Abstract
Lignocellulose is considered as one of the most copious biopolymer accessible on this planet. Lignocellulosic hydrolysis which yields sugar and phenolics is a must for fermentation processes and pilot scale production of value added products. Cellulases are the class of enzymes which are mainly produced by fungi and bacteria and help in cellulose hydrolysis by acting on the β-1,4 linkages of cellulosic chains. The microbial cellulases have been found to be used in several industries such as biofuel, food, brewing, textile and laundry. Recently, functional metagenomics have been found to be an important strategy for the discovery of cellulose genes. However, the efficiency of such techniques for enzyme discovery from environmental metagenomes is not sufficient to meet the increasing industrial demands. Scientific and industrial advancements, role of metagenomics and future scenario related to the application of several cellulase pertaining to different industries will be discussed in this chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
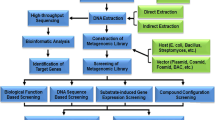
The genomic examination of a microbial population is called metagenomics which was termed by Handelsman et al. (1998) with an idea of collective analysis of similar but not identical genomes. The thought of exploring and investigating the ecological microbiome has unlocked new prospects with respect to utilization of uncultivated microbial populations. Metagenomics is a comprehensive approach which has provided a path breaking process for recovery of unculturable microbes. One of the most thriving fields in biotechnology is the application of microorganisms for the production of value added products such as antibiotics, enzymes etc. Also, there is a huge requirement for large scale production of enzymes in industries for commercial purpose.
A wider range of biochemical properties and higher growth rate attributed to microorganisms, allow them to become potential candidates to be employed as industrial enzymes (Adrio and Demain 2014). One of the most important commercial applications of metagenomics includes innovations related to antibiotics, bioremediation, and detection of biocatalysts (Gilbert and Dupont 2011). Majority of the industrial catalysts are produced in European Union (60%), whereas the rest of the enzymes production is attributed by USA and oriental nations. However, south east Asian countries such as India, China, Korea are emerging as enzyme production hubs with efficient research and development sector in global catalyst market (World Enzymes 2011).
Cellulase Classification
Cellulose has compact polymeric arrangement which remain enclosed in a highly complex matrix of hemicellulose and pectin (Demain et al. 2005). Majority of enzymes have been observed to take part in cellulosic degradation through microbial genome sequencing. Microbial enzymes are found in huge variety possessing diversified properties, yields, structure and functions, thus differing catalytic properties. Diversified forms of enzyme may be produced as a result of horizontal gene transfer from microbes residing in similar conditions and ecological niche (Jorgensen 2007).
Cellulose degrading enzymes widely known as cellulases are capable of hydrolyzing the cellulosic biomass by attacking the β-1, 4 glycosidic bonds of the polymers (Juturu and Wu 2014). The degradation of cellulosic biomass requires three distinct factors related to extracellular cellulase enzyme system (Acharya and Chaudhury 2012). The components in the enzyme system include β-1, 4 glucosidase, Exo-1, 4-β-glucanase and Endo-1, 4-β-glucanase. The endo-1, 4-β-glucanase also known as carboxymethyl cellulase catalyzes the breakdown of cellulose chain polymers into shorter ones, whereas exo-1, 4-β-glucanase widely known as cellobiohydrolase acts on the non-reducing end of the polymeric chain and β-1, 4 glucosidase disrupts the glucosidic bond of cellodextrins and cellobiose to release free glucose molecules. Endoglucanase attacks at the random sites of the cellulose polymeric chain, hence producing varying lengths of oligosaccharides (Sharada et al. 2014). Exoglucanase however, attacks on mostly reducing ends of cellulose polymeric chain producing glucose and cellobiose as key products.
Results reported by Schallmey and coworkers suggest identification of polyhydroxyalkanoate synthase encoding genes (Schallmey et al. 2011). The sequence analysis of the cloned samples revealed homology to the sequences (≈61–67%), however significant difference in functions from the PHA (phaC and phaA) encoding genes. Similar study by Cheema et al. (2012) revealed nine novel PHA synthase genes in a fosmid metagenomic library from oil contaminated soil.
One of the fosmid clones from the metagenomic library showed 76% sequence similarity with Alcaligenes sp. synthase. Enzymes like cellulases, xylanases, proteases, amylases and many more have been unclocked from genetically untapped resources via metagenomics.
The present review highlights the latest development in the field of metagenomics related to industrial biotechnology with respect to cellulase.
Role of Metagenomics in Harnessing Cellulases
Cellulose is considered to be one of the most obtained biopolymer on earth. The breakdown of cellulose is catalyzed by cellulose enzyme, hence finding its utilization in industries related to detergent, paper recycling, and juice extraction. Cellulase stands as one of the largest enzymes w.r.t economic turnover. Based on the oxygen availability to the microorganisms, cellulase enzyme can be classified into complexed and non-complexed forms (Lynd et al. 2002). The complexed forms of these enzymes mainly consist of cellulosome which is mostly found in anaerobic cellulose degraders. The cellulosomes are firmly attached to the cell wall and its flexible nature allows it to bind cellulose (Schwarz 2001). However, in non-complexed cellulase system, the enzyme subunits are flexible and can be collected from the supernatant of the aerobic microbial culture (Rapp and Beerman 1991). Presently, non-complexed cellulase enzyme systems are widely utilized for most of the industrial applications (Acharya and Chaudhary 2012). One of the most important industrial producers of cellulase is Hypocrea jecorina which hydrolyzes plant biomass to sugars (Kubicek et al. 2009). Such microbes can thrive the unfavorable conditions and produce stable enzyme which potentially help in catalyzing bioconversion reactions (Knapp 1985). Reports from Kanafusa-Shinkai et al. (2013) suggest that the cellulase enzyme system of Caldicelluloseruptor bescil are 2× active than Hypocrea jecorina.
The technique metagenomics has been employed to discover new cellulose from different environments such as soil samples from cold zones, compost and rumen by preparing metagenomic library clones (Yeh et al. 2013; Lee et al. 2006; Gong et al. 2013; Table 2). These enzymes have achieved special mention in industrial sector as they have potential capacity to convert biomass to renewable energy. Approximately 27.755 potential genes having significant match to catalytic domain were obtained by Hess et al. (2011). The study suggested presence of cellulolytic genes in large numbers which can be utilized to prepare genome drafts of uncultivated microorganism involved in biomass hydrolysis. In a study by Alvarez et al. (2013), identified and characterized novel cellulase (CelE1), which showed maximum catalytic properties at pH 7.0 and 50 °C and significant activity in alkaline conditions. The ruminal metagenomic library revealed 61 clones expressing cellulase activities were isolated (Duan et al. 2009). The amino acid sequences matched via SMART bioinformatics tool revealed 14 cellulase genes with signal peptide and glycosyl hydrolase family 5 catalytic domain. However, for genes DM1–1 and M8–2 signal peptides were not available (Duan et al. 2009).
In another study by Voget et al. (2006), the biochemical characterization of cellulase (Cel5 A, endoglucanase) from soil sample. The study reported presence of novel cellulase Cel5A extracted from soil samples through biochemcial identification. The obtained sequence of 363 amino acid had functional similarity of 77% with cellulase activity of Cellvibrio mixtus. The Cel5A (1092 bp ORF) when cloned in E. coli and the related protein (42.1 kDa) was purified employing chromatography which was found to be active against cellulose contents with −1,4 linkages. The enzyme showed optimum activity at pH 6.5 in an activity range of pH 5.5–9.0. The unusual properties of the enzyme cellulase allow it to be potential candidate in industrial sector for bioprocess and its added values. In a recent study by Wong et al. (2017), metagenomic analysis of gut microbiome of Castor canadensis and Alces americanus revealed presence of cellulose degrading microbes having phylogenetic origins derived from Firmicutes, Bcateroidetes, and Proteobacteria. Sequences belonging to class Clostridia and Bacteroidia contributed the highest hits (23–52%) for the carbohydrate active enzymes across both the metagenomes.
Study by Ransom-Jones et al. (2017) investigated the lignocellulose-degrading microbial diversity from landfill site. Metagenomic analysis suggested the dominance of Firmicutes, Bacteroidetes, Fibrobacteres and Spirochaetes. Functional analysis revealed presence of ≈3385–4223 CAZymes for Firmicutes and Bacteroidetes respectively. Six distinct CAZyme families were obtained via functional annotation for Spirochaetes. Recent analytical investigation made by Wilhelm et al. (2019) reported the microbial community in forest soil through quantitative stable isotope probing and metagenomic genome assembling analysis across North America. The active cellulolytic populations observed were Deltaproteobacteria, Gammaproteobacteria, Planctomyces, and Ascomycota. Cellulose degrading microbes observed in the functional annotation were Caulobacter, Janthinobacter, and Salinibacterium.
Study by Kanokratana et al. (2015) revealed various types of glycosyl hydrolase from sugarcane bagasse collection site through fosmid metagnomic library preparation. Bioinformatic analysis of the xylan positive fosmids suggested presence of endo- β−1,4-xylanase of GH11 family. Additionally, two genes (cel9 and xyn11) were observed to be expressed in E. coli. The enzymes expressed were found to be active at thermophilic temperatures (75–80 °C) and acidic pH.
Reports from recent study by Wang et al. (2016) suggested involvement of Actinobacteria in lignocellulosic decomposition from compost. Metagenomic analysis reported dominance of Actinobacteria, however, presence of Proteobacteria, Bacteroidetes, Firmicutes were also observed. Actinomycetes were observed to have high CAZyme gene disribution (46.1%) which retained enzymes like β-glucosidase, cellobiohydrolase, and ligninase genes (Table 1).
Application of Microbial Cellulases
Cellulosic breakdown occurs in both oxic and anoxic conditions. Various anaerobic microbes as cellulosic degraders have been reported in several studies (Freier et al. 1988; Hamilton-Brehm et al. 2010; Kato et al. 2004; Table 2). The non-complexed cellulose enzyme system subunits are reported from aerobic microbial degraders such as, fungi and bacteria with special mention to fungal cellulases (Resch et al. 2013; Table 2).
Agriculture
Several fungal genus such as Trichoderma, Penicillium, Chaetomium have been observed to act significant in agriculture by increasing crop production, fecilitating plant growth and allowing enhanced seed germination (Phitsuwan et al. 2012). Additionally, it has been reported that few fungal enzymes are potential candidate for attacking the pathogenic strains. Enzymes such as B-1, 3-d-glucanase and N-acetyl glucosaminidase have been reported to degrade the spore germination of B. cinerea (Howell 2003). Several microorganisms die due to scanty nutrient available to them due to the presence of Trichoderma sp. (Waghunge et al. 2016). Hence, nutrient competition to Trichoderma sp. is the most common phenomenon responsible for pathogen death. This fungal candidate also helps to promote induced resistance in host plant body by initiating certain chemical production. They promote plant development by initiating an endophytic mode. Fungal cellulases also help in restoring soil quality by degrading the lignocellulosic biomass inside the soil.
Food Processing
Cellulase has managed a strong position in food and feed industry. It is an integral component of maceration unit utilized for extraction and processing of juice and pulp for juice and puree productions (Rai et al. 2007). Cellulase helps in preventing pigment oxidation by generating stable protein bound pigments (Table 3). Report from Kuhad et al. (2011) suggest that cellulase with some more enzymes has been utilized to enhance the taste of citrus fruits.
Study by Çinar (2005) indicated the combination of cellulase with other cell wall degrading enzymes such as pectinase and hemicellulase are employed to improve nutritional level of forages. Another study by Kung et al. (1997) suggested an improved and enhanced digestion of animal fodder and feed in combination with cellulase.
Brewery Industry
Preparation and production of ethanol is enhanced with the help of utilization of cellulase to hydrolyze polymeric substances to simple sugars. Beer quality is dependent on the enzyme activity during malting and subsequent fermentation phases. Seed reserve hydrolysis by B-glucanase and other enzymes (A and B amylase and carboxypeptidase) influence better seed germination which in turn influences malting (Bhat 2000). Though in practice, brewers employ low quality substrate which has poor activity of endo-β-glucanase due to climatic variations. This allows formation of non starch polysaccharide (up to 10% β-glucan), hence gel like formation during brewing process resulting in low level wort filtration and poor alcohol yield.
Textile Industry
Enzyme cellulase has been widely accepted in textile industry for its capability to enhance softness and fabric quality. It has been been employed for biopolishing of cotton fabrics and biostoning of denims to provide the new generation stonewash appearances. The enzyme employed, hydrolyzes the fibre lumps from the fabric surface and removes the dye attached to such protrusions, hence imparting the faded look and development of color gradient to the denim (Arja 2007). This process is called biostoning. Cellulase is widely known for its use in wet processing in textile industry to enhance the appearance of cotton clothings. Cellulase has also been utilized in preparation of detergents to improve the softness, and color appearnaces of fabrics.
Conclusion
Metagenomics imparts a scope to explore the undiscovered microbial biodiversity from a microbiome and utilize the untapped potential of the microbes to generate value added products and processes. Recently, the industrial sector has also gained economically by major innovations such as unlocking of novel enzyme functions made via microbial metagenomics. Recent reports analyzing the sequences via cloning of metagenomic inserts has resulted in isolation and identification of untapped microbial communities for value added products such as significant fermentation processes and enzyme production. In spite of novel investigations and reports from several studies, a lot remains to be understood about the cellulase enzyme alongwith the microbial mechanisms involved. Metagenomic studies have till date helped in identification of novel cellulolytic biocatalysts amongst other enzyme varieties. However, a vast reserve of enzymes is yet to be characterized. Furthermore, approaches like metatranscriptomic and stable isotope probing should be employed for innovations of enzyme systems. Significant bioprocesses should be developed so that the cellulosic wastes can be efficiently treated and used as cost-effective carbon source.
References
Acharya S, Chaudhary A (2012) Bioprospecting thermophiles for cellulase production: a review. Braz J Microbiol 843:44–856
Adrio JL, Demain AL (2014) Microbial enzymes: tools for biotechnological processes. Biomol Ther 4:117–139
Alvarez TM, Paiva JH, Ruiz DM et al (2013) Structure and function of a novel cellulase 5 from sugarcane soil metagenome. PLoS One 8:e83635
Arja MO (2007) Cellulases in the textile industry. In: Polaina J, MacCabe AP (eds) Industrial enzymes. Springer, Dordrecht, pp 51–63
Bhat MK (2000) Cellulases and related enzymes in biotechnology. Biotechnol Adv 18:355–383
Cheema S, Bassas-Galia M, Sarma PM, Lal B, Arias S (2012) Exploiting metagenomic diversity for novel polyhydroxyalkanoate synthases: production of a terpolymer poly (3-hydroxybutyrate-co-3-hydroxyhexanoate-co-3-hydroxyoctanoate) with a recombinant Pseudomonas putida strain. Bioresour Technol 103:322–328
Çinar I (2005) Effects of cellulase and pectinase concentrations on the colour yield of enzyme extracted plant carotenoids. Process Biochem 40:945–949
De Faveri D, Aliakbarian B, Avogadro M, Perego P, Converti A (2008) Improvement of olive oil phenolics content by means of enzyme formulations: effect of different enzyme activities and levels. Biochem Eng J 41:149–156
De Vries M, Schöler A, Ertl J, Xu Z, Schloter M (2015) Metagenomic analyses reveal no differences in genes involved in cellulose degradation under different tillage treatments. FEMS Microbiol Eco 91:69
Demain AL, Newcomb M, Wu JHD (2005) Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154
Dienes D, Egyházi A, Réczey K (2004) Treatment of recycled fiber with Trichoderma cellulases. Ind Crop Prod 20:11–21
Duan CJ, Xian L, Zhao GC et al (2009) Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol 107:245–256
Freier D, Mothershed CP, Wiegel J (1988) Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol 54:204–211
Gilbert JA, Dupont CL (2011) Microbial metagenomics: beyond the genome. Ann Rev Marine Sci 3:347–371
Gong X, Gruniniger RJ, Forster RJ, Teather RM, McAllister TA (2013) Biochemical analysis of a highly specific, pH stable xylanase gene identified from a bovine rumen-derived metagenomic library. Appl Microbiol Biotechnol 97:2423–2431
Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M et al (2010) Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl Environ Microbiol 76:1014–1020
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:R245–R249
Hess M, Sczyrba A, Egan R et al (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467
Howell CR (2003) Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis 87:4–10
Jiménez DJ, de Lima Brossi MJ, Schückel J, Kračun SK, Willats WGT, van Elsas JD (2016) Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches. Appl Microbiol Biotechnol 100:10463–10477
Jorgensen KS (2007) In situ bioremediation. Adv Appl Microbiol 61:285–305
Juturu V, Wu JC (2014) Microbial cellulases: engineering, production and applications. Renew Sust Energ Rev 33:188–120
Kanafusa-Shinkai S, Wakayama J, Tsukamoto K, Hayashi N, Miyazaki Y, Ohmori H, Tajima K, Yokoyama H (2013) Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium. J Biosci Bioeng 115:64–70
Kanokratana P, Eurwilaichitr L, Pootanakit K, Champreda V (2015) Identification of glycosyl hydrolases from a metagenomic library of microflora in sugarcane bagasse collection site and their cooperative action on cellulose degradation. J Biosci Bioeng 119:384–391
Karmakar M, Ray RR (2011) Current trends in research and application of microbial cellulases. Res J Microbiol 6:41–53
Kato S, Haruta S, Cui ZJ, Ishii M, Yokota A, Igarashi Y (2004) Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int J Syst Evol Microbiol 54:2043–2047
Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2005) Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol Ecol 51:133–142
Knapp JS (1985) Biodegradation of cellulose and lignins. Comprehen Biotechnol 4:835
Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B (2009) Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels 2:19
Kuhad RC, Gupta R, Singh A (2011) Microbial cellulases and their industrial applications. Enzyme Res 2011:280696
Kung L Jr, Kreck EM, Tung RS, Hession AO, Sheperd AC, Cohen MA, Swain HE Leedle JA (1997) Effects of a live yeast culture and enzymes on in vitro ruminal fermentation and milk production of dairy cows. J Dairy Sci 80:2045–2051
Lee M, Lee C, Oh T, Song JK, Yoon J (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409
López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P (2016) Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279
Lynd LR, Weimer PJ, Van Zyl WH, Isak S, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
McDonald JE, Houghton JNI, Rooks D, Allison HE, McCarthy AJ (2012) The microbial ecology of anaerobic cellulose degradation in municipal waste landfill sites: evidence of a role for fibrobacters. Environ Microbiol 14:1077–1087
Milala MA, Shugaba A, Gidado A, ENe AC, Wafar JA (2005) Studies on the use of agricultural wastes for cellulase enzyme production by Aspergillus niger. Res J Agric Biol Sci 1:325–328
Phitsuwan P, Laohakunjit N, Kerdchoechuen O, Kyu KL, Ratanakhanokchai K (2012) Present and potential applications of cellulases in agriculture, biotechnology, and bioenergy. Folia Microbiol 58:163–176
Pottkämper J, Barthen P, Ilmberger N, Schwaneberg U, Schenk A, Schulte M, Streit WR (2009) Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green Chem 11:957
Rai P, Majumdar G, Gupta SD, De S (2007) Effect of various pretreatment methods on permeate flux and quality during ultrafiltration of mosambi juice. J Food Eng 78:561–568
Ransom-Jones E, McCarthy AJ, Haldenby S, Doonan J, McDonald JE (2017) Lignocellulose-degrading microbial communities in landfill sites represent a repository of unexplored biomass-degrading diversity. mSphere 2:e00300–e00317
Rapp P, Beerman (1991) Bacterial cellulases. In: Weimer CH (ed) Biosynthesis and degradation of cellulose. Marcel Dekker, New York, pp 535–595
Resch MG, Donohoe BS, Baker JO, Decker SR, Bayer EA, Beckham GT, Himmel ME (2013) Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ Sci 6:1858–1867
Schallmey M, Ly A, Wang C et al (2011) Harvesting of novel polyhydroxyalkanaote (PHA) synthase encoding genes from a soil metagenome library using phenotypic screening. FEMS Microbiol Lett 321:150–156
Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol 56:634–649
Sharada R, Venkateswarlu G, Venkateswar S, AnandRao M (2014) Applications of cellulases—review. Int J Pharm Chem Biol Sci 4:424–437
Singh A, Kuhad RC, Ward OP (2007) Industrial application of microbial cellulases. In: Kuhad RC, Singh A (eds) Lignocellulose biotechnology: future prospects. I.K. International Publishing House, New Delhi, pp 345–358
Sizova MV, Izquierdo JA, Panikov NS, Lynd LR (2011) Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 77:2282–2291
Thornbury M, Sicheri J, Slaine P, Getz LJ, Finlayson-Trick E, Cook J et al (2019) Characterization of novel lignocellulose-degrading enzymes from the porcupine microbiome using synthetic metagenomics. PLoS One 14:e0209221
Voget S, Steele HL, Streit WR (2006) Characterization of a metagenome-derived halotolerant cellulose. J Biotechnol 126:26–36
Waghunge RR, Rahul MS, Ambalal NS (2016) Trichoderma: a significant fungus for agriculture and environment. Afr J Agr Res 11:1952–1965
Wang C, Dong D, Wang H, Müller K, Qin Y, Wang H, Wu W (2016) Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels 9:22
Wang L, Jiang X, Xu H, Zhang Y (2018) Metagenomic analysis of cellulose and volatile fatty acids metabolism by microorganisms in cow rumen. BioRxiv 414961
Wilhelm RC, Singh R, Eltis LD, Mohn WW (2019) Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J 13:413–429
Wong MT, Wang W, Couturier M et al (2017) Comparative metagenomics of cellulose- and poplar hydrolysate-degrading microcosms from gut microflora of the Canadian Beaver (Castor canadensis) and North American Moose (Alces americanus) after long-term enrichment. Front Microbiol 8:2504
World Enzymes (2011) Industry study with forecasts for 2015 & 2020. The Freedonia Group, USA, pp 1–376
Yeh Y, Chang SC, Kuo H, Tong C, Yu S, Ho TD (2013) A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 519:360–366
Zhang L, Chung J, Jiang Q, Sun R, Zhang J, Zhong Y, Ren N (2017) Characteristics of rumen microorganisms involved in anaerobic degradation of cellulose at various pH values. RSC Adv 7:40303–40310
Zverlov VV, Schwarz WH (2008) Bacterial cellulose hydrolysis in anaerobic environmental subsystems- Clostridium thermocellum and Clostridium stercorarium, thermophilic plant fiber degraders. Ann NY Acad Sci 1125:298–307
Acknowledgement
PJ is thankful to Amity University, Kolkata for providing the infrastructure facilities. VK would also like to acknowledge Department of Science and Technology (DST), SERB, GoI for the financial support under Startup Research Scheme (File No.: SRG/2019/001279).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, V., Jha, P. (2020). Role of Metagenomics in Discovery of Industrially Important Cellulase. In: Shrivastava, S. (eds) Industrial Applications of Glycoside Hydrolases . Springer, Singapore. https://doi.org/10.1007/978-981-15-4767-6_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-4767-6_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4766-9
Online ISBN: 978-981-15-4767-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)