to grow high value fishes including ornamental fishes and is the most expensive culture system.
Basic requirements of any aquaculture operation are the continuous availability of good quality water and its maintenance, seed of the species to be cultured and a balanced feed/diet for the cultivable species. Additionally, maintenance of the health of the cultivable species is equally important to obtain optimum production from any culture system. About 179 million metric tons of fish were produced in the year 2018, out of which 31 million metric ton was from sea aquaculture and the larger contribution of 51 million metric tons was from freshwater aquaculture. At present, China contributes 35% of the world fish production. Of the 179 million metric ton fish produced worldwide, 156 million metric tons are consumed at a rate of 20.5 kg per capita by humans and the remaining 22 million metric tons are used as fish meal to produce feed for fish and other animals and also for oil extraction. It is known that capture fisheries, both freshwater and seawater has stagnated since 1990, whereas aquaculture production, both fresh and sea water has steadily increased although at different rates. It is known that freshwater comprises only 0.01% of the total water body on earth where 12% of the animals live; therefore, water bodies of freshwater, although rich in diversity, find themselves as more threatened. At present, human population has crossed seven billion and any further increase in this number will add more pressure on land and freshwater resources. As mentioned earlier, the rate of consumption of fish has been increasing steadily with an increase in the number of consumers. To keep pace with the increasing demand of fish, there is a need to innovate in order to produce more fish from less water and space.
Production of fish can be increased through conventional methods of superior stock production using a combination of classical breeding and selection method, genomic ploidy manipulation, hybridization and sex reversal along with good water quality maintenance and an optimum balanced feed. In the present section, the discussion will be restricted to the use of modern technologies; some of them are yet to be used on the commercial level. These include transgenesis, gene editing and micro RNA and their role in higher productivity of aquaculture.
1.2 Transgenesis
It is the process of integration of a foreign gene into a living organism that confers upon the organism a new property (phenotype) that it will transmit unaltered to its descendants. The foreign gene integration can be of different types: wide type—where transfer may occur between two different kingdoms (e.g. bacteria to plant or animal): close transfer—where gene transfer may happen within the same kingdom (e.g. one animal species to another animal): tweaking—where gene is already present but its expression may be altered (Tester 1999). For our purpose close transfer is more important and may be subdivided as follows: heterologous—where gene transfer occurs between different phyla (e.g. fish to amphibians); homologous—where transfer occurs between two genera or species and cisgenic, where the organisms are distinct species but may interbreed in certain specialized conditions. The heterologous transfer may again be subdivided into (1) same group transfer—where transfer occurs within the genus, (2) auto-transgenic—where the transfer occurs within the same species and (3) soma transgenic—where the transgene remains and expresses exclusively in the somatic tissue(s).
Before embarking upon the production of transgenic fish, one needs to decide the purpose of such experimentation. Depending upon the aim of the experiment, whether for understanding the basic biological phenomenon or for commercial purpose, the organism and the downstream processes are chosen accordingly. Once the purpose is decided, the steps to be taken to obtain a transgenic individual are as follows: engineer the transgene construct, deliver the transgene, obtain the founder population and finally produce a transgenic homozygous stock. (For a comprehensive account on transgenesis, please see Chen et al. 1995, Devlin et al. 2009.)
1.2.1 Engineering the Transgene Construct
First, the gene to be introduced into the fish has to be identified (depending on the purpose of transgenesis); the other components required are 5′ and 3′ regulatory sequences which will regulate the function of the introduced gene. Ideally the 5′ regulatory sequence is a promoter which may also be chosen on the basis of its own functionality i.e. constitutive or inducible, which may be either strong or weak. In general, the promoter chosen is not necessarily the same promoter of the selected gene to be used in transgene construct. Selection of strong/weak or constitutive/inducible promoter depends on how the transgene function will be utilized in the transgenic organism. As many as 13 different promoters are used for transgenesis in fishes (Devlin et al. 2009). The 3′ regulatory sequence comprises the poly-adenylation signal and termination signal of the transcript. Once all the components of the transgene are selected, it needs to be isolated and cloned individually in a suitable vector so that it is able to grow in large quantities for future use. The final transgene construct is prepared in the following sequence, 5″ promoter—the gene of choice—3′ regulatory sequence and the whole construct is inserted in a suitable vector for maintenance as a stock. Once such a construct is prepared, the functionality of each component can be checked in an in vitro system using suitable cell lines.
1.2.2 Transgene Delivery
Once transgene construct is engineered it needs to be introduced into the (un)fertilized eggs of the fish for further propagation. Several methods have been adopted for this purpose with varying success.
1.2.2.1 Microinjection
Microinjection into the (unfertilized) eggs is most popular among these methods. For microinjection, DNA is taken in a fine micro capillary tube and delivered into the nucleus immediately after fertilization. In the mouse, both pronuclei are visible after fertilization under an interference microscope. The male pronucleus is much larger, hence, easier to microinject. About 1–2 nL of DNA solution with equivalent copy number 106 to 107 is injected into each egg. In most fishes the pronuclei is not visible in the fertilized eggs. The DNA (106 to 107 copy numbers in 1–2 nL volume) is injected directly into the cytoplasm. Normally, the eggs of fishes are surrounded by chorion which gets hardened after the fertilization resulting in difficulty to micro inject. Removal of chorion either manually, by chemical means or even by drilling can be successfully done before micro injection. The fish eggs have a micropyle that is clearly visible, through which the sperm enters for fertilization. Microinjection into the cytoplasm can be done through the micropyle of the egg where nuclei after fertilization reside. One advantage of microinjection in fish is that a large number of eggs can be obtained by the induced breeding technique. The disadvantages are: (1) need trained personnel to perform microinjection at a reasonably good speed, (2) short time duration for injection—around 30 min from fertilization to the first cell division which is critical if done at the two-cell stage it increases the chances of getting a mosaic, individual, (3) rate of integration is only around 20%, (4) at times the transgene can get circularized, forming a concatamer and remain as such for a few cell divisions as a free molecule without integration and (5) occurrence of a high number of mosaic individuals.
1.2.2.2 Electroporation
This method is successfully used for gene transfer in fish eggs. Initially, it was used for transforming different types of cells under in vitro and in vivo conditions. In this method, the eggs are kept in a DNA solution and high voltage is applied for a short period of time, giving multiple pulses. During this process, a temporary pore formation occurs in the membrane which allows the DNA (and other macromolecules) to enter the cell by simple diffusion. Once it enters, the DNA finally moves into the nuclei and integrates into the genome. The advantage of this method is that a large number of eggs can be treated simultaneously and it does not require highly trained manpower. Due to the presence of a chorion, this method is not very successful in the case of fish eggs but upon removal of chorion, higher success rates have been reported in carps, catfishes and zebra fish. Transfer of DNA by this method is very successful for molluscan eggs which are small and have either a very thin chorion or no chorion at all. The method of electroporation is easy to perform and the rate of integration is around 20% or higher, which is similar or slightly higher than the rate achieved with the microinjection technique (Buono and Linser 1992, Lu et al. 1992, Powers et al. 1992,).
1.2.2.3 Sperm Mediated Gene Transfer
It is another method used successfully in transgenesis. The fish sperm cells can remain dormant in the seminal fluid or in an artificial medium for long period and can be activated when right conditions are provided without reducing the rate of fertilization of the eggs. It is also known that DNA can bind to the sperm when incubated. Such sperm cells, when used for fertilization, can result in transgenic fish. Thus, the sperm is acting as a vehicle to carry the transgene into the egg. The frequency of such transfers is unfortunately low. Interestingly, however, sperm cells when electroporated in the presence of DNA results in a dramatic increase in the frequency of transgenic individuals (Müller et al. 1992; Powers et al. 1992; Symonds et al. 1994; Tsai et al. 1995a, b). This procedure also does not require highly trained manpower and can be used in the field conditions on a mass scale. Electroporation (sperm mediated or otherwise) is, therefore, considered as an efficient and versatile gene transfer technology (Powers et al. 1992; Chen et al. 1995; Rajesh and Majumdar 2005).
1.2.2.4 Several Other Methods
Gene transfer including lipofection, biolistics (gene gun) and retroviral infection have been used in fishes with varying success rates. Spermatogonial stem cell modification and transplantation also have the potential to revolutionize the field of fish transgenesis (Tonelli et al. 2017).
1.2.3 Maintenance of Injected Eggs
After gene transfer, the eggs are allowed to grow under sterile conditions, sometimes in the presence of antibiotics to prevent bacterial infection. Generally, tropical fishes hatch within 24 h whereas temperate species take a much longer time to hatch, therefore, requiring more precaution during this period of their development. Once yolk is absorbed, the larvae are nurtured with natural/artificial feed till they are ready for further experimentation.
1.2.4 Transgene Integration
Once the transgene is transferred into the nucleus/cytoplasm of the fertilized eggs, it needs to be observed whether it has integrated into the chromosome of the recipient or not. In most of the cases, it is seen that the transgene does not get integrated immediately rather it replicates and only during the first cleavage stage it gets integrated. Most of the time, this results in G0 transgenic individuals that are mosaic in nature i.e. all cells do not possess the transgene (Ozato et al. 1986; Dunham et al. 1987, 1992; Stuart et al. 1988; Zhang et al. 1990; Lu et al. 1992). As the rate of integration of transgene is low, various methods are adopted, including (1) a nuclear localization signal protein with transgene (2) a pseudotyped retrovirus with special integration protein or (3) use of transposases along with transgene to enhance the rate of integration. It is also seen that a low percentage of transgenic individual has the transgene integrated in multiple places. The other problem with transgenesis is that the integration happens randomly in the genome rather than at a specific site. A eukaryotic chromosome has both heterochromatic and euchromatic regions, the heterochromatic regions are found to be transcriptionally silent whereas the euchromatic regions are active. It is known that a transcriptionally active gene from the euchromatic region, when transposed to heterochromatic region, gets silenced whereas a gene from the heterochromatic region when transposed to euchromatic region becomes transcriptionally active. Similarly, the transgene gets silenced if integrated near the heterochromatic region of the chromosome.
1.2.5 Detection of Integrated Transgene
Mainly three techniques are followed: (1) dot blot hybridization, (2) PCR and (3) inverse PCR, to locate the transgene in the genome of the putative genetically engineered fish. For these methods, DNA is first isolated from the fish. In dot blot analysis, isolated DNA is denatured, immobilized on a nitrocellulose or equivalent membrane and hybridized with a portion of the transgene previously labelled with a radioactive or a fluorescent nucleotide. In PCR, primers specific for the transgene are used to amplify it, using the transgenic fish DNA as template. In the case of inverse PCR (Ochman et al. 1988), the DNA from the putative transgenic fish is digested such that the transgene remains intact as a single fragment. The fragment is circularized by ligation. Primers are then selected from the transgene such that the amplification products include the end region of the integrated site. In the first two cases, it is only possible to infer the presence of a transgene in the individuals which show positive hybridization or a specific amplification of the transgene after PCR which may present as a free circular/linear DNA moiety. To confirm the integration of the transgene, the best method would be Southern hybridization. DNA isolated from the putative transgenic fish is digested with restriction enzyme(s) which will not split the transgene. When Southern hybridization is done on such DNA with a labelled transgene probe, hybridization will always be seen at a higher molecular region than the transgene. Further, using inverse PCR, Rajesh and Majumdar (2005) have shown that the transgene is integrated in the SINE sequences of genome of transgenic Labeo rohita. The transgenic fish can be further analyzed by dot blot hybridization with serial dilution of the DNA to determine the transgene copy number in the genome.
1.2.6 Homozygous Transgenic Fish
When the positive transgenic fish is identified and the integration of the transgene is known, testing the performance with respect to the transgene can be done. To obtain a homozygous stock of individuals, transgenic fish is crossed with a non-transgenic fish. This is expected to produce 50% transgenic progeny in a typical Mendelian cross, if the F0 individual is heterozygous with respect to the transgene. Unfortunately, in many cases, F1 progeny falls short of the desired 50% heterozygote transgenic fish, indicating that the F0 is mosaic in nature. F1 heterozygous progenies, when crossed finally, results in a homozygous stock. This stock can be used to determine the expected performance of the transgene in a homozygous condition.
1.2.7 Applications of Transgenic Fish
In 1983, the first report on the successful production of transgenic fish called guanil or crown carp, a cyprinid fish, was made by a Chinese scientist (China.org.cn, Wang Yaping, a researcher at the Chinese Academy of Sciences, Institute of Hydrobiology), with an aim to enhance the growth rate, the primary aim of any commercial aquaculture operation. Growth hormone gene from a fast-growing grass eating carp was transferred into an omnivorous carp native to the Yellow river; it was shown to grow to marketable size within a year, which is half the time required to grow a non-transgenic fish. Besides this, transgenic fish are made for several applications such as, making a model system to determine biological/cellular function, for identification of regulatory elements in gene expression, to study the role of gene in development of other organisms or its role in specific organs, in drug development and testing, to human disease models or models for environmental monitoring, in modulation of phenotypes in aquarium fishes for commercial use, production of therapeutics for human use, in the development of disease resistant fishes and to study production enhancement through higher growth rate in several commercially important fishes.
1.2.7.1 Disease Resistance
The cecropins, small cationic peptides found originally in the moth Hyalophora cecropia (Steiner et al. 1981), show antibacterial properties against many Gram negative bacteria and is nontoxic to the eukaryotic cell (Jaynes et al. 1989; Kjuul et al. 1999). Transgenic catfish with cecropin B construct has shown significant resistance to bacterial infection (Dunham et al. 2002). Similar results were obtained in transgenic Medaka, having a cecropin transgene. When challenged separately with Pseudomonas fluorescens and Vibrio anguillarium the transgenic Medaka showed 0–10% and 10–30% mortality, respectively when compared to non-transgenic controls (Sarmasik et al. 2002).
1.2.7.2 Production of Therapeutics for Human Use
The human coagulation factor VII a vitamin K-dependent glycoprotein is used to treat patients with haemophilia A or B and severe thrombocytopenia. Transgenes, having CMV promoter and human factor VII cDNA injected into fertilized eggs of Zebra fish (Danio rerio), Tilapia (Oreochromis niloticus) and African catfish (Clarias gariepinus) have shown that the human factor VII could be detected in the embryo of all the three species, indicating the possibility of using fish as bioreactors (Hwang et al. 2004) similar to other bovine species.
1.2.7.3 Colour Variation in Aquarium Fish Species
Transgene constructs, having Xenopus elongation factor I alpha enhancer-promoter with cDNA of GFP (Green fluorescent protein), when injected into a zebra fish embryo, showed expression of GFP (without any inducer) within 4 h of initiation of development. The F2 transgenic fish possessing this construct showed GFP expression even when only one copy of the transgene was present (Amsterdam et al. 1995). Similar results on the expression of GFP was seen in transgenic zebra fish, when promoters such as type II cytokeratin, muscle creatine kinase and acidic ribosomal phosphoprotein gene of the same fish were used separately, but in conjunction with GFP (Ju et al. 1999).
1.2.7.4 Environmental Monitoring and Biosensor
Zebra fish and Medaka are used extensively in detecting and monitoring toxic substances including teratogens in the environment that affect vertebrate development. With transgenic technology this can be done in vivo and in real time. Transgene constructs are prepared with promoters from genes such as Cytochrome P450, metallothionein, small heat shock protein hsp 11 and oestrogen receptor, individually with the GFP gene. When these transgenic fish come in contact with dioxins, heavy metals, pesticides or oestradiol/xenoestrogen, they express GFP, indicating the presence of such substances in the environment (Lee et al. 2015; Bambino and Chu 2017). Interestingly, Lee et al. (2014) have shown that transgenic zebra fish with human CHOP promoter with GFP as transgene detects heavy metals and endocrine disruptors in the environment. It is also shown that treatment with Cu2+ triggers GFP expression in the skin and muscle tissue, whereas Cd2+ induces GFP expression in the skin, olfactory epithelium and pronephric ducts. When such treated fishes are returned to normal water, the GFP expression diminishes without any morphological defects.
1.2.7.5 Growth Manipulation
Since 1983, many groups have been working on growth manipulation in several fishes including commercially important food fishes through transgenic technology. In most of the cases, growth hormone gene has been used in combination with different constitutive or inducible promoters/enhancers as a transgene. Earlier, non-fish growth hormone gene was used in the transgene construct because of non-availability of the fish growth hormone gene. Similarly, viral/bacterial promoters were used for driving the growth hormone expression. Later on, as the data on sequence of many fish species became known, all-fish constructs (both promoter and growth hormone gene derived from fish) were used to obtain transgenic fishes with higher growth rate (for details see Beardmore and Porter 2003; Devlin et al. 2009).
1.3 Clearance from Regulatory Authorities
Since mid-1980s, experiments on growth enhancement in fish through transgenesis have successfully been conducted in more than 30 species, including some commercially important fishes. Unfortunately, none of the transgenic GM stocks could be used in commercial aquaculture as regulatory authorities throughout the world refused permission due to an unknown risk to human safety upon consumption of GM food. The other risk factor under consideration was genetic contamination if these GM fish escape and breed with natural stock in the environment. One exception was mentioned by Beardmore and Porter (2003) that at the other extreme the use of “autotransgenics” must be seen as a risk which were in orders of magnitude lower than that for “allotransgenics” and probably negligible (Beardmore 1997; Nam et al. 2008).
In 2015, after more than 20 years of review, the US FDA approved a genetically engineered Atlantic salmon, “AquAdvantage” for commercial production, the first genetically engineered animal to get this approval for human consumption. AquAdvantage is a genetically modified Atlantic salmon that has a transgene constructed from a promoter of anti-freeze protein from ocean pout (Zoarces americanus) hooked to the growth hormone gene of Chinook salmon (Oncorhynchus tshawytscha), a typical transgenic and triploid female fish, generated by standard gynogenesis to make it sterile. This fish takes half the time (about 18 months) to reach marketable size with reduced FCR, indicating shortened growth time with less food consumption. In 1989, this fish was developed by AquaBounty Technologies Inc., a company located in Massachusetts, USA and is now cultivated in artificial tanks on land in Canada and Panama. Though approval to grow the fish was given by the US FDA, the US Congress later stopped commercial sale of the same. In 2016, Health Canada and Canadian Food Inspection Agency approved the sale of AquAdvantage salmon in their country. This is the first genetically engineered animal for consumption in Canada and also in the entire world. The market acceptance of this transgenic fish can be ascertained from the fact that on 4th August 2017 in an open market 4.5 tons of fish was sold at a price of $11.7/kg (Waltz 2017) and to reach this stage the company had to wait for more than 25 years. Finally in March 2019, the US FDA approved the import of AquAdvantage salmon embryo into their containment facility in Indiana for cultivation and sale in the USA as food, labelled as “Bioengineered” as mentioned by Scott Gottlieb, Commissioner of the US FDA. He also said that “the fish is safe to eat, the genetic construct added to the fish genome is safe for the animal and that the manufacturer’s claims that it reaches a growth marker important to aquaculture more rapidly than its non-GM farm-raised Atlantic salmon counterpart, has been confirmed”. It was also mentioned in a press release that the FDA has determined that the approval of the AquAdvantage salmon would not have any significant environmental impact because of the multiple and redundant measures being taken to contain the fish and prevent their escape and establishment in the environment (US FDA press release April 08, 2019). With this development, a big hurdle of transgenic fish cultivation for commercialization has been hopefully overcome and other countries can take this as precedence for developing their own transgenic fish for human consumption.
1.4 Gene Editing
Site-specific alterations were contemplated ever since DNA double helical structure was deciphered. Since then, our knowledge has increased substantially with respect to site-specific recognition of the DNA by several compounds, mechanism of recombination and DNA repair which eliminates or reduces lethal effects of DNA damage in different organisms. This knowledge allows us to perform modifications targeted to a specific locus in the genome to understand its function. In the early years, several methods were employed like, (1) base pair recognition by small molecules oligonucleotides, (2) triple helix formation with oligonucleotides and their chemical cleavage, (3) chemical cleavage of oligonucleotides, (4) site-specific modification by cross linkers and cleavage agents like bleomycin or psoralen, (5) recognition by peptide nucleic acid and breakage by a cleaving agent like bleomycin or (6) use of self-splicing introns through base pairing with the DNA/RNA. None of these got wide acceptability because of the low efficiency with which it could do specific targeting of DNA.
The discovery of ZFNs (zinc-finger nuclease) and TALENs (transcription activator-like effector nucleases) overcame such deficiencies. These two systems possess a sequence specific DNA-binding domain, fused to a nonspecific DNA cleavage module. The altered DNA-binding domains derived from zincfinger and transcription activator-like effector proteins provides the versatility for the system. Theses enzymes introduce targeted DNA double stranded breaks (DSBs) inducing cellular DNA repair mechanisms like error-prone non-homologous end joining (NHEJ) and homology-directed repair (HDR). Simplicity and flexibility were brought in by zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) to the forefront of gene modification (See review Gaj et al. 2013). The only problem with these systems that exist is that for every site to be altered one pair of enzymes had to be generated, therefore, making it a lengthy process. The discovery of RNA programmable DNA endonuclease, CRISPR/Cas9 (Clustered Regularly Interspaced Palindromic Repeats/CRISPR associated 9) revolutionized the gene editing system.
The following discussion is based broadly on the views of Cong et al. (2013), Gaj et al. (2013), Mali et al. (2013), Doudna and Charpentier (2014), Kim and Kim (2014), Barrangou and Doudna (2016), Lander (2016). Therefore, individual references are not provided. Besides, the controversies regarding the heroes of CRISPR/Cas system has now been resolved by the award of the Nobel Prize for Chemistry (2020) to Emmanuelle Charpentier and Jennifer A. Doudna.
CRISPR locus, identified in E. coli by Ishino et al. (1987), was later detected in many bacteria and archaea. It has multiple repeats of ~30 bp with a spacer of ~36 bp in between the repeats. The spacer sequences are unique whereas the repeats are the same and palindromic in nature. CRISPR in association with other adjacent loci including Cas9 (CRISPR/Cas9) carries out the gene editing function. This acts as an adaptive immune system (Jinek et al. 2012), protecting the microbes from the invasion of virus and plasmids through Cas9, a DNA endonuclease digesting the invading DNA, similar to the spacer sequence in the CRISPR locus (for convenience Streptococcus pyogenes CRISPR/Cas9 is discussed as a model). The Cas9 endonuclease has two DNA cleavage domains, HNH and RuvC responsible for double strand breaks in DNA. For specificity of recognition, DNA requires two RNAs, crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA). crRNA is transcribed from the CRISPR locus as precursor crRNA whereas the tracrRNA is transcribed from an adjacent locus. When the crRNA and tracrRNA bind to their complementary regions, it is processed by RNAse III and Cas9 to make it a shorter gRNA (guide RNA). Interestingly, the crRNA has NGG sequence at the 3′ end, called protospacer adjustment motif (PAM) with a 20 bp upstream sequence. When the gRNA (crRNA + tracrRNA) and Cas9 complex binds to the complementary 20 + NGG sequence in the DNA, the Cas9 nicks at 3 bp before the PAM by HNH domain in the complementary strand, whereas the RuvC domain cleaves the opposite strand in the same position, producing blunt ends. Both domains of the Cas9 nicks at the 3′ end of the DNA. CRISPR/Cas9 can be used to generate double strand DNA breaks provided the complementary sequence 20 + NGG is known. Precise breakage points may also be determined, as nicking by the Cas9 is exactly 3 bp upstream of the PAM i.e. NGG. Later, it has been shown that crRNA and tracrRNA can be linked together to form a single guide RNA (sgRNA), possessing two critical structures, one at the 5′ end of the DNA target and the second being a duplex structure that binds to the Cas9 at the 3′ end. Further, it has also been shown that rather than allowing transcription of the individual RNAs in vivo it is now possible to synthesize the sgRNA of ~100 bp lengths with same function asgRNA. In the late 1980s, it has been shown that DNA segments can be transferred to a desired location by homologous recombination using embryonic stem cell technology (review by Capecchi 2005). However, the frequency of such transformations is found to be very low. In the mid-1990s, results from studies in yeast and mammalian systems have shown that the frequency of homology-directed repair (HRD) and non-homologous end joining (NHEJ) could be increased several folds if the desired region of DNA has a double strand break (Haber 2000; Jasin and Rothstein 2013). The gRNA-Cas9 causes double strand breakage which triggers the DNA repair mechanisms thereby resulting in (1) complete error free joining of the strand or (2) addition or deletion of base pairs equivalent to NHEJ or (3) HDR in the presence of suitable homology containing replacement. Besides, when two gRNA-Cas9 complexes are used, large-scale deletions can be generated. If the complexes are used along with a segment of DNA, an inversion or replacement of the desired segment may be possible. The replacement of a segment of DNA (gene) is the basis of gene repair/correction/therapy which has now completely revolutionized the field of molecular genetics.
The question that now arises is whether such a tool can be used universally for eukaryotic organisms or not. In zebra fish, it has been shown that gRNA and Cas9 encoding RNA when co-injected into fertilized eggs, result in target specific mutations with a frequency that varied between 10% and 53% (Hwang et al. 2013), On the other hand, Gratacap et al. (2020) successfully used CRISPR/Cas9 for integration at two loci, EGFP and RIG-I separately in CHSE-214 cell line derived from Chinook salmon (Oncorhynchus tshawytscha) using lentivirus vector delivery system. The frequency of integration varied between 43% and 70% under different conditions. Zhang and his group modified the Cas9 amino acid codons with mice specific codons and showed that gene editing could be done in mice with such modified Cas9 and gRNA (Cong et al. 2013). Similarly, Church and his group used CRISPR technology in human cell lines to demonstrate proof of principle of gene editing (Mali et al. 2013). Adult mice with a mutated liver enzyme were corrected through targeted delivery by hydrodynamic tail vein injection of sgRNA and Cas9 RNA conjugated with a liver specific protein (Yin et al. 2014). Transgenic mice carrying a human DMD (Duchene muscular dystrophy) gene could be corrected by Adeno virus associated CRISPR/Cas9 gene editing system delivered by injecting via the intra-peritoneal (post natal Day 1) intra-muscular (post natal Day 12) or retro-orbital (post natal Day 18) routes (Long et al. 2015).
sgRNA-Cas9 complex can be used for various purposes and only a few are listed below which may be useful for the aquaculture industry.
-
1.
Cas9 is a 160 kDa protein that requires both its HNH and RuvC domains for its function of double strand breakage of DNA. Even if both domains are mutated, the sgRNA-Cas9 complex can still bind and remain bound to the specific DNA without creating DNA breakage. This property can be used to determine gene function, as RNA polymerase will be stalled at that site.
-
2.
Single strand breakage can be generated by mutating any of the domains of Cas9. If HNH domain is mutated, the non-complementary strand of the DNA will be nicked and the opposite result may be obtained if RuvC is mutated. Therefore, single strand breakages at specific regions can be generated in a known DNA segment.
-
3.
Visualization of a gene function is possible in vivo by the localization of fluorescently labelled sgRNA-Cas9 binding to DNA.
-
4.
Interestingly, by using several gRNA and Cas9 simultaneously i.e. by multiplexing, it is possible to edit several genes in the same cell/organism.
-
5.
Mutagenic chain reactions can be done to produce homozygous mutation from heterozygous conditions (Gantz and Bier 2015).
-
6.
Gene drive in insects (Gantz et al. 2015) and mammals Grunwald et al. (2019) can be studied.
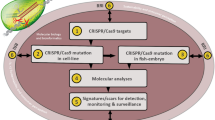
1.5 Genome Editing in Aquaculture Species
CRISPR/Cas9-based genome editing has been successfully applied in several major food fish species and in Pacific oyster for changing certain commercial characteristics viz., growth, and pigmentation, control of reproduction, sterility, immunogenicity enhancement, disease resistance and pigment formation (Gratacap et al. 2019). The species studied were Rainbow trout, Atlantic salmon, Tilapia, Channel catfish, Southern cat fish, Sea bream, Common carp, Indian and Chinese carps, Northern Chinese lamprey, Shrimp species and Oyster following the methods and studies that were initiated in the model fish (zebra fish) to establish proof of principle. Typically, mRNA encoding the Cas9 protein was injected together with the sgRNA at a single cell stage of development, to demonstrate high efficiency editing in various fish species. The Cas9 protein (instead of the mRNA) along with the gRNA was shown to be equally effective in gene editing is fishes.
1.5.1 Growth Enhancement
A mention is made of only a few genes whose functions when disrupted cause promotion of growth, which is one of the major criteria of aquaculture production. Myostatin (MSTN), also known as growth differentiation factor 8 (GDF8), is a member of the transforming growth factor-beta (TGF-beta) family that functions as a negative regulator of skeletal muscle development and thereby the overall growth of the fish (Lee 2004). Because of this role of MSTN, any natural mutations rendering it inactive and disrupting the physiological function of the protein are associated with robust muscle growth as seen in the Belgian “double bull” muscle phenotype in cattle (McPherron and Lee 1997) and in mice (McPherron et al. 1997). The conservation of the regulatory function of MSTN in muscle growth from fish to mammals (Khalil et al. 2017) is very promising to design strategies capable of inhibiting the expression of this gene for enhancing growth as shown in numerous studies on aquaculture production (Proudfoot et al. 2020).
Gao et al. (2016) used TALEN in zebra fish to generate homozygous MSTNb deficiency which resulted in 21% higher weight in comparison with the control group. Histological studies indicated increased number of muscle fibres in the MSTNb deficient fish. Wang et al. (2018) mutated both MSTNa and MSTNb by CRISPR/Cas9 in zebra fish. Mutant fish that were homozygous for MSTNb showed 30% higher body weight when compared to the control, whereas mutations in MSTNa has no effect on the body weight. However, when both genes were mutated, immunity of the fish was affected.
Common carp (Cyprinus carpio) is a very widely cultured aquaculture species. It has four MSTN genes as it is a tetraploid species. Of these four genes, only MSTNa is expressed in most of the tissues. sgRNA specific for MSTNa along with Cas9 mRNA when injected into the embryo, the mutation rate was found to be more than 70%. Growth with respect to body weight and length parameters was measured at 1, 2 and 3 months of the age. Both parameters showed increase at all-time points of measurement. Histological analysis indicated both hyperplasia and hypertrophy of the muscle cells which correlated with higher growth (Zhong et al. 2016).
In Channel catfish (Ictalurus punctatus), the exon one of the MSTN gene was targeted for CRISPR/Cas9 mutagenesis by Khalil et al. (2017). The hatchability after injection was found to be 42%, survivability at feeding stage was 90% and mutation frequency varied between 88% and 100%. The growth and the length of the fry on 40th day was 30% and 7% higher, respectively in the mutated group when compared to the control group.
Growth hormone (GH) secreted from the anterior pituitary, controls growth in vertebrates including fishes. Secretion of GH is a highly regulated process. Somatostatin (SST), a tetra-decapeptide discovered in sheep hypothalamus, was found to be a physiological inhibitor of GH secretion. Mammals have a single SST gene whereas teleost possess six SST encoding genes which exhibit differential tissue expression pattern. Zebra fish with SST4 mutants were obtained by the CRISPR/Cas9, showed its effects on growth and gonadal development. The SST4 knockout zebrafish grew significantly faster and was heavier at the onset of gonadal maturation when compared to the wild type (Sui et al. 2019).
1.5.2 Colour Modification
In aquaculture species, body colour is an important phenotype both from evolutionary and commercial points of view. For example, the Tilapia a group of commercially important fish is cultivated in many tropical countries. Normally their body colour is dark, though red/pale coloured strains (Majumdar et al. 1997) are also available which fetch a price that is at least three times more than the dark coloured one. Therefore, colour alteration in fishes is a good commercial proposition.
Jao et al. (2013) analyzed pigmentation morphology in zebra fish. They chose three genes, tyrosinase (tyr), golden (gol-the putative cation exchanger Slc24a5) and mitfa (required for neural crest-derived melanocyte formation) for manipulation. The tyr gene produces pigmentation in the skin, whereas the gol gene shows hypopigmentation in both retina and skin. CRISPR/Cas9 gene editing in tyr and gol gene showed more than 90% mutation and mitfa gene had 80% mutation which was confirmed by sequence analysis. The phenotype in both tyr and gol mutated groups showed hypopigmentation although mosaic in its distribution but the effect was retained throughout its life, indicating that it was not an epigenetic effect. No alteration was seen in pigmentation pattern in mitfa-mutated group.
Edvardsen et al. (2014) selected two pigment forming genes for mutation by CRISPR/Cas9 gene targeting method in Atlantic salmon (Salmo salar). These were tyrosinase (tyr) and solute carrier family 45, member 2 (slc45a2). Mutation screening with respect to skin pigmentation was done at the 17 somite stage. Embryo injected with slc45a2 and tyr showed 40% and 22% mutations, respectively. Hatched embryos without pigmentation were subjected to PCR and sequencing to confirm the presence of mutation in the respective genes, slc45a2 and tyr.
Fang et al. (2018) mutated tyrosinase gene by CRISPR/Cas9 (sgRNA-specific tyrosinase and mRNA of Cas9 were injected into fertilized eggs) method in orange red medaka (Oryzias latipes), an aquarium fish used as a model organism. They found more than 90% mutation in F0 and F1 progenies leading to an albino phenotype.
In China, the white variety of Crucian carp (Carassius auratus cuvieri) is a highly prized fish. Liu et al. (2019) mutated tyrosinase gene in Carassius auratus cuvieri and the hybrid (Carassius auratus cuvieri female X Carassius auratus male—red variety) by CRISPR/Cas9, resulting in 60–90% mutation. The phenotype varied from complete albino to mosaic pigmentation of melanin. The mutated fish had reduced tyrosinase concentration. It was also noticed that several pivotal genes in the pathway of melanin formation (e.g. tyrp1, mitfa, mitfb, dct and sox10) were down regulated in the mutated fishes, thus causing reduced pigment formation.
1.5.3 Control of Reproduction
Fish is the most specious group of vertebrates. They possess various forms of sexuality e.g. hermaphroditic, gonochoristic, protandrous, protogynous, alpha male or female. Similarly, the sex determination mechanism also varies considerably e.g. by genotypic (monogenic or polygenic), chromosomal, environmental conditions or by social interactions (mainly dominance). Interestingly, the sex phenotype can be modulated by environmental or chemical means (especially hormones), as the sex is not always determined at the time of fertilization as in the case of mammals, therefore, it has plasticity which can be used for beneficial purposes. Besides, many fish species show differential growth rates and sexual dimorphisms. From a commercial point of view, this can be exploited, provided sex phenotypes can be modulated by external means. A few such examples will be discussed below where gene editing technique has been successfully used to change the sex of individuals in a species.
It is known that gonadal steroids play an important role in sex determination of fish. Aromatase plays an important role in steroidogenesis by converting androgens to estrogens, which plays a critical role in the development of the ovary. By inhibiting aromatase, it was shown that the fish changed its sex from female to male. Treatment of zebra fish with an aromatase inhibitor during gonadal differentiation resulted in small-male population (Fenske and Segner 2004). Sex reversal of adult male zebra fish to female was also possible by treating it with an aromatase inhibitor (Takatsu et al. 2013). There are two aromatase genes (CYP19) in fish, gonad-specific cyp19a1a that expresses in the ovary and brain-specific cyp19a1b (Zhang et al. 2014). In zebrafish, Lau et al. (2016) generated two different mutants in the exon 1 of cyp19a1a by CRISPR/Cas9. When a comparison of sex ratios was made between homozygous wild, heterozygous mutant and homozygous mutants, no difference was seen in the sex ratios in the homozygous wild and the heterozygous mutant which was about 50% males: 50% females, whereas in both the homozygous mutants, it was all male. Histological analysis confirmed the gonadal morphology. When homozygous mutant males were treated with estradiol they reverted back to the female phenotype.
Chinese half tongue sole (Cynoglossus semilaevis), a marine flat fish cultured commercially in China, has a ZZ male and ZW female sex determination mechanism. Though this fish has a distinct sex determining chromosome, it was seen that around 14% of ZW females changed to physiological males under normal conditions, the percentage could increase up to 73% when they were grown at higher temperature like 28 °C. It is known that dmrt1 is a male determining gene, located in the Z chromosome. Cui et al. (2017) used TALEN derived gene editing technique to mutate exon I of dmrt1 gene in this fish, resulting in a mutation rate of 55%. The ZZ males turned to females and the ZW females became intersex with respect to their sex phenotype. The sex reversed females showed higher expression of female specific genes foxl2 and cyp 19a1a and a lower expression of male specific genes Sox9a and Amh. In this fish, the females were larger in size and grew faster than the males. The dmrt1 mutated sex changed females grew faster than the males and were almost equivalent to wild type females in their rate of growth. Therefore, it can be seen from these examples that sex change has an added advantage of higher growth rates in a particular sex, resulting in commercial benefits to the cultivators.
Li et al. (2014) generated mutations in Oreochromis niloticus by CRISPR/Cas9 in nanos2 (38%), nanos3 (49%), foxl2 (42%) and dmrt1 (22%) genes. The mutated G0 population of foxl2 and dmrt1 genes transmit the mutations to the F1 generation. Nanos3 mutation in the XX female Tilapia shows germ cell deficiency and sex reversal. These individuals do not express ovary specific cyp19a1a gene. Nanos2 mutation in XY male shows germ cell deficiency in the testis but does not indicate sex reversal. Dmrt1 mutation causes male to female sex reversal and induces aromatase expression. Similarly, foxl2 mutation induces reduction of aromatase and causes sex reversal in XX females, similar to the mutation generated in the same gene using TALEN by Li et al. (2013).
1.5.4 CRISPR/Cas9 in Shrimp and Oyster
In shrimp species Exopalaemon carinicauda, Zhang et al. (2018) isolated moult inhibiting hormone (MIH) possessing three exons and two introns, which is a negative regulator for suppressing the moulting process, thus reducing the growth rate. This gene is expressed mainly in the eyestalks. Mutation at the exon two in the MIH gene is generated by CRISPR/Cas9 with a frequency of 4.8%. The mutated larvae show body length increase and shortening of metamorphosis time from mysis to post larva stage, without any ill effects on health or death rate, indicating the possibility of using this technique in the breeding of this shrimp. In the case of a bivalve mollusc—the Pacific oyster, Crassostrea gigas mutations were generated successfully by CRISPR/Cas9 in two genes, myostatin and twist (Yu et al. 2019). This shows the versatility of the CRISPR/Cas9 gene editing technology.
1.5.5 Disease Resistance
Around 40% loss occurs in aquaculture production because of infectious diseases. Disease resistance phenotypes are generally polygeneic in nature and do not show high heritability. To overcome these obstacles, Gratacap et al. (2019) provided a detailed flow chart to combine gene editing and classical selective breeding to obtain disease resistant aquatic species of commercial importance.
1.5.6 Clearance from Regulatory Authority
The time taken to obtain regulatory clearance for transgenic fish is rather long. Fortunately, gene edited plants and fish have been given regulatory clearances much sooner than is usually the case, as these are classified as non-GMO. It is clear that in-del mutation, generated by CRISPR/Cas9 in an individual, will be considered as non-GMO. It will be interesting to see whether the replacement of a gene or part of it, obtained through CRISPR technology, may follow the same path or not.
Button mushroom (Agaricus bisporus) after harvest gets discoloured from white to brown within a few days, thus, losing market value. This browning is caused by a group of enzymes called polyphenol oxidases (PPO). Yinong Yang, a plant pathologist of Pennsylvania state university, USA mutated one of the six genes of PPO by CRISPR/Cas9 gene editing tool. This reduced the activity of the PPO enzyme by 30% preventing browning and thereby, increasing the shelf life of the mushroom. Waltz (2016) reported in “Nature” that “the USDA will not regulate a mushroom that has been genetically modified with gene editing tool CRISPR/Cas9 and it can be cultivated and sold without passing through the agency’s regulatory process-making it the first CRISPR edited organism to receive a green light from the US government”. It was also mentioned that Yang’s mushroom did not trigger USDA oversight because it did not contain foreign DNA such as viruses or bacteria (Waltz 2016), therefore, the crop does not pose any health risk. In response to this development Dr Joyce Van Eck, Assistant Professor at Boyce Thompson Institute mentioned if USDA decided that this did not require regulation, it would definitely encourage many people already using CRISPR technology.
In December 2018, in a press release from two companies, Intrexon and Aquabounty; it was reported that the Tilapia strain FLT 01 developed by them by a gene editing tool would not be considered as GMO by National Advisory Commission on Agricultural Biotechnology, Argentina (CONABIA). This was the first gene edited animal to get a regulatory clearance after it was designated as non-GMO. The researcher claimed that the fish has 70% more fillets and showed a 16% higher growth rate with a 14% increase in food conversion ratio. Dr Martin Lema (Chairman of CONABIA, Biotechnology Directorate, State Secretariat of Foodstuff and Bioeconomy in Argentina) mentioned in a paper presented in the symposium at the International Society for Biosafety Research held in Terragona, Spain in 2019, that Gene edited Tilapia line FLT 01 trait, improved fillet yield through loss of function mutation; microinjection with nuclease mRNA (no RNA involved); small deletion created an early stop codon; homologous at final product; cero off target site by design; seven low probability off target sites were verified to be non-modified by sequencing; and no risk hypothesis reported.
However, with regard to gene editing in the case of humans, several unanswered questions remain. The organizing committee on First International Summit on Human Gene Editing mentioned that CRISPR and other gene editing techniques were to be used for somatic cell based therapies that were “intended to affect only the individuals who receive them”. It would be irresponsible to proceed with any clinical use of germ line editing unless and until (1) the relevant safety and efficacy issues have been resolved, based on appropriate understanding and balancing of risks, potential benefits and alternatives, and (2) there is broad societal consensus about appropriateness of the proposed application (Thrasher et al. 2016).
1.6 Micro RNA
The discussion below on micro RNA is based on the following articles and therefore, individual citations have not been made in several places in the text: Wahid et al. (2010), Bizuayehu and Babiak (2014), Rasal et al. (2016), Schier and Giraldez (2016), Andreassen and Hoyheim (2017), Herkenhoff et al. (2018).
In 1993, a small RNA lin4, isolated from C. elegans (Lee et al. 1993) was shown to regulate the function of lin14 gene (Wightman et al. 1993). This initiated the isolation and characterization of a group of small RNAs, called microRNA (miRNA) from various species of plants, animals and viruses whose numbers are increasing exponentially. The database called miRBASE (Released 22 December 2018) has data from 271 species, including nine teleost fish with 38,589 entries of hairpin precursor miRNAs which may give 48,885 mature miRNA. miRNAs are shown to function in the developmental processes, neuronal pathway formation, cell proliferation, cell death, muscle formation and growth, regulation of metabolic pathways, haematopoietic differentiation, immunity, reproduction and response to environmental stimuli in an organism. Thousands of miRNAs have been isolated and characterized and they have shown to be conserved between species. Sometimes the genes for the miRNAs may occur in clusters and such clusters also show conservation in different vertebrates. For example, 26 miRNA gene clusters are conserved in Atlantic salmon, Atlantic Cod, Zebra fish and humans.
Among the teleost fishes, miRNA was first discovered in zebra fish (Lim et al. 2003). The miRNA is about 21–25 nucleotides long single stranded RNA, produced from a precursor RNA. The transcribed products of these miRNA genes have the capacity to form a hairpin like structure. Initially, miRNA genes were identified in the inter-generic regions but later it was also found in the intronic regions of the gene. Generally, these genes are clustered in a region of the genome that is transcribed as a polycistronic message by the same promoter; however, single miRNA genes have also been located in areas that give monocistronic messages. Depending on their location in the genome, miRNAs are classified into the following categories: (a) intronic miRNA in coding transcription unit (TU) e.g. miRNA 101-2 cluster which is found in the intron of a coding RNA gene HSPC338 (b) intronic miRNA in noncoding TU e.g. miRNA 135a-2 cluster in the gene NCRMS (c) exonic miRNA in coding TU e.g. miRNA985 in the gene CACNG8 (d) exonic miRNA in noncoding TU e.g. miRMA 206 in the gene 7H4. Most of the miRNA are transcribed by RNA pol II with the exception of a small group in Alu repeats transcribed by RNA pol III in the nucleus. The transcripts, produced by both the polymerases, may go up to several Kb in length and are called primary miRNA (Pri-miRNA), possessing a stem-loop-stem structure with the following three defining features: terminal loop, internal bulges and a double stranded stem. The pri-miRNA is cleaved by Drosha in the presence of cofactor DiGeorge Syndrome Critical region gene 8 (DGCR8) in human and Pasha in Drosophila and worm, to get a strand of about 60–70 nucleotides long comprising of about 33 base pairs of the stem loop structure, a terminal loop and single strand RNA as a precursor miRNA (Pre-miRNA) with a 2–3 nucleotide overhang in the 3′ region. Interestingly, the Drosha marks the 5′ region of the mature miRNA. The pre-miRNA is transported from the nucleus to the cytoplasm through the nuclear pore by RanGTP dependent exportin-5 (EXP5), a nuclear transport receptor. In the cytoplasm, RNase III and Dicer a conserved enzyme cleaves the Pre-miRNA near the terminal loop of the Drosha cutting site giving rise to ~22 nucleotides long double stranded miRNA. This results in two mature miRNA molecules, one from the 5′ and the other from the 3′ end of the pre-miRNA. Dicer along with Argonaute and other proteins viz TRBP and PACT are responsible for miRNA processing and finally loading into the RNA induced silencing complex (RISC) by forming miRNA-RISC. A helicase yet to be discovered will be found to be responsible for the separation of the duplex RNA. One of the strands, the mature miRNA strand remains with Argonaute whereas the other strand, the passenger miRNA is degraded. It is also shown that endonucleolytic enzyme activity of Argonaute is responsible for the removal of the passenger miRNA. There are exceptions where both strands are found to be functional. Of the two strands, which one will be functional is to a certain extant determined by the thermodynamic stability of the strand. The miRNA function can only be initiated once the miRNA-RISC is formed which either (1) inhibits the expression or (2) causes degradation of the mRNA transcript in the cytoplasm. To perform these functions, the miRNA-RISC may bind in the 3′UTR of the mRNA where in most of the cases the miRNA recognition elements (MRE) are predominantly located. It is also shown that it may bind to the exonic region or the 5′UTR of the mRNA. The mRNA regulation depends on the amount of complementarity of the miRNA seed region, a domain spanning nucleotide positions 2–7 at the 5′ of the miRNA and the MRE of the mRNA. Therefore, a small number of miRNA can regulate many mRNAs. Based on this property, it is calculated that 60% of the protein coding genes in mammals may be regulated by miRNAs and its final role may be gene function regulation. This has both an advantage and a disadvantage while determining the assignment of a specific miRNA. The easy sequencing strategies available at present for RNA and DNA help in identifying a large number of miRNA in a short span of time. Once identified, one needs to find out the site where these will bind on the mRNAs. Identification of such sites is possible using bioinformatics tools. The whole process depends on the availability of genome sequence and transcriptome data of a particular species. As we are aware that more than 25,000 species of fish are known whereas genome sequence data and transcriptome data are not available, even for all important food fish species, which is one of the drawbacks for studies on miRNA biology in fishes. As mentioned earlier, single miRNA may have complementary sequences in several places in a single mRNA. It is also seen that the same miRNA may have complementary sequences in different mRNAs, originating from different genes/species. To find out the correct miRNA and its true binding site, binding analysis and inhibition of the gene as the regulatory function of a particular miRNA need to be done in an in vitro analysis which is a time consuming exercise.
Only a few examples have been chosen in this chapter where the role of miRNA is evident on a few genes affecting particular phenotypes, although a huge amount of information available in literature.
1.6.1 miRNA Usage, Colour Modulation
Experimental data from mice and Drosophila established the role of miRNA in body colour pattern. Common carp (Cyprinus carpio) has several body colours viz, red, white, orange and black which follows Mendelian inheritance. Similarly, Nile tilapia (Oreochromis niloticus) also has red and white body colours. It is shown that four miRNAs (miRNA 25, miRNA 137, miRNA 203a and miRNA 429) are strongly expressed in the skin of both the fishes. When miRNA 429 antagomir was injected to red coloured live fishes (5 g body weight) each day for 30 days the miR 429 got silenced and a reduction of the melanin content was seen in the skin. In silico analysis showed that 3′UTR region of Foxd3 has a potential target site for miRNA 429. In vitro study showed that Foxd3 gene activity was reduced in HEK2BT cells when transfected with miRNA 429. In vivo study with miRNA 429 antagomir injected into fishes showed an increase of Foxd3 activity. These studies indicated that Foxd3 gene was regulated by the interaction of miRNA 429 and the 3′UTR of Foxd3 mRNA. It is known that melanocyte inducing transcription factor (MITF) gene regulates TYR, TRP1 and TRP2 genes which are involved in melanin production. Foxd3 product binds to the promoter of MITF gene and represses its function. Down regulation of miRNA 429 increases Foxd3 gene expression and will down regulate MITF which in turn will down regulate the downstream genes, thereby, regulating melanin formation in the skin. Hence, the miRNA 429 was shown to regulate melanin formation in two different species of the fish (Yan et al. 2013a).
1.6.2 Growth Studies
MyoD, a member of the helix loop helix transcription factors, is known to play a great role in controlling myogenic regulatory pathway. Yan et al. (2013b) isolated several miRNAs (miRNA 23b, miRNA190, miRNA190b, miRNA 203b) from the skeletal muscle of Nile tilapia (Oreochromis niloticus). MyoD is highly expressed in juvenile fish, it is hardly detectable in adult fish but expressed again moderately during the senile phase of the fish. However, miRNA 203b expression is lowest in the juvenile fish, highest in the adult fish and moderate in the senile fish. When fishes were treated with miRNA 203b antagomir, it significantly reduced miRNA 203b expression. Besides, the miRNA 203b has a potential target in the 3′UTR of MyoD. These data indicate that miRNA 203b regulates negatively the function of MyoD gene, thereby, the myogenic pathway in this fish (Yan et al. 2013b).
Myostatin (mstn) gene negatively regulates the development of muscle in most vertebrates including fish. In Nile Tilapia (Oreochromis niloticus) two variations of mstn are found viz. mstna and mstnb. The mstnb gene is expressed in different tissues including the muscle, whereas mstna is expressed only in the brain. It is shown that miRNA-181b-5p is expressed in different tissues including white muscle. It binds to the 3′UTR of the mstnb, specifically in two regions, 307–313 bp and 1022–1029 bp. Overexpression of miRNA 181b-5p decreases MSTNb protein, whereas knockdown of the same causes upregulation of the protein. In conclusion, it was mentioned by Zhao et al. (2019) that miRNA-181b-5p decreased MSTN b protein production, thereby promoting muscle growth in Tilapia.
1.6.3 Cold Adaptation
Litopenaeus vannamei, a tropical shrimp, when grown in water at lower temperatures, parameters like survivability and growth are affected. He et al. (2018) analyzed miRNAs related to cold adaptation in this species. In the cold treated group, 34 miRNAs (21 up regulated and 13 down regulated) were differentially expressed in comparison to the shrimps grown at a warmer temperature (28 °C). In another group, where the shrimps were kept first at 16 °C (cold adapted) for 6 days and then adapted back to 28 °C showed 31 miRNAs (16 up regulated and 15 down regulated) differentially expressed, when compared to the shrimp grown at 28 °C. Validation of five miRNAs (three known miRNAs, mja-miR-6491, mja-miR-6494, bta-miR-2478 and two newly identified, novel-68 and novel-5) was done that were differentially expressed significantly in the hepatopancreas and muscle tissues by RT-qPCR. Of these three miRNAs, mja-miR6491, mja-miR-6494 and novel-5 were upregulated in both hepatopancreas and muscle, whereas bta-miR-2478 and novel-68 were expressed more in the muscle in the cold treated group. Pathway analysis of these miRNAs showed that target genes were mostly related to fatty acid metabolism which was expected in the cold adapted individual for regulating membrane fluidity.
To identify miRNAs involved in cold adaptation, Ji et al. (2020) grew zebra fish embryonic cells (ZF4) at 18 °C for 30 days. Cells grown in cold had 24 up regulated (19 known and five novel) and 23 down regulated (nine known and 14 novel) miRNAs in comparison to the cells grown at 28 °C. It was noted that the selected four up regulated miRNAs (dre-miR-16b, dre-miR-729, dre-miR-338-5p, dre-miR-193b-5p) and four down regulated miRNAs (dre-miR-100-3p, dre-miR-100-5p, dre-miR-2184, dre-miR-19a-3p), reverted back to their normal expression when cells were shifted back to 28 °C culture condition, indicating the adaptive nature of these miRNAs. miR-100-3p down regulated genes atad5a, cyp2ae1 and lamp1, whereas miR-16b targets genes rilp, atxn7, tnika and btbd9 by reducing their function, indicating their role in cold adaptation (Ji et al. 2020).
1.6.4 Control of Reproduction
In many vertebrate species, miR-202 is expressed predominantly in the gonads of both the sexes. Gay et al. (2018) have shown in medaka that mature miR-202-5p is present in large quantities in granulosa cells of the ovary and in unfertilized eggs. When they knocked out the miR-202 gene by CRISPR-Cas9 method to determine its role in gonadal function, the mutated females showed no egg production or produced dramatically reduced number of eggs that could not be fertilized, thereby, causing sterility in the female medaka.
1.6.5 Clearance from Regulatory Authority
Research activities in miRNA are moving at a very fast pace, especially, in the areas of diagnostics and therapeutics in humans. The first miRNA-based therapeutic drug was discovered by Lindow and Kauppinen (2012); a locked nucleic acid (LNA) modified antisense oligonucleotide “Miravirsen” which targets miRNA 122 expressed in the liver. Clinical trials are currently going on for the treatment of hepatitis C virus infection by Miravirsen in humans (Chakraborty et al. 2017). In 2015, Inter space Biosciences Inc, USA launched for the first time, a microRNA based gene expression classifier, “Thyromir” to diagnose benign and malignant thyroid cancer in humans. Unfortunately, as mentioned in Bonneau et al. (2019) “all the miRNA based drugs are currently in clinical trials and none have yet reached the regulatory approval stage for pharmaceutical breakthrough”. In such a scenario, regulatory approval of diagnostic and therapeutic usage of miRNA-based technology in aquaculture may not happen in the near future.
1.7 Conclusion
It is abundantly clear that aquaculture productivity has increased several folds in the last few decades due to advancement in classical technologies like improved water quality management, food/feed development and better stock development through selective breeding. However, the application of modern molecular techniques like transgenesis, gene targeting and microRNA can revolutionize productivity in intensive aquaculture (both fresh and salt water) by reducing the time period for growth, improving feed intake, lowering the feed conversion ratio and enhancement of stock by directly modifying the gene(s) responsible for growth, disease resistance or control of reproduction. All these technologies will reduce the production cost substantially and thereby provide a cheaper protein even for the economically weaker sections of the population. It is also seen that important pharmaceutical products can be made using aquatic organisms as bioreactors. Considering all these potentially beneficial aspects, it is desirable for the aqua culturists to adopt newer technologies that are being developed by the scientists and technologists to enhance aquaculture productivity and production.
References
Amsterdam AS, Lin S, Hopkins N (1995) The Aequorea victoria green fluorescent protein can be used as a reporter in live zebra fish embryos. Dev Biol 171:123–129
Andreassen R, Hoyheim B (2017) miRNA associated with immune response in teleost fish. Dev Comp Immunol 75:77–85
Bambino K, Chu J (2017) Zebra fish in toxicology and environmental health. Curr Top Dev Biol 124:331–367. https://doi.org/10.1016/bs.ctdb.2016.10.007
Barrangou R, Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34:933–941. https://doi.org/10.1038/nbt.3659
Beardmore JA (1997) Transgenics: autotransgenics and allotransgenics. Transgenic Res 6:107–108
Beardmore JA, Porter JS (2003) Genetically modified organisms and aquaculture. In: FAO fisheries circular No. 989. FAO, Rome. 38 p
Bizuayehu TT, Babiak I (2014) MicroRNA in teleost fish. Genome Biol Evol 6:1911–1937
Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V (2019) How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. JIFCC 30:114–127
Buono RJ, Linser PJ (1992) Transient expression of RSVCAT in transgenic zebrafish made by electroporation. Mol Mar Biol Biotechnol 1:271–275. PMID: 1339227
Capecchi MR (2005) Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 6:507–512
Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee S-S (2017) Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucl Acids 8:132–143
Chen TT, Lu JK, Shamblott MJ, Chench CM, Lin CM, Burns JC, Reimschuessel R, Chatakondi N, Dunham RA (1995) Transgenic fish: ideal models for basic research and biotechnological applications. Zool Stud 34:215–234
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Cui Z, Liu Y, Wang W, Wang Q, Zhang N, Lin F, Wang N, Shao C, Dong Z, Li Y, Yang Y, Hu M, Li H, Gao F, Wei Z, Meng L, Liu Y, Wei M, Zhu Y, Guo H, Cheng CHK, Schartl M, Chen S (2017) Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep 7:42213. https://doi.org/10.1038/srep42213
Devlin RH, Raven PA, Sundstrdnt LF, Uh M (2009) Issues and methodology for development of transgenic fish for aquaculture with a focus on growth enhancement. In: Overturf K (ed) Molecular research in aquaculture. Wiley-Blackwell, London, pp 217–260
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346:1258096
Dunham RA, Eash J, Askins J, Townes TM (1987) Transfer of the metallothionein–human growth hormone fusion gene into channel catfish. Trans Am Fish Soc 116:87–91
Dunham RA, Ramboux AC, Duncan PL, Hayat M, Chen TT, Lin CM, Kight K, Gonzalez-Villasenor I, Powers DA (1992) Transfer, expression, and inheritance of salmonid growth hormone genes in channel catfish, Ictalurus punctatus, and effects on performance traits. Mol Mar Biol Biotechnol 1:380–389
Dunham RA, Warr G, Nichols A, Duncan PL, Argue B, Middleton D, Liu Z (2002) Enhanced bacterial disease resistance of transgenic channel catfish, Ictalurus punctatus, possessing cecropin genes. Mar Biotechnol 4:338–344
Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A (2014) Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS One 9:e108622. https://doi.org/10.1371/journal.pone.0108622
Fang J, Chen T, Pan Q, Wang Q (2018) Generation of albino medaka (Oryzias latipes) by CRISPR/Cas9. J Expt Zool B Mol Dev Evol 330:242–246
FAO (2020) The State of World Fisheries and Aquaculture 2020. Sustainability in action. FAO, Rome. https://doi.org/10.4060/ca9229en
Fenske M, Segner H (2004) Aromatase modulation alters gonadal differentiation in developing zebrafish (Danio rerio). Aquat Toxicol 67:105–126
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405. https://doi.org/10.1016/j.tibtech.2013.04.004
Gantz VM, Bier E (2015) The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348:442–444
Gantz VM, Jasinskieneb N, Tatarenkovab O, Fazekasb A, Maciasb VM, Biera E, Jamesb AA (2015) Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A 112:E6736–E6743
Gao Y, Dai Z, Shi C, Zhai G, Jin X, He J, Lou Q, Yin Z (2016) Depletion of myostatin b promotes somatic growth and lipid metabolism in zebrafish. Front Endocrinol 7:88., 1–10. https://doi.org/10.3389/fendo.2016.00088
Gay S, Bugeon J, Bouchareb A, Henry L, Delahaye C, Legeai F, Montfort J, Le Cam A, Siegel A, Bobe J, Thermes V (2018) MiR-202 controls female fecundity by regulating medaka oogenesis. PLoS Genet 14:e1007593. https://doi.org/10.1371/journal.pgen.1007593
Gratacap RL, Wargelius A, Edvardson RB, Houston RD (2019) Potential of genome editing to improve aquaculture breeding and production. Trends Genet 35:672–684
Gratacap RL, Regan T, Dehler CE, Martin SAM, Boudinot P, Collet B, Ross D, Houston RD (2020) Efficient CRISPR/Cas9 genome editing in a salmonid fish cell line using a lentivirus delivery system. BMC Biotechnol 20:35–44
Grunwald HA, Gantz VM, Poplawski G, Xu X-r S, Bier E, Cooper KL (2019) Super-Mendelian inheritance mediated by CRISPR/Cas9 in the female mouse germline. Nature 566:105–109
Haber JE (2000) Lucky breaks: analysis of recombination in Saccharomyces. Mutat Res 451:53–69
He P, Wei P, Zhang B, Zhao Y, Li Q, Chen X, Zeng D, Peng M, Yang C, Peng J, Chen X (2018) Identification of microRNAs involved in cold adaptation of Litopenaeus vannamei by high-throughput sequencing. Gene 677:24–31
Herkenhoff ME, Oliveira AC, Nachtigall PG, Costa JM, Campos VF, Hilsdorf AWS, Pinhal D (2018) Fishing into the microRNA transcriptome. Front Genet 9:1–15
Hwang G, Müller F, Rahman MA, Williams DW, Murdock PJ, Pasi KJ, Goldspink G, Farahmand H, Maclean N (2004) Fish as bioreactors: transgene expression of human coagulation factor VII in fish embryos. Mar Biotechnol 6:485–492. https://doi.org/10.1007/s10126-004-3121-2
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Jao L-E, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 34:13904–13909
Jasin M, Rothstein R (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5:a012740
Jaynes JM, Julian GR, Jeffers GW, White KL, Enright FM (1989) In vitro cytocidal effect of lytic peptides on several transformed mammalian cell lines. Pept Res 2:157–160
Ji X, Jiang P, Luo J, Li M, Bai Y, Zhang J, Han B (2020) Identification and characterization of miRNAs involved in cold acclimation of zebra fish ZF4 cells. PLoS One 15:e0226905. https://doi.org/10.1371/journal.pone.0226905
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Ju B, Xu Y, He J, Liao J, Yan T, Hew CL, Lam TJ, Gong Z (1999) Faithful expression of green fluorescent protein (GFP) in transgenic zebra fish embryos under control of zebra fish gene promoters. Dev Genet 25:158–167. https://doi.org/10.1002/(SICI)1520-6408(1999)25:2<158::AID-DVG10>3.0.CO;2-6
Khalil K, Elayat M, Khalifa E, Daghash S, Elaswad A, Miller M, Abdelrahman H, Ye Z, Odin R, Drescher D, Vo K, Gosh K, Bugg W, Robinson D, Dunham R (2017) Generation of Myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 System. Sci Rep 7:7301. https://doi.org/10.1038/s41598-017-07223-7
Kim H, Kim JS (2014) A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–334
Kjuul AK, Bullesbach EE, Espelid S, Dunham RA, Jorgensen TO, Warr GW, Styrvold OB (1999) Effects of cecropin peptides on bacteria pathogenic to fish. J Fish Dis 22:387–394
Lander ES (2016) The heroes of CRISPR. Cell 164:18–28
Lau ES-W, Zhang Z, Qin M, Ge W (2016) Knockout of zebra fish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci Rep 6:37357. https://doi.org/10.1038/srep37357
Lee S-J (2004) Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol 20:61–86
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lee H-C, Lu P-N, Huang H-L, Chu C, Li H-P, Tsai H-J (2014) Zebrafish transgenic line huORFZ is an effective living bioindicator for detecting environmental toxicants. PLoS One 9:e90160. https://doi.org/10.1371/journal.pone.0090160
Lee O, Green JM, Tyler CR (2015) Transgenic fish systems and their application in ecotoxicology. Crit Rev Toxicol 45:124–141. PubMed: 25394772
Li MH, Yang HH, Li MR, Sun YL, Jiang XL, Xie Q-P, Wang T-R, Shi H-J, Sun L-N, Zhou L-Y, Wang D-S (2013) Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 154:4814–4825
Li M, Yang H, Zhao J, Fang L, Shi H, Li M, Sun Y, Zhang X, Jiang D, Zhou L, Wang D (2014) Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 197:591–599
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP (2003) Vertebrate microRNA genes. Science 299:1540
Lindow M, Kauppinen S (2012) Discovering the first microRNA-targeted drug. J Cell Biol 199:407–412. https://doi.org/10.1083/jcb.201208082
Liu Q, Qi Y, Liang Q, Song Q, Liu J, Li W, Shu Y, Tau M, Zhang C, Qin Q, Wang J, Liu S (2019) Targeted disruption of tyrosinase causes melanin reduction in Carassius auratus cuvieri and its hybrid progeny. Sci China Life Sci 62:1194–1202. https://doi.org/10.1007/s11427-018-9404-7
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN (2015) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351:400–403
Lu JK, Chen TT, Chrisman CL, Andrisani OM, Dixon JE (1992) Integration, expression and germ-line transmission of foreign growth hormone genes in medaka (Oryzias latipes). Mol Mar Biol Biotechnol 1:366–375. PMID: 1285009
Majumdar KC, Nasaruddin K, Ravinder K (1997) Pink body colour in Tilapia shows single gene inheritance. Aquac Res 28:581–589
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
McPherron AC, Lee S-J (1997) Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A 94:12457–12461
McPherron AC, Lawler AM, Lee S-J (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387:83–90
Müller F, Ivics Z, Erdélyi F, Papp T, Váradi L, Horváth L, Maclean N, Orbán L (1992) Introducing foreign genes into fish eggs with electroporated sperm as a carrier. Mol Mar Biol Biotechnol 1:276–281
Nam YK, Maclean N, Hwang G, Kim DS (2008) Autotransgenic and allotransgenic manipulation of growth traits in fish for aquaculture: a review. J Fish Biol 72:1–26
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Ozato K, Kondoh H, Inohara H, Iwamatsu T, Wakamatsu Y, Okada TS (1986) Production of transgenic fish: introduction and expression of chickenδ-crystallin gene in medaka embryos. Cell Differ Dev 19:237–244
Powers DA, Cole T, Creech K, Chen TT, Lin CM, Kight K, Dunham R (1992) Electroporation: a method for transferring genes into the gametes of zebrafish, Brachydanio rerio, channel catfish, Ictalurus punctatus, and common carp, Cyprinus carpio. Mol Mar Biol Biotechnol 1:301–309
Proudfoot C, McFarlane G, Whitelaw B, Lillico S (2020) Livestock breeding for the 21st century: the promise of the editing revolution. Front Agric Sci Eng 7:129–135
Rajesh R, Majumdar KC (2005) Transgene integration- an analysis in Auto transgenic Labeo rohita Hamilton (Pisces: Cyprinidae). Fish Physiol Biochem 31:281–287
Rasal KD, Nandanpawar PC, Swain P, Badhe MR, Sundaray JK, Jayasankar P (2016) MicroRNA in aquaculture fishes: a way forward with high-throughput sequencing and a computational approach. Rev Fish Biol Fish 26:199–212. https://doi.org/10.1007/s11160-016-9421-6
Sarmasik A, Warr G, Chen TT (2002) Production of transgenic medaka with increased resistance to bacterial pathogens. Mar Biotechnol 4:310–322
Schier AF, Giraldez AJ (2016) MicroRNA function and mechanism: insights from zebra fish. Cold Spring Harb Symp Quant Biol 71:195–203
Smith K (2019) Time to start intervening in the human germline? A utilitarian perspective. Bioethics 34:90–104. https://doi.org/10.1111/bioe.12691
Steiner H, Hultmark D, Engstrom A, Bennick H, Boman HG (1981) Sequence and specificity of two antibacterial proteins involved in antibacterial immunity. Nature 292:246–248
Stuart GW, McMurray JV, Westerfield M (1988) Replication, integration and stable germ-line transmission of foreign sequences injected into early zebra fish embryos. Development 103:403–412
Sui C, Chena J, Ma J, Zhaoa W, Canárioa AVM, Martinsd RST (2019) Somatostatin 4 regulates growth and modulates gametogenesis in zebrafish. Aquac Fisher 4:239–246. https://doi.org/10.1016/j.aaf.2019.05.002
Symonds JE, Walker SP, Sin FYT (1994) Electroporation of salmon sperm with plasmid DNA: evidence of enhanced sperm/DNA association. Aquaculture 119:313–327
Takatsu K, Miyaoku K, Roy SR, Murono Y, Sago T, Itagaki H, Nakamura M, Tokumoto T (2013) Induction of female-to-male sex change in adult zebrafish by aromatase inhibitor treatment. Sci Rep 3:3400
Tester M (1999) Seeking clarity in the debate over the safety of GM foods. Nature 402:575
Thrasher A, Baltimore D, Pei D, Lander ES, Winnacker E-L, Baylis F, Daley GQ, Doudna JA, Berg P, Ossorio P, Zhou Q, Lovell-Badge R (2016) On human gene editing: international summit statement by the organizing committee. Issue Sci Technol 32(3)
Tonelli FMP, Lacerda SMSN, Tonelli FCP, Costa GMJ, França LR, Resende RR (2017) Progress and biotechnological prospects in fish transgenesis. Biotechnol Adv 35:832–844
Tsai HJ, Tseng TS, Liao IC (1995a) Electroporation of sperm to introduce foreign DNA into the genome of loach (Misgurnus anguillicaudatus). Can J Fish Aquat Sci 52:776–787
Tsai HJ, Wang SH, Inoue K, Takagi S, Kimura M, Wakamatsu Y, Ozato K (1995b) Initiation of the transgenic lacZ gene expression in medaka (Oryzias latipes) embryos. Mol Mar Biol Biotechnol 4:1–9
Wahid F, Shehzad A, Khan T, Kim YY (2010) MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta Mol Cell Res 1803:1231–1243
Waltz E (2016) Gene-edited CRISPR mushroom escapes US regulation. Nature 532:293
Waltz E (2017) First genetically engineered salmon sold in Canada. Nature 548:148
Wang C, Chen Y-L, Bian W-P, Xie S-L, Qi G-L, Liu L, Strauss PR, Zou J-X, Pei D-S (2018) Deletion of mstna and mstnb impairs the immune system and affects growth performance in Zebra fish. Fish Shellfish Immunol 72:572–580
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862
Yan B, Liu B, Zhu C-D, Li K-L, Yue L-J, Zhao J-L, Gong X-L, Wang C-H (2013a) microRNA regulation of skin pigmentation in fish. J Cell Sci 126:3401–3408. https://doi.org/10.1242/jcs.125831
Yan B, Guo J-T, Zhu C-D, Zhao L-H, Zhao J-L (2013b) miR-203b: a novel regulator of MyoD expression in tilapia skeletal muscle. J Exp Biol 216:447–451. https://doi.org/10.1242/jeb.076315
Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32:551–553
Yu H, Li H, Li Q, Xu R, Yue C, Du S (2019) Targeted gene disruption in Pacific oyster based on CRISPR/Cas9 ribonucleoprotein complexes. Mar Biotechnol 21:301–309. https://doi.org/10.1007/s10126-019-09885-y
Zhang PJ, Hayat M, Joyce C, Gonzalez VL, Lin CM, Dunham RA, Chen TT, Powers DA (1990) Gene transfer, expression and inheritance of pRSV-rainbow trout-GH cDNA in the common carp, Cyprinus carpio (Linnaeus). Mol Reprod Dev 25:3–13
Zhang Y, Zhang S, Lu H, Zhang L, Zhang W (2014) Genes encoding aromatases in teleosts: evolution and expression regulation. Gen Comp Endocrinol 205:151–158
Zhang J, Song F, Sun Y, Yu K, Xiang J (2018) CRISPR/Cas9-mediated deletion of EcMIH shortens metamorphosis time from mysis larva to postlarva of Exopalaemon carinicauda. Fish Shellfish Immunol 77:244–251
Zhao Z, Yu X, Jia J, Yang G, Sun C, Li W (2019) miR-181b-5p may regulate muscle growth in Tilapia by targeting Myostatin b. Front Endocrinol 10:812. https://doi.org/10.3389/fendo.2019.00812
Zhong Z, Niu P, Wang M, Huang G, Xu S, Sun Y, Xu X, Hou Y, Sun X, Yan Y, Wang H (2016) Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci Rep 6(22953):2016. https://doi.org/10.1038/srep22953
Acknowledgement
We are indebted to Dr Uma Devi Komath for correcting the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Majumdar, K.C., Ramachandran, R. (2021). Aquaculture Productivity Enhancement Through Advanced Technologies. In: Pandey, P.K., Parhi, J. (eds) Advances in Fisheries Biotechnology. Springer, Singapore. https://doi.org/10.1007/978-981-16-3215-0_1
Download citation
DOI: https://doi.org/10.1007/978-981-16-3215-0_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-3214-3
Online ISBN: 978-981-16-3215-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)