Abstract
The first light-sensing proteins used in optogenetics were rhodopsins. The word “rhodopsin” originates from the Greek words “rhodo” and “opsis,” indicating rose and sight, respectively. Although the classical meaning of rhodopsin is the red-colored pigment in our eyes, the modern meaning of rhodopsin encompasses photoactive proteins containing a retinal chromophore in animals and microbes. Animal and microbial rhodopsins possess 11-cis and all-trans retinal, respectively, to capture light in seven transmembrane α-helices, and photoisomerizations into all-trans and 13-cis forms, respectively, initiate each function. We are able to find ion-transporting proteins in microbial rhodopsins, such as light-gated channels and light-driven pumps, which are the main tools in optogenetics. In this chapter, historical aspects and molecular properties of rhodopsins are introduced. In the first part, “what is rhodopsin?”, general introduction of rhodopsin is presented. Then, molecular mechanism of bacteriorodopsin, a light-driven proton pump and the best-studied microbial rhodopsin, is described. In the section of channelrhodopsin, the light-gated ion channel, molecular properties, and several variants are introduced. As the history has proven, understanding the molecular mechanism of microbial rhodopsins is a prerequisite for useful functional design of optogenetics tools in future.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Animal rhodopsin
- Microbial rhodopsin
- Retinal
- Photoisomerization
- Pump
- Channel
- Photocycle
- Proton transfer
- Hydrogen bond
- Structural change
1 What Is Rhodopsin?
The word “rhodopsin” originates from the Greek words “rhodo” and “opsis,” which indicate rose and sight, respectively. Thus, the classical meaning of rhodopsin is the red-colored pigment in the retinal rods of eyes. The chromophore molecule that absorbs light is retinal, which is the origin of red color. Retinal, the aldehyde of vitamin A, is derived from β-carotene and is bound to the protein in the shape of 11-cis forms (Fig. 1.1) (Shichida and Matsuyama 2009; Hofmann et al. 2009; Palczewski 2012; Koyanagi and Terakita 2014; Ernst et al. 2014). Then, similar colored retinal-binding proteins were found in microbes, largely expanding the definition of the word rhodopsin (Lanyi 2004; Grote et al. 2014; Brown 2014; Ernst et al. 2014; Inoue et al. 2015; Govorunova et al. 2017). In case of microbial rhodopsins, all-trans retinal is bound to the protein (Fig. 1.1). The modern meaning of rhodopsin encompasses photoactive proteins containing a retinal chromophore in animals and microbes (Ernst et al. 2014). Rhodopsins are now found in all domains of life and are classified into two groups. While lower organisms utilize the family of microbial rhodopsins for various functions including light-driven ion pump and light-gated ion channel, animals use the photosensory functions of a different family of rhodopsins (animal rhodopsin), a specialized subset of G-protein-coupled receptors (GPCRs). Microbial and animal rhodopsins share a common architecture of seven transmembrane α-helices (TM1–7) with the N- and C-terminus facing outwards from and inside of the cell, respectively, but have almost no sequence homology and differ largely in their functions (Fig. 1.2a) (Ernst et al. 2014). Retinal is attached by a Schiff base linkage to the ε-amino group of a lysine side chain in the middle of the seventh helix, and this retinal Schiff base (RSB) is protonated (RSBH+) in most cases (Fig. 1.1).
(a) Architecture of rhodopsins. In the rhodopsin field, by convention the cytoplasmic and extracellular sides are drawn at the upper and lower sides of figure, respectively, which is opposite to the conventional drawing of membrane proteins such as G-protein-coupled receptors and transporters. (b) Ion transports of ion pump and channel are unidirectional and bidirectional, respectively
In this chapter, I introduce ion-transporting rhodopsins as they were first used in optogenetics. In living cells, the lipid bilayer effectively repels electrically charged ions that the cell must transport in order to live. Transport of ions is carried out by ion-transporting proteins, which are classified into two types (Fig. 1.2b) (Kandori et al. 2018). In ion channels, ion transport is bidirectional and dissipates energy, leading to a decrease in the ion’s electrochemical potential (free energy). The second type, ion pumps, gains free energy of the system by ion transport, for which energy input is a prerequisite. This is the ion pump. Ion pathways are necessary for both channels and pumps, and each pathway is fully connected between both sides of the membrane when the channel is open. The process by which a channel is opened and closed is referred to as “gating” in this field of study, and channels need at least one gate (Fig. 1.2b) (Hille 1984). A lot of ions are transported upon opening of the gate. For pumps, in contrast, ion pathways cannot be fully connected at one time between both sides of the membrane because the formed ion gradient will easily collapse. This is an important aspect when distinguishing pumps from channels. In other words, pumps require at least two gates, whose regulated opening requires energy input (Fig. 1.2b). To explain complex mechanism of ion pumps, alternating access model and Panama Canal model are used, where transported ion(s) are bound in the resting state (Tanford 1983; Kandori et al. 2018). Only the bound ion(s) are transported during the activation cycle, and consequently, much less ions can be transported in pumps than in channels.
Microbial rhodopsins were first found in the Archaea (Halobacterium salinarum) (Oesterhelt and Stoeckenius 1971) and were therefore initially termed archaeal rhodopsins. H. salinarum contains bacteriorhodopsin (BR) (Oesterhelt and Stoeckenius 1971) and halorhodopsin (HR) (Matsuno-Yagi and Mukohata 1977) that act as a light-driven outward proton pump or an inward Cl— ion pump, respectively. BR and HR contribute to the formation of a membrane potential and thus function in light-energy conversion. The two other H. salinarum rhodopsins are sensory rhodopsins I and II (SRI and SRII) (Spudich and Bogomolni 1984; Jung et al. 2003), which act as positive and negative phototaxis sensors, respectively. For the first 30 years since the early 1970s, microbial rhodopsins were epitomized by haloarchaeal proteins, the first-discovered and best-studied light-driven proton pump BR and its close relatives (HR, SRI, and SRII). However, over the past 15 years, thousands of related photoactive proteins with similar or different functions were identified in Archaea, Eubacteria, and Eukaryota (Brown 2014; Govorunova et al. 2017). Since the original discovery of BR in H. salinarum, similar rhodopsins have been found in Eubacteria and lower Eukaryota, and they are now called microbial rhodopsins. Channelrhodopsins (ChRs), another group of microbial rhodopsins, were discovered in green algae where they function as light-gated cation channels within the algal eye to depolarize the plasma membrane upon light absorption (Fig. 1.2) (Nagel et al. 2002, 2003). Thus, ChRs naturally function as signaling photoreceptors as well. Discovery of ChR led to the emergence of a new field, optogenetics (Miesenbock 2011), in which light-gated ion channels and light-driven ion pumps are used to depolarize and hyperpolarize selected cells of neuronal networks. This new method is highly expected to understand the circuitry of the brain (Deisseroth 2011; Diester et al. 2011).
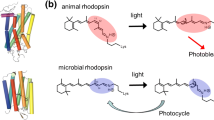
It should be noted that microbial rhodopsins are used in optogenetics in which animal brain functions are studied by incorporating microbial rhodopsins, but not animal rhodopsins, into the animal brain. There are three reasons for this. One is the isomeric structure of the chromophore. An 11-cis retinal (Fig. 1.1), the chromophore molecule of animal rhodopsins, is not generally abundant in animal cells. In contrast, endogenous all-trans retinal, the chromophore molecule of microbial rhodopsins, is sufficient for optogenetics in animal cells. The second reason is “bleaching.” Upon light absorption, animal and microbial rhodopsins exhibit retinal isomerization from the 11-cis to all-trans, and all-trans to 13-cis forms, respectively (Fig. 1.3). While the isomerization reaction initiates each function, the end of the photoreaction differs between animal and microbial rhodopsins. The isomerized all-trans retinal is released in our visual pigments and does not return to the 11-cis form, and is thus called “photobleaching” (Fig. 1.3) (Ernst et al. 2014). This is not a problem in human visual cells because enzymatically isomerized 11-cis retinal is newly supplied, but this is not the case in other animal cells. In contrast, the 13-cis form is thermally isomerized into the all-trans form, and the spontaneous return is termed the “photocycle” in microbial rhodopsins (Fig. 1.3). The third reason is that ion-transporting properties of light-gated ion channels and light-driven ion pumps enable the direct manipulation of membrane potentials of selected cells in neuronal networks without additional elements. Due to the existence of naturally abundant all-trans retinal and its photocycle feature, ion-transporting microbial rhodopsins have become a tool in optogenetics.
2 Bacteriorhodopsin, the First Ion Transport by Light
BR from H. salinarum, the first-discovered microbial rhodopsin in 1971 (Oesterhelt and Stoeckenius 1971), is the first membrane protein whose structure was found to be composed of seven helices by electron microscopy (Henderson and Unwin 1975), and was also the first membrane protein to have its amino acid sequence determined (Khorana et al. 1979). As the best-studied microbial rhodopsin, it serves as a paradigm of a light-driven retinal-binding ion pump and aids in studies of novel rhodopsins (Lanyi 2004; Ernst et al. 2014). Archaerhodopsin 3 (Arch), a homologous proton-pump protein, is the best used in optogenetics as a neural silencer (Chow et al. 2010), sharing 58% amino acid identity with BR. Similar molecular mechanisms were assumed for the light-dependent ion transport function between Arch and BR.
Figure 1.4 illustrates the overall structure of BR, highlighting the conserved aromatic amino acids with an important function. The retinal-binding pocket is the most conserved common element of the structure. Strongly conserved Trp86 and Trp182 constitute an important part of the chromophore binding site by sandwiching all-trans-retinal vertically (Fig. 1.4). The presence of these bulky groups possibly determines the isomerization pathway from the all-trans to the 13-cis form (Gozem et al. 2017). Moreover, the interaction of photoisomerized retinal with Trp182 may serve as the mechanical transducer for passing the energy stored in retinal deformation into the functionally important changes of the helical tilts necessary for function (Weidlich et al. 1996). Another important position occupied by aromatic amino acids in the retinal-binding pocket is that of Tyr185 in BR (Fig. 1.4), which participates in hydrogen-bonding stabilization of the Schiff base counterion for many rhodopsins. This is replaced by a Phe in ChRs, suggesting that the lack of a hydrogen-bonding interaction at this position is important for channel function.
Structure of bacteriorhodopsin (BR) with conserved aromatic residues (PDB: 1QM8). Tyr83, Trp86, and Trp182 are strongly conserved among microbial rhodopsins (orange). Aromatic residues are strongly conserved at the Tyr185, Trp189, and Phe219 positions (yellow). In BR, Trp86, Trp182, Tyr185, and Trp189 constitute the chromophore binding pocket for all-trans retinal (red)
In addition to the aromatic sidechain rings, electrostatic and hydrogen-bonding interactions in the proximal part of retinal are crucial in defining the functionality of microbial rhodopsins (Lanyi 2004; Ernst et al. 2014). The sidechain of BR Lys216 (or its homologs in other microbial rhodopsins) forms a covalent bond with the retinal molecule through the Schiff base (Fig. 1.1). Since the Schiff base is usually protonated, Lys216 and super-conserved Arg82 of helix C in BR provide two positive charges within the protein (Fig. 1.5), which requires two negative charges for electrostatic stabilization. This dictates the most common configuration of the Schiff base counterions made by two carboxylic acids (Asp85 and Asp212 in BR), which are perfectly conserved for proton-pumping microbial rhodopsins (Fig. 1.5) (Ernst et al. 2014; Kandori 2015).
Structure of the Schiff base region in bacteriorhodopsin (BR). This is the side view of the Protein Data Bank structure 1C3W, which has a resolution of 1.55 Å (Luecke et al. 1999). The membrane normal is approximately in the vertical direction of this figure. Hydrogen atoms and hydrogen bonds (dashed lines) are supposed from the structure, while the numbers indicate hydrogen-bonding distances in Å. Arg, Asp, and Lys at positions 82, 212, and 216, respectively, are fully conserved in ChR and HR, while Asp85, Thr89, and Tyr185 in BR are replaced in ChR and HR, as shown in the figure
The proton pathway across the membrane from the cytoplasmic to the extracellular side in BR is shown in Fig. 1.6, together with protonatable groups and the order of respective proton transfers (Kandori 2015; Gerwert et al. 2014). A summary of the photocycle is shown in Fig. 1.7, which illustrates key intermediate states for most microbial rhodopsins. Although the photocycle of BR contains six intermediates, namely J, K, L, M, N, and O states that are named alphabetically, only three states (K, M, and N) are shown in Fig. 1.7 to show the mechanism clearly. After light absorption, photoisomerization occurs from the all-trans- to 13-cis-form in 10−13 s (Ernst et al. 2014; Gozem et al. 2017). The ultrafast retinal isomerization yields the formation of red-shifted J and K intermediates in which J is the precursor of the K state. As the protein cavity, which accommodates retinal, cannot change its shape promptly, the K intermediate contains twisted 13-cis retinal and an altered hydrogen-bonding network in the Schiff base region, which yield higher free energy in K than in the original state. This leads to subsequent protein structural changes that accompany relaxation.
Typical photocycle of microbial rhodopsins showing isomeric and protonation state of retinal. Names of the photocycle intermediates are for BR, while those in parenthesis are for ChR. X− represents the Schiff base counterion, and Asp85 in BR also acts as the proton acceptor from the Schiff base. X− is Glu123 in ChR2, whereas the proton acceptor of the Schiff base is not well established. In the chloride pump HR, X− is a chloride ion so that the M intermediate is not formed because the Schiff base is not deprotonated. Instead, the chloride ion is transported upwards (in this figure) in HR. CP and EC indicate cytoplasmic and extracellular domains, respectively. In the unphotolyzed state of microbial rhodopsins, the EC side is open through a hydrogen-bonding network, but the CP side is closed. While this is persistent in the K (P1) and M (P2) states, the CP side is open in the N (P3) state. When the EC side is closed (black), the CP side is open, as is the case for an ion pump, as occurs in the N intermediate of BR. When the EC side is open (red), the CP side is open, as is the case for an ion channel, as occurs for the P3 intermediate of ChR
In the case of BR, relaxation of the K intermediate leads to the formation of the blue-shifted L intermediate. For proton-pumping (and some of the photosensory) rhodopsins, the L intermediate serves as the precursor of the proton transfer reaction from the Schiff base to its primary carboxylic proton acceptor, by which the M intermediate is formed. This is a key step in proton transport. Since the M intermediate has a deprotonated 13-cis chromophore, it exhibits a characteristically strong blue-shifted absorption (λ max at 360–410 nm), and is well isolated from other intermediates. In BR, the proton acceptor (X− in Fig. 1.7) is Asp85 so that the primary proton transfer takes place from the Schiff base to Asp85. In the case of chloride pump HR, the Schiff base does not deprotonate during the photocycle because Asp85 in BR is replaced by Thr. In HR, X− in Fig. 1.7 is a chloride ion, which is directly translocated upon decay of the L intermediate.
If the Schiff base of M is reprotonated from Asp85 in BR, no proton transport occurs. In reality, the Schiff base is reprotonated from Asp96 in the cytoplasmic region (Fig. 1.6), by which the N intermediate is formed. The molecular mechanism of unidirectional transport of protons in BR has attracted the attention of many researchers, and it is believed that the primary proton transfer from the Schiff base to Asp85 and the subsequent proton transfer from Asp96 to the Schiff base determine the unidirectionality from the cytoplasmic to the extracellular region. The crystal structure of BR exhibits an asymmetric pattern of hydration, while seven internal water molecules are found in the extracellular half, only two are observed in the cytoplasmic half (Fig. 1.6). Such asymmetry makes sense in view of BR’s function, as the water molecules build a hydrogen-bonding network on the extracellular side for fast proton release while the cytoplasmic side is likely inaccessible in the dark and allows proton uptake only after the light-induced accessibility switch (Ernst et al. 2014). Such asymmetric access (EC open and CP closed) is not only the case for the unphotolyzed state but also the case for the K and M intermediates, as shown in Fig. 1.7.
To make proton conduction in the cytoplasmic region possible (CP open), an additional conformational change allowing the entrance of water into the vicinity of Asp96 should take place. Such a conformational alteration is realized mainly by changes in helical tilts (especially of the cytoplasmic half of helix F), and the N intermediate is often characterized by the largest changes in the protein backbone conformation, most notably, outward tilts of the cytoplasmic end of helix F. Such helical motions are functionally significant both for ion transport and interactions with transducers of sensory rhodopsins. The photocycle usually ends with another red-shifted intermediate, known as the O intermediate, serving as a last step in resetting the original unphotolyzed conformation.
In the chloride-pumping HR, the negatively charged Asp85 in BR is replaced by Thr (Fig. 1.5), and the lack of a negative charge is compensated by the binding of Cl− near the Schiff base region. Consequently, Cl− stabilizes the protonated Schiff base as the counterion (X− in Fig. 1.7). During the photocycle of HR, protonation of the Schiff base is maintained and Cl− is translocated, unlike H+-pumping BR (Kandori 2015). This is also the case in eubacterial light-driven Cl− pumps. Resemblance of light-driven H+ and Cl− pumps has been demonstrated by functional conversion by mutation. In 1995, BR was successfully converted into a Cl− pump by replacement of a single amino acid (Asp85 to Thr) (Sasaki et al. 1995). In contrast, HR was not converted into a H+ pump by mutation, where protein-bound water molecules play crucial role (Muroda et al. 2012). In case of eubacterial pumps, Cl− pump was converted into H+ pump (Hasemi et al. 2016; Inoue et al. 2016a), but H+ pump could not be converted into Cl− pump (Inoue et al. 2016a). Asymmetric functional conversion was observed for both archaeal and eubacterial light-driven pumps, whereas the direction of successful conversion was opposite between them.
3 Channelrhodopsin, the Light-Gated Ion Channel by Light
The molecular mechanism of the light-driven proton pump in BR is well established although the detailed directionality remains uncertain. In pumps, the transport pathways between the two sides of the membrane cannot be fully connected because the gradient formed by active transport will collapse. This view is clearly explained in the case of BR. This is an important aspect when distinguishing pumps from channels. The former needs energy input, which ensures the unidirectionality of transport across the membrane. The protein architecture of BR in Figs. 1.4 and 1.6 effectively explains the unidirectional transport in which the extracellular side contains many water molecules while the cytoplasmic side has no pathway (EC open and CP closed in Fig. 1.7). Therefore, an alternative pump-specific conformation must work, in which the protein releases a proton first on one side, and takes up a proton from the other side (EC closed and CP open in Fig. 1.7). In contrast, a channel needs a fully connected ion pathway for passive transport of ions upon opening (EC and CP open). From the structure of BR (Figs. 1.4 and 1.6), the channel function is most likely impossible for microbial rhodopsins.
In 2002–2003, three groups independently identified novel DNA sequences that encode microbial-type rhodopsins in Chlamydomonas (Nagel et al. 2002; Sineshchekov et al. 2002; Suzuki et al. 2003). Furthermore, a light-gated ion channel function was proved for two proteins, channelrhodopsin-1 (ChR1) and channelrhodopsin-2 (ChR2), in Xenopus oocytes using two electrode voltage clamp measurements (Nagel et al. 2002, 2003). ChR2 was also shown to be expressed and used to depolarize mammalian cells in response to light (Boyden et al. 2005; Ishizuka et al. 2006). Then, several groups began to work with ChRs, primarily with a truncated version of ChR2 that expressed better than the full-length protein and much better than ChR1 (Ernst et al. 2014; Deisseroth 2011). This was the dawn of optogenetics. Surprisingly, the mammalian brain contains sufficient retinoid levels to allow wild-type ChR2 to function without the addition of exogenous retinoid cofactors.
ChR is a light-gated ion channel, implying that the channel is closed in the dark, and opens upon light absorption. Initially, researchers suspected that the channel pathway lay inside the seven transmembrane helices as well as the proton pathway in BR (Fig. 1.6). Instead, it is more likely that ChR forms a dimer and that the intra-dimer cavity constitutes the ion conduction pathway. However, mutation studies of ChR suggested that the ion conduction pathway lay inside the seven transmembrane helices (Ernst et al. 2014). In addition, X-ray crystallography of a chimeric protein of ChR1 and ChR2 showed the presence of the channel cavity at the extracellular domain, strongly suggesting that the ion conduction pathway lay inside the seven transmembrane helices (Kato et al. 2012). It is now believed that a monomer is the functional unit of ChR. The crystal structure of ChR showed that the protein architecture is common to all microbial rhodopsins (Kato et al. 2012; Volkov et al. 2017), but that some structural modification must be linked to their unique function. For instance, in all microbial rhodopsins, the hydrogen-bonding acceptor of the Schiff base is a water molecule (Fig. 1.5), whereas crystal structure and FTIR spectroscopy revealed the direct interaction of the Schiff base with the counterion (Glu at the position of Asp85 in BR; Fig. 1.5) (Kato et al. 2012; Ito et al. 2014).
Like BR, the ChR photocycle has been studied by various methods, and it is now established that ChR has a photocycle similar to other microbial rhodopsins (Ernst et al. 2014; Lórenz-Fonfría and Heberle 2014; Schneider et al. 2015). After the absorption of light, photoisomerization occurs from the all-trans- to the 13-cis-form very rapidly, and forms the red-shifted K-like intermediate (P1 in Fig. 1.7). As the protein cavity, which accommodates retinal, cannot respond promptly, the P1 intermediate contains twisted 13-cis retinal and an altered hydrogen-bonding network in the Schiff base region, which possesses higher free energy than in the original state. This yields subsequent protein structural changes that accompany relaxation as well as the case in BR. Then, the proton is transferred from the Schiff base, forming the M-like P2 intermediate. The P2 intermediate has a deprotonated 13-cis chromophore whose absorption is strongly blue-shifted (λ max at 380 nm). The N-like P3 intermediate is formed by reprotonation of the Schiff base, and it is believed to be the ion-conducting state. It is reasonable to assume that the N-like state exhibits the largest conformational changes in microbial rhodopsins. When the CP side is open, the EC side must also be open in ChR so that a transient ion conduction pathway is created. Therefore, in Fig. 1.7, pumps and channels show the N-like intermediate in CP open/EC closed and CP open/EC open conformations, respectively.
The channel property of ChR is important for optogenetic applications. In particular, absorption color, ion selectivity, and open/close dynamics should be taken into account. Thus far, various mutants have been shown to improve the properties of ChR. How then were these variants designed before structural determination? Knowledge of BR has contributed significantly to the design of mutants. For instance, two important Asp in BR, Asp85 and Asp96, correspond to Glu123 and His134 in ChR2, respectively, to which mutations were attempted. In the case of E123T/A, channel closure is faster than that of the wild type, allowing for a 10-times higher repetition rate (200 Hz) than the wild type in optogenetics (Fig. 1.8) (Gunaydin et al. 2010). The E123T/A mutant also shows red-shifted absorption, caused by neutralization of the Schiff base counterion (Glu123; X− in Fig. 1.7) (Ernst et al. 2014). In the case of H134R, channel closure is slower than that of the wild type, improving conductivity and changing ion selectivity (Nagel et al. 2005). Asp96 and His134 in BR and ChR2, respectively, are located at important positions of the cytoplasmic domain (Figs. 1.6 and 1.8), presumably influencing ion transport. In BR, Thr90, Asp115, and Met118 (Cys128, Asp156, and Thr159 in ChR2, respectively) are located near the retinal chromophore (Fig. 1.8). D156A/C128S exhibits an extremely slow photocycle, leading to a permanently open channel that can be photoconverted to the original state by orange light (Berndt et al. 2011). Since ion conductance was increased for T159C, it was used under weak light conditions (Berndt et al. 2009). These examples show how specific variants of ChR2 have been applied in optogenetics (Zhang et al. 2011).
Key amino acids for ChR2 channel activity based on the BR structure, which is modified from Hegemann and Möglich (2011). Red and black characters correspond to the residue numbers in BR and ChR2, respectively, and stick drawings represent the residues in ChR2. The residues conserved or different in ChR1 and ChR2 from Chlamydomonas reinhardtii and Volvox carteri are colored blue or gray, respectively. Mutations of these residues substantially influence absorption, conductance, kinetics, and ion selectivity, as indicated for each residue. In particular, four mutations, E123T, H134R, D156A/C128S, and T159C, described in the text, are highlighted by green circles
Despite the creation of useful ChR2 variants, there are still many requirements for channel function. Limited conductance of ChR2 needs to be improved. However, unlike other channel proteins, ion pathway is not straight as ions are conducted inside seven transmembrane helices in ChR2. This is the substantial limitation of low conductance of ChR2, and creation of wider pores through molecular engineering may be a challenge. Ion selectivity of ChR2 is much higher for H+ than for Na+ (the permeability ratio, PH/PNa, is about 106) (Nagel et al. 2003), indicating that ChR2 should be recognized as a light-gated H+ channel rather than an Na+ channel, and if pH differs between inside and outside of cells, ChR2 acts as a proton channel. ChR2 has some permeability of Ca2+. No conduction or increased conduction of Ca2+ is highly demanded, and Ca2+-permeable channelrhodopsin CatCh (L132C ChR2) was reported, though it does not remarkably increase Ca2+ permeability (Kleinlogel et al. 2011). Absorbing color is another problem of ChR2, which absorbs blue light maximally (470–480 nm). Blue light is harmful to organisms and also penetrates them less than red light. Therefore, red-absorbing ChRs have been reported such as ReaChR and Chrimson (Zhang et al. 2008; Lin et al. 2013; Klapoetke et al. 2014).
While ChR2 was used for neural excitation by depolarizing cells, neural silencing tools were highly demanded from the beginning. Light-driven ion pumps such as a light-driven H+-pump BR and a light-driven Cl−-pump HR were the candidates as they can hyperpolarize cells by light. However, at that time, I was skeptical about the practical usage of these pumps because of efficiency. In light-driven pumps, a single ion is translocated by a single photocycle (Figs. 1.4 and 1.5), while multiple ions can be transported when light-gated ion channel is activated by a single photocycle. Nevertheless, a light-driven Cl− pump HR was successfully used for neural silencing by hyperpolarizing cells (Zhang et al. 2007). Optogenetics was actually established by neural excitation and silencing, using ChR2 and HR, respectively, in 2007. Then, various light-driven H+ pumps were tested as a tool of neural silencing, among which Arch has been best used (Chow et al. 2010).
4 Novel Ion-Transporting Rhodopsins as Optogenetics Tools
A light-gated channel was discovered in 2002, and optogenetics began in 2005 by using ChR2 as a neural excitation tool. Optogenetic was fully established a few years later by using light-driven H+ and Cl− pumps as neural silencing tools, which were discovered in 1970s. During this decade, new microbial rhodopsins have been discovered and actively applied to optogenetics. They are summarized in Figs. 1.9 and 1.10, and briefly introduced below.
Function of rhodopsins; light-driven Cl− pump, light-driven H+ pump, light-driven Na+ pump, light-driven inward H+ pump, light-gated cation channel, light-gated anion channel, light sensor with transmembrane transducer and soluble transducer, and light-activated enzyme. Purple or orange arrows indicate unidirectional or bidirectional transport of ions in pumps or channels, respectively. On the other hand, pale orange arrows show the signal transduction from rhodopsins to either soluble or transmembrane transducer proteins
Role of photoreceptive proteins in optogenetics. Channels and pumps perform passive and active transports of ions, respectively. At the beginning of optogenetics, light-gated cation channel (ChR2) depolarizes neural cells by influx of Na+, leading to generation of action potential (neural excitation), while light-driven Cl− pump (HR) hyperpolarizes neural cells by influx of Cl−, leading to inhibition of action potential (neural silencing). More optogenetics tools are now at hand. Light-driven inward H+ pumps depolarize cells, while light-driven (outward) H+ and Na+ pumps and light-gated anion channels hyperpolarize cells. Recently discovered enzyme rhodopsins, one family of microbial rhodopsins, are used to control concentrations of cAMP and cGMP. In addition, animal rhodopsins are also used for optogenetics
Existence of Na+-pumping rhodopsin was a surprise in the field. In active transporter, substrate is bound to the resting state, and the transporting mechanism is described by alternating access model or Panama Canal model (Kandori et al. 2018). In rhodopsins, positively charged protonated Schiff base is a key element for the usage of light, and light-driven H+ and Cl− pumps bind the substrates H+ and Cl−, respectively, in this region (Figs. 1.5 and 1.7). In contrast, we believed Na+-binding unlikely because of electrostatic repulsion with the protonated Schiff base. However, nature created light-driven Na+ pump rhodopsin (Fig. 1.9) (Inoue et al. 2013). Interestingly, this protein does not need Na+ to be bound to the resting state, indicating that light-driven Na+ pumps are a unique active transporter. Unidirectional transport of Na+ is controlled by passive diffusion of Na+ from the cytoplasmic side. Ion selectivity filter is located at the intracellular surface, whose modification led to the creation of light-driven pumps of K+, Rb+, and Cs+ (Kato et al. 2015; Konno et al. 2016). Light-driven Na+ pumps are also used as neural silencing tools in optogenetics (Fig. 1.10) (Tsunoda et al. 2017; Grimm et al. 2018).
Existence of light-driven inward H+ pump was another surprise because inward H+ transport competes with ATP synthase function. In 2009, we successfully created inward H+ transport using a mutant of a photochromic sensor, Anabaena sensory rhodopsin (D217E ASR) (Kawanabe et al. 2009). While this was an example of engineering success, we reported that nature also created an inward H+ pump in 2016 (Fig. 1.9) (Inoue et al. 2016b). A rhodopsin from Parvularcula oceani (PoXeR) is in the same family of ASR, and the mechanistic analyses revealed that small differences in interactions at the active center determine the direction of primary H+ transfer between outward and inward H+ pumps, even though the retinal chromophore structure and primary photoisomerization are identical (Inoue et al. 2016b; Shevchenko et al. 2017). In view of optogenetics, inward H+ pump functions for depolarizing cells (Fig. 1.10), but cation channels are more superior as pump transports one proton per one photon. On the other hand, inward H+ pump is more beneficial for intracellular organelle optogenetics. While intracellular organelle can be acidified by light-driven H+ pump, alkalization tool of intracellular organelle has been demanded. In fact, D217E ASR was used to alkalize AMPA-type glutamate receptor endocytosis in the study of long-term depression of AMPA-type glutamate receptor (AMPA receptor)-mediated synaptic transmission for learning and memory (Kakegawa et al. 2018).
Cellular inside is negatively charged (−70 mV), but Cl− ions are uptaken upon opening of anion channel because concentration of Cl− is much higher in extracellular side than in cytoplasm (Fig. 1.10). In general, channels are more efficient tool than pumps because of multiple ion permeation. Thus, light-gated anion channels have been strongly required in optogenetics, and in fact, they were engineered from cation channels (Wietek et al. 2014; Berndt et al. 2014). On the other hand, natural anion channel rhodopsin (ACR) was discovered from a cryptophyte Guillardia theta in 2015 (Fig. 1.9) (Govorunova et al. 2015). Conduction of ACR is more than cation channel rhodopsins, and natural ACR is considered better neural silencing tools than light-driven ion pumps. Structures of ACR and engineered anion-channel rhodopsin were determined (Kim et al. 2018; Kato et al. 2018), and knowledge of structure and mechanism of ACR will be used for the improvement of tools, and contributes to neural silencing (Wiegert et al. 2017), as was the case for ChR2.
Guillardia theta possesses more than 40 genes of rhodopsins, and light-gated cation channels from Guillardia theta are also noted. A light-driven H+ pump BR contains a DTD motif at positions 85, 89, and 96, which is unique to archaeal H+ pumps. Recently, two groups independently reported that the DTD rhodopsins from Guillardia theta are light-gated cation channel, not pump (Govorunova et al. 2016; Yamauchi et al. 2017). One of these proteins, GtCCR4, permeates little protons and Ca2+ (Shigemura et al. 2019), which is a remarkable contrast to those of ChR2. If influx of Ca2+ is problematic in optogenetic experiments, GtCCR4 is a suitable tool. A recently reported new cation channel, named ChRmine (Marshel et al. 2019), is in the same family of the DTD cation channels.
5 Conclusion and Perspectives
The original meaning of rhodopsin is the red-colored pigment for vision, but similar colored retinal proteins were found in microbes. Consequently the definition of the word rhodopsin is largely expanded, and the modern meaning of rhodopsin encompasses photoactive proteins containing a retinal chromophore in animals and microbes. While the meaning of rhodopsin has been altered in history, it is also the case on the word “optogenetics.” Optogenetics is originally the combination of “optics” and “genetics,” which describes optical control of neural activity by use of ion-transporting microbial rhodopsins such as channelrhodopsin and light-driven Cl− pump (Fig. 1.10). This allows rapid temporal response, which was required in the brain function researches. On the other hand, extensive studies largely expanded the definition of the word optogenetics.
Function of no ion-transporting rhodopsins was recognized as light sensor, such as SRI and SRII, which activate membrane-bound transducer proteins. In case of a photochromic sensor ASR, its transducer is a soluble protein. Animal rhodopsins are G-protein-coupled receptors, whose transducers are also soluble (heterotrimeric G-protein) that are anchored to the membrane. As various signaling processes in mammalian cells are regulated by GPCR, animal rhodopsins are potential candidate in optogenetics, where photobleaching property has to be improved. In this sense, bistable pigments of animal rhodopsin, such as melanopsin (Ye et al. 2011) and opn3 (Koyanagi et al. 2013), are useful, and microbial rhodopsin can be used as a template for light activation of G-protein (Sasaki et al. 2014). To control intracellular second messengers, enzyme rhodopsins should be noted, which are composed of a membrane-embedded rhodopsin domain and a C-terminal cytoplasmic enzyme domain that are activated when light is absorbed by the all- trans-retinal chromophore (Fig. 1.9). A fungal light-activated guanylyl cyclase (Rh-GC) and light-activated phosphodiesterase (Rh-PDE) from a unicellular and colonial single flagellate eukaryote were reported in 2014 and 2017, respectively (Avelar et al. 2014; Yoshida et al. 2017). Rh-GC increases centration of cGMP by light, while Rh-PDE decreases the concentrations of cGMP and cAMP by light. As cAMP and cGMP are known to be the second messengers in intracellular signaling, they are new optogenetic tools to control concentration of the second messengers (Fig. 1.10). I introduced various rhodopsins in this chapter, and heliorhodopsin (HeR) should be finally noted. It has been believed that microbial rhodopsin (also called type-1 rhodopsin) and animal rhodopsin (also called type-2 rhodopsin) are the only rhodopsins. Nevertheless, a previously unrecognized diverse family, heliorhodopsins (HeRs), was recently discovered through the use of functional metagenomics (Pushkarev et al. 2018). HeRs have inverted membrane topology compared to other retinal proteins (Shihoya et al. 2019), have no ion-transport activity, and their slow photocycle suggests a light-sensor function. Therefore, HeRs will be a new optogenetics tool in future.
Abbreviations
- ACR:
-
Anion channel rhodopsin
- Arch:
-
Archaerhodopsin 3
- ASR:
-
Anabaena sensory rhodopsin
- BR:
-
Bacteriorhodopsin
- ChR:
-
Channelrhodopsin
- CP:
-
Cytoplasmic
- EC:
-
Extracellular
- GPCR:
-
G-protein-coupled receptors
- GtCCR4:
-
Cation channel rhodopsin from Guillardia theta
- HeR:
-
Heliorhodopsin
- HR:
-
Halorhodopsin
- PoXeR:
-
Rhodopsin from Parvularcula oceani
- Rh-GC:
-
Light-activated guanylyl cyclase rhodopsin
- Rh-PDE:
-
Light-activated phosphodiesterase rhodopsin
- RSB:
-
Retinal Schiff base
- SRI:
-
Sensory rhodopsin I
- SRII:
-
Sensory rhodopsin II
References
Avelar GM, Schumacher RI, Zaini PA, Leonard G, Richards TA, Gomes SL (2014) A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr Biol 24:1234–1240
Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K (2009) Bistable neural state switches. Nat Neurosci 12:229–234
Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, Oertner TG (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci U S A 108:7595–7600
Berndt A, Lee SY, Ramakrishnan C, Deisseroth K (2014) Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344:420–424
Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8:1263–1268
Brown LS (2014) Eubacterial rhodopsins - Unique photosensors and diverse ion pumps. Biochim Biophys Acta 1837:553–561
Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463:98–102
Deisseroth K (2011) Optogenetics. Nat Methods 8:26–29
Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, Yizhar O, Ramakrishnan C, Deisseroth K, Shenoy KV (2011) An optogenetic toolbox designed for primates. Nat Neurosci 14:387–397
Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev 114:126–163
Gerwert K, Freier E, Wolf S (2014) The role of protein-bound water molecules in microbial rhodopsins. Biochim Biophys Acta 1837:606–613
Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL (2015) Neuroscience. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349:647–650
Govorunova EG, Sineshchekov OA, Spudich JL (2016) Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J 110:2302–2304
Govorunova EG, Sineshchekov OA, Li H, Spudich JL (2017) Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu Rev Biochem 86:845–872
Gozem S, Luk HL, Schapiro I, Olivucci M (2017) Theory and simulation of the ultrafast double-bond isomerization of biological chromophores. Chem Rev 117:13502–13565
Grimm C, Silapetere A, Vogt A, Bernal Sierra YA, Hegemann P (2018) Electrical properties, substrate specificity and optogenetic potential of the engineered light-driven sodium pump eKR2. Sci Rep 8:9316
Grote M, Engelhard M, Hegemann P (2014) Of ion pumps, sensors and channels - perspectives on microbial rhodopsins between science and history. Biochim Biophys Acta 1837:533–545
Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P (2010) Ultrafast optogenetic control. Nat Neurosci 13:387–392
Hasemi T, Kikukawa T, Kamo N, Demura M (2016) Characterization of a cyanobacterial chloride-pumping rhodopsin and its conversion into a proton pump. J Biol Chem 291:355–362
Hegemann P, Möglich A (2011) Channelrhodopsin engineering and exploration of new optogenetic tools. Nat Methods 8:39–42
Henderson R, Unwin PN (1975) Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257:28–32
Hille B (1984) Ion channels and excitable membranes. Oxford University Press, Oxford
Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP (2009) A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci 34:540–552
Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, Kandori H (2013) A light-driven sodium ion pump in marine bacteria. Nat Commun 4:1678
Inoue K, Kato Y, Kandori H (2015) Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol 23:91–98
Inoue K, Nomura Y, Kandori H (2016a) Asymmetric functional conversion of eubacterial light-driven ion pumps. J Biol Chem 291(19):9883–9893
Inoue K, Ito S, Kato Y, Nomura Y, Shibata M, Uchihashi T, Tsunoda SP, Kandori H (2016b) A natural light-driven inward proton pump. Nat Commun 7:13415
Ishizuka T, Kakuda M, Araki R, Yawo H (2006) Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res 54:85–94
Ito S, Kato HE, Taniguchi R, Iwata T, Nureki O, Kandori H (2014) Water-containing hydrogen-bonding network in the active center of channelrhodopsin. J Am Chem Soc 136:3475–3482
Jung KH, Trivedi VD, Spudich JL (2003) Demonstration of a sensory rhodopsin in eubacteria. Mol Microbiol 47:1513–1522
Kakegawa W, Katoh A, Narumi S, Miura E, Motohashi J, Takahashi A, Kohda K, Fukazawa Y, Yuzaki M, Matsuda S (2018) Optogenetic control of synaptic AMPA receptor endocytosis reveals roles of LTD in motor learning. Neuron 99:985–998
Kandori H (2015) Ion-pumping microbial rhodopsins. Front Mol Sci 2:52
Kandori H, Inoue K, Tsunoda SP (2018) Light-driven sodium-pumping rhodopsin: a new concept of active transport. Chem Rev 118:10646–10658
Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, Hegemann P, Maturana AD, Ishitani R, Deisseroth K, Nureki O (2012) Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482:369–374
Kato HE, Inoue K, Abe-Yoshizumi R, Kato Y, Ono H, Konno M, Hososhima S, Ishizuka T, Hoque MR, Kunitomo H, Ito J, Yoshizawa S, Yamashita K, Takemoto M, Nishizawa T, Taniguchi R, Kogure K, Maturana AD, Iino Y, Yawo H, Ishitani R, Kandori H, Nureki O (2015) Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521:48–53
Kato HE, Kim YS, Paggi JM, Evans KE, Allen WE, Richardson C, Inoue K, Ito S, Ramakrishnan C, Fenno LE, Yamashita K, Hilger D, Lee SY, Berndt A, Shen K, Kandori H, Dror RO, Kobilka BK, Deisseroth K (2018) Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature 561:349–354
Kawanabe A, Furutani Y, Jung KH, Kandori H (2009) Engineering an inward proton transport from a bacterial sensor rhodopsin. J Am Chem Soc 131:16439–16444
Khorana HG, Gerber GE, Herlihy WC, Gray CP, Anderegg RJ, Nihei K, Biemann K (1979) Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci U S A 76:5046–5050
Kim YS, Kato HE, Yamashita K, Ito S, Inoue K, Ramakrishnan C, Fenno LE, Evans KE, Paggi JM, Dror RO, Kandori H, Kobilka BK, Deisseroth K (2018) Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature 561:343–348
Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK, Boyden ES (2014) Indepeendent optical excitation of distinct neural populations. Nat Methods 11:338–346
Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C, Bamberg E (2011) Ultra light-sensitive and fast neuronal activation with the Ca2+-permiable channelrhodopsin CatCh. Nat Neurosci 14:513–518
Konno M, Kato Y, Kato HE, Inoue K, Nureki O, Kandori H (2016) Mutant of a light-driven sodium ion pump can transport cesium ions. J Phys Chem Lett 7:51–55
Koyanagi M, Terakita A (2014) Diversity of animal opsin-based pigments and their optogenetic potential. Biochim Biophys Acta 1837:710–716
Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A (2013) Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc Natl Acad Sci U S A 110:4998–5003
Lanyi JK (2004) Bacteriorhodopsin. Annu Rev Physiol 66:665–688
Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY (2013) ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci 16:1499–1508
Lórenz-Fonfría VA, Heberle J (2014) Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim Biophys Acta 1837:626–642
Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK (1999) Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol 291:899–911
Marshel JH, Kim YS, Machado TA, Quirin S, Benson B, Kadmon J, Raja C, Chibukhchyan A, Ramakrishnan C, Inoue M, Shane JC, McKnight DJ, Yoshizawa S, Kato HE, Ganguli S, Deisseroth K (2019) Cortical layer-specific critical dynamics triggering perception. Science 365:6453
Matsuno-Yagi A, Mukohata Y (1977) Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun 78:237–243
Miesenbock G (2011) Optogenetic control of cells and circuits. Annu Rev Cell Dev Biol 27:731–758
Muroda K, Nakashima K, Shibata M, Demura M, Kandori H (2012) Protein-bound water as the determinant of asymmetric functional conversion between light-driven proton and chloride pumps. Biochemistry 51:4677–4684
Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296:2395–2398
Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A 100:13940–13945
Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A (2005) Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol 15:2279–2284
Oesterhelt D, Stoeckenius W (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233:149–152
Palczewski K (2012) Chemistry and biology of vision. J Biol Chem 287:1612–1619
Pushkarev A, Inoue K, Larom S, Flores-Uribe J, Singh M, Konno M, Tomida S, Ito S, Nakamura R, Tsunoda SP, Philosof A, Sharon I, Yutin N, Koonin EV, Kandori H, Béjà O (2018) A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558:595–599
Sasaki J, Brown LS, Chon YS, Kandori H, Maeda A, Needleman R, Lanyi J (1995) Conversion of bacteriorhodopsin into a chloride ion pump. Science 269:73–75
Sasaki K, Yamashita T, Yoshida K, Inoue K, Shichida Y, Kandori H (2014) Chimeric proton-pumping rhodopsins containing the cytoplasmic loop of bovine rhodopsin. PLoS One 9:e91323
Schneider F, Grimm C, Hegemann P (2015) Biophysics of Channelrhodopsin. Annu Rev Biophys 44:167–186
Shevchenko V, Mager T, Kovalev K, Polovinkin V, Alekseev A, Juettner J, Chizhov I, Bamann C, Vavourakis C, Ghai R, Gushchin I, Borshchevskiy V, Rogachev A, Melnikov I, Popov A, Balandin T, Rodriguez-Valera F, Manstein DJ, Bueldt G, Bamberg E, Gordeliy V (2017) Inward H+ pump xenorhodopsin: mechanism and alternative optogenetic approach. Sci Adv 3:e1603187
Shichida Y, Matsuyama T (2009) Evolution of opsins and phototransduction. Philos Trans R Soc Lond Ser B Biol Sci 364:2881–2895
Shigemura S, Hososhima S, Kandori H, Tsunoda SP (2019) Ion channel properties of a cation channelrhodopsin, Gt_CCR4. Appl Sci 9:3440
Shihoya W, Inoue K, Singh M, Konno M, Hososhima S, Yamashita K, Ikeda K, Higuchi A, Izume T, Okazaki S, Hashimoto M, Mizutori R, Tomida S, Yamauchi Y, Abe-Yoshizumi R, Katayama K, Tsunoda SP, Shibata M, Furutani Y, Pushkarev A, Béjà O, Uchihashi T, Kandori H, Nureki O (2019) Crystal structure of heliorhodopsin. Nature 572:132–136
Sineshchekov OA, Jung KH, Spudich JL (2002) Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 99:8689–8694
Spudich JL, Bogomolni RA (1984) Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature 312:509–513
Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, Tabata S, Miura K, Fukuzawa H, Nakamura S, Takahashi T (2003) Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem Biophys Res Commun 301:711–717
Tanford C (1983) Mechanism of free energy coupling in active transport. Annu Rev Biochem 52:379–409
Tsunoda SP, Prigge M, Abe-Yoshizumi R, Inoue K, Kozaki Y, Ishizuka T, Yawo H, Yizhar O, Kandori H (2017) Functional characterization of sodium-pumping rhodopsins with different pumping properties. PLoS One 12:e0179232
Volkov O, Kovalev K, Polovinkin V, Borshchevskiy V, Bamann C, Astashkin R, Marin E, Popov A, Balandin T, Willbold D, Büldt G, Bamberg E, Gordeliy V (2017) Structural insights into ion conduction by channelrhodopsin 2. Science 358:6366
Weidlich O, Schalt B, Friedman N, Sheves M, Lanyi JK, Brown LS, Siebert F (1996) Steric interaction between the 9-methyl group of the retinal and tryptophan 182 controls 13-cis to all-trans reisomerization and proton uptake in the bacteriorhodopsin photocycle. Biochemistry 35:10807–10814
Wiegert JS, Mahn M, Prigge M, Printz Y, Yizhar O (2017) Silencing neurons: tools, applications, and experimental constraints. Neuron 95:504–529
Wietek J, Wiegert JS, Adeeishvili N, Schneider F, Watanabe H, Tsunoda SP, Vogt A, Elstner M, Oertner TG, Hegemann P (2014) Conversion of channelrhodopsin into a light-gated chloride channel. Science 344:409–412
Yamauchi Y, Konno M, Ito S, Tsunoda SP, Inoue K, Kandori H (2017) Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys Physicobiol 14:57–66
Ye H, Daoud-El Baba M, Peng RW, Fussenegger M (2011) A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332:1565–1568
Yoshida K, Tsunoda SP, Brown LS, Kandori H (2017) A unique choanoflagellate enzyme rhodopsin exhibits light-dependent cyclic nucleotide phosphodiesterase activity. J Biol Chem 292:7531–7541
Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446:633–639
Zhang F, Prigge M, Beyrière F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K (2008) Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci 11:631–633
Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, Magnuson J, Hegemann P, Deisseroth K (2011) The microbial opsin family of optogenetic tools. Cell 147:1446–1457
Acknowledgments
This work was supported by Japanese Ministry of Education, Culture, Sports and Technology Grants 18H03986, 19H04959, and JST CREST Grant JPMJCR1753.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kandori, H. (2021). History and Perspectives of Ion-Transporting Rhodopsins. In: Yawo, H., Kandori, H., Koizumi, A., Kageyama, R. (eds) Optogenetics. Advances in Experimental Medicine and Biology, vol 1293. Springer, Singapore. https://doi.org/10.1007/978-981-15-8763-4_1
Download citation
DOI: https://doi.org/10.1007/978-981-15-8763-4_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-8762-7
Online ISBN: 978-981-15-8763-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)