Abstract
The gut is an enormously complex environment in which microbiota, nutrients, and host cells interact widely, a process significant for the gut homeostasis and host health. Gut microbial organization plays many roles in the host body, including metabolic, barrier effect, and trophic functions. Many diseases have henceforth been connected to gut microbiota impairment. This part provides in depth view of the close relationships between gut microbiota, host health, and disease.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Gut Microbiota
2.1.1 Introduction
Microbiota is a sophisticated community of microorganisms comprising of bacteria, viruses, protozoa, and fungi, dwelling in various zones of human body, for example, mouth, respiratory framework, skin, gastroenteric tube, and vagina [1]. More than 70% of microbiota resides within the gastrointestinal (GI) tract in a mutually beneficial association with its host, spreading continuously from gastric lumen to colon/rectum, where it arrives at its most severe concentration.
The human gastrointestinal tract (GIT) constitutes the largest interfaces (250–400 m2) among the host, ecological elements, and antigens within the human body. Approximately, 60 tons of food runs through the human GIT in an average lifespan, along with an abundance of environmental microorganisms that pose a major threat to the integrity of gut [2]. Assortment of bacteria, eukarya, and archaea occupying the GIT is named as “gut microbiota” and has co-developed with the host to establish a complex, and mutually beneficial connection [3, 4]. The mammalian GIT has higher and varied amount of microbes, best-known as intestinal microbiota. Archaea, bacteria, protozoa, fungi, and viruses live together and associate with the host, especially immune and epithelial cells [5]. The quantity of microorganisms living in GIT has been evaluated to surpass 1014 that include approximately 10 times more bacterial cells than the quantity of human cells and more than 100 times the quantity of genomic material (microbiome) as the human genome [3, 6]. Nevertheless, an amended estimate has recommended that the proportion of bacterial: human cells is probably close to 1:1 [1]. Because of the immense quantity of bacterial cells in the body, the host and microorganisms occupying it are often mentioned as a “superorganism” [6, 7].
Microbiota provides numerous advantages for the host, by means of physiological roles, for example, reinforcing the integrity of gut or forming the epithelium of intestine [8], extracting energy [9], guarding from pathogens [10], and controlling immunity of host [11]. Because of a modified microbial composition, known as dysbiosis, there is possibility for the disruption of above-mentioned mechanisms. With the development of progressively advanced methods to characterize sophisticated biological systems, a function of the microbiota in an enormous number of intestinal and extra-intestinal diseases has become consistently evident [12, 13]. This chapter summarizes our present comprehension of the human GI microbiota composition and development, and its effect on host health and gut integrity.
2.1.2 Structure and Composition of the Human GI Microbiota
An adult gut microbiota contains 10 to 100 trillion microbes, which is 10 times the quantity of total somatic and germ cells of humans [14]. Gut microbiome contain 100- to 150-times more genes than human genome [15]. The gut microbiota has co-developed with humans and has demonstrated significant consequences for different host reactions. The modified composition of gut microbiota has been connected to metabolic diseases, like obesity, diabetes, or non-alcoholic fatty liver diseases. Such studies have shown the significance of gut in modulating metabolic disorders and host metabolism.
Intestinal microbiota comprises autochthonous individuals occupying the gut mucosa, as well as transitory microbiota that is component of the food consumed. Gut microbiota has been assessed to include more than 100 distinct species in every organism. Around 1500 unique species were described as component of the human gut microbiota. Intestinal microbiota is established by a total of 1013–1014 microbial cells and is generally expected to represent ten times more cells than eukaryotic cells of humans. Large intestine is the site of the body with highest abundance of microbes, with 1011–1012 cells/g of intestinal matter [16]. Bacteria rule the gut microbiota, which is mainly portrayed by Firmicutes and Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, Synergistetes, and Verrucomicrobia [17]. Fungi and archaea account for up to 1% of the human gut microbiota species [18]. Among the major typical genera of the above-mentioned phyla, Bacteroides sp., Prevotella sp., Blautia sp., Clostridium sp., Ruminococcus sp., Faecalibacterium sp., and Bifidobacterium sp., (in breast-fed infants) are important because of their high abundance [15, 19].
It has been recently suggested that all of the inter-individual variation of intestinal microbiota could be categorized into enterotypes, characterized as a system of co-abundant microbial communities controlled by the salient existence of one of these three genera: Bacteroides, Ruminococcus, and Prevotella [20]. Some authors found enterotypes to be a very simplified theory, thus, decreasing the complexity of intestinal microbiota into three groups [21]. For example, only two of these enterotypes [22] have been identified by some authors, as two perpetual clusters of microbiota configurations isolated by a gradient of bacterial species with varied abundances [23]. Classifying the intestinal microbiota into enterotypes or other classes, having strong connections with dietary patterns, could be very useful in customizing the cure of diseases continuing with microbial dysbiosis [24]. This will necessitate the advancement of mathematical models capable of consolidate the entire complexity, and subsequently more experimental data will be required [25].
Evolution of next-generation DNA sequencing technologies over the last 10 years has permitted a profound comprehension of microbial composition of species living in the gut, upper airways of the respiratory tract, vagina, skin, or mouth. Research was conducted to study about the improvement of diversity of gut microbiota because of the advent of culture-independent methodologies, for example, low-cost and high-throughput sequencing strategies. Focusing on 16S ribosomal RNA (rRNA) gene of bacteria is a well-known methodology [26, 27] as this gene occurs in all archaea and bacteria and comprises nine highly variable (V1–V9) regions, thus permitting the easy recognition of species. Previous strategies focused on sequencing the whole 16S rRNA gene. By utilizing this strategy, the strong insensitivity and bias of culturing techniques were featured in an early investigation, as 76% of the sequences of rRNA acquired from an adult male fecal sample belonged to new and uncharacterized species [28]. Lately, the focal point of 16S rRNA sequencing has moved towards more prominent depth investigation of shorter subregions of gene [27]; even so, the usage of shorter read lengths will lead to errors [26]. More accurate estimation of microbiota composition and diversity might be given by entire genome shotgun metagenomics because of the sensitivity, and high resolution of these methods [26]. The most detailed perspective of human-related microbial selection to date has been provided by combined knowledge from the human microbiome project and MetaHit [29, 30]. Accumulated information from these investigations grouped 2172 species, isolated from humans, into twelve separate phyla, out of which 93.5% species belonged to Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. Three of the twelve distinguished phyla enclosed just a single species isolated from humans, along with an intestinal species, Akkermansia muciniphila, the sole recognized representative of Verrucomicrobia phyla. 386 species known in humans are anaerobic and are located mostly in mucosal habitats, for example, GIT and oral cavity [29].
2.1.3 Metabolic Roles of Microbiota
Mammals have a restricted inherent ability to process polysaccharides, but they can assimilate simple sugars in the small intestine. The primary substratum for the growth and maintenance of intestinal flora is all the indigestible elements, which represents the main energy source in colon [31, 32]. Since the microbiota’s genetic and species diversity gives various host-related enzymatic, metabolic, and biochemical pathways, the outcome is energy extraction, digestible substrates for the host, and an energy and nutrients supply for the expansion of particular inhabitant species of bacteria [33]. Thus, the microbiota is known as an important metabolic organ [34].
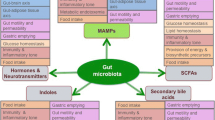
Intestinal bacteria, primarily Firmicutes, Bacteroidetes, and Actinobacteria, obtain energy from the transformation and fermentation of indigestible food substrates, especially from carbohydrate fermentation. Indigestible polysaccharides break down into monosaccharides, and later into bacterial fermentation products, particularly gases (CO2 and H2) and short-chain fatty acids (SCFAs) [35, 36]. For adults, the average supply of substrates is around 5–20 g of carbohydrates and 20–60 g of proteins. The fermentation process achieves high levels with an abundant generation of SCFAs in the ascending colon and cecum, where the pH is relatively acidic (in the range of 5 and 6) and the growth of bacteria is rapid. The supply of substrates reduces in the distal colon (having neutral pH), where the activity of bacterial community reduces dramatically and putrefactive procedures become quantitatively more crucial. Therefore, the generation of SCFAs (butyrate, propionate, acetate in the proportion 15:25:60) portrayed metabolic endpoint, which employ a strong trophic and energetic activity in the intestinal lumen [37]. Bacteroidetes generates acetate and propionate by degrading the undigested polysaccharides, and Firmicutes creates butyrate [38]. Acetate is ingested and afterward transferred to the peripheral level, and there it serves as a substratum for cholesterol synthesis, while propionate takes an active part in gluconeogenesis. Butyrate, as a primary energy source for colonocytes, enhances the sensitivity to insulin in mice and has a potential anti-obesogenic activity and also an anti-inflammatory effect [39]. Butyrate and different SCFAs have a major role in controlling intestinal cell proliferation and growth of obesity [40]. Butyrate encourages the constancy of cellular heritage, preferring the transformation of cells from neoplastic to non-neoplastic phenotype. Production of SCFAs is also induced by the anaerobic metabolism of protein substrates and/or peptides that may produce harmful components such as ammonia, thiols, amines, indoles, and phenols. SCFAs are responsible for performing various biological activities, such as modulation of glycemia [41], action on glucose homeostasis [42], inhibitory control of excessive production of cholesterol [36], regulation of satiety through peptides [43], increasing intake of energy without increasing the peptide YY or glucagon-like peptide 1 concentration in humans and rodents [44, 45], management of bowel kinetic activity, transport of fluid, muco-protective action [46], anti-carcinogenic action [47], and anti-inflammatory action [48] (Fig. 2.1).
Functions of intestinal microbiota [49]
Microbiota can influence its own composition as well. The production of SCFAs differs depending on the fermentable carbohydrates existing in the bowel lumen [50, 51] that can alter the microbiota composition itself. Furthermore, starch resistant to digestion has been reported to directly enhance levels of butyrate in humans [52], and arabinoxylan, formed by the prebiotic arabinoxylan oligosaccharides, enhances levels of propionate in transversal colon [53]. Also, the microbiota conducts another significant metabolic functions, for instance, at the intestinal level it is necessary for synthesis of certain enzymatic co-factors and vitamins (folic acid, pantothenic acid, vitamin B1, B2, B6, B12, PP, H, K) and for the assimilation of iron, calcium, and magnesium [38]. It is additionally accountable for bile acids deconjugation in the liver catalyzed by an enzyme bile salt hydrolase that exists in numerous species of bacteria. Hydrolysis hinders the reuptake of these molecules by enterocytes while promoting their elimination and blocking their enterohepatic recirculation [54]. The interference of intestinal bacteria in hepatic transformation of cholesterol into bile acids, with significant implications in fat assimilation, has therefore been proven (Table 2.1).
2.1.4 Development of the Human GI Microbiota
The human GIT begins from the mouth, spreading through the anatomical regions—the esophagus, stomach, small intestine, colon, rectum, and terminating at the anus [69]. The structural and functional growth of GIT is a pivotal component of human growth, since the gut must harbor the heterogeneity of dietary inputs and external antigens which are incorporated along with food into human body across various phases of life [70]. Human GIT maturation begins in utero and proceeds after birth with certain roles, for example, epithelial barrier systems, intestinal immune system, and accessory structures [70]. The primitive gut is formed about 22 days after conception from the dorsal portion of yolk sac, directing towards the emergence of foregut, midgut, and hindgut, around 25 days after conception [71]. The midgut increases quickly in length so far that it cannot fit within the developing abdominal cavity and herniates into the vitelline sac before experiencing complex turns and coming back to the abdominal cavity after gestation period of around 10 to 12 weeks [71].
It is assumed that the production of microbiota starts from birth, despite the fact that this dogma is confronted by a confined various investigations in which microorganisms have been found in womb tissues, such as placenta [72, 73]. GIT is quickly colonized after birth, with life events, for example, sickness, changes in diet, and antibiotic treatment causing disordered microbiota shifts [73, 74]. Mode of delivery seems to affect the microbiota composition, with microbiota of infants delivered vaginally possessing higher number of Lactobacilli during the initial days, as a result of elevated Lactobacilli load in the flora of vagina [75, 76]. The microbiota of infants born by C-section is insufficient and deferred in the colonization of Bacteroides genus, but are colonized by facultative anaerobes like Clostridium species [77,78,79]. The microbiota is commonly low in diversity in the initial stages of development, and is governed by two fundamental phyla, Actinobacteria and Proteobacteria [73, 80]. Microbial abundance increases during the first year of development, and the composition of microbiota changes to adult-like microbial profile with time-related patterns specific to each newborn child [81]. At around 2.5 years old, the newborn child microbiota’s composition, diversity, and functional capabilities are close to those of adult microbiota [73, 74]. Despite the fact that the composition of gut microbiota is generally steady in adulthood, it remains exposed to perturbation by life events [82]. The microbial community shifts in people aged over 65 years, with an elevated prevalence of Bacteroidetes and Clostridium cluster IV, in comparison to young individuals with more prevalent cluster XIVa [83]. Another report discovered the similarity of microbiota of young generation and an elderly population (70 years), and a significant decline of microbiota diversity from a cohort of centenarians [84]. A notable relationship among diversity and living arrangements has been identified in the older population, like group dwelling or long-term residential care [85]. Microbiota’s ability to perform metabolic processes, such as SCFA synthesis, and amylolysis, is typically decreased in elders, while there is an increase in proteolytic activity [86]. With increasing evidence of the role of SCFAs as metabolic and immune mediators, the decline in SCFAs was believed to support the inflammation-ageing process in aged people’s intestine [87].
Advances in metagenomic technologies have revealed the composition of human gut microbiota from early infancy [81] to old age [88]. The human intestine after birth is quickly occupied by a variety of factors and microbes considered to impact colonization which involves gestational age, delivery mode, sanitation, diet, and antibiotic treatment [89, 90]. Facultative anaerobes are the first colonizers, which builds a new environment promoting the colonization of anaerobes such as Bacteroides, Bifidobacterium, and Clostridium sp. Low diversity and relative abundance of Proteobacteria and Actinobacteria define the intestinal microbiota of neonates, which becomes more complicated with the growth and abundance of Firmicutes and Bacteroidetes as time period after birth increases [91,92,93]. At the end of first year of development, infants have an individually defined microbial profile, converging towards the distinctive microbiota of an adult, so that by the age of 25, the microbiota completely matches the composition and diversity of an adult [74, 81, 94]. The initial three years of life serves as the most important phase for dietary interventions to promote child growth and development. At this time, the intestinal microbiota, a crucial tool for health and neuro-development [95] is developed and its modification during this phase can significantly influence health and development of host. Development of gut microbiota is influenced by various factors such as delivery mode, genetics, diet, health status, gestational age, etc. (Fig. 2.2).
Factors that affect the development of infant, adult, and elderly gut microbiota [96]
2.1.5 Biogeography of the Human Microbiota in GIT
The microbiota composition in GIT represents the physiological properties of a particular part and is formed on both a longitudinal and transverse axis [97]. Chemical, metabolic, and immunological gradients along the intestine affect the microbiota density and composition. There are usually elevated concentrations of acids, oxygen and anti-microbials in the small intestine, and a limited transition time [98]. These characteristics restrict the development of bacteria to such an extent that only quickly growing, facultative anaerobes having the capacity to bind to mucus/epithelia are thought to be enduring [98]. Lactobacillaceae dominates the microbial community of small intestine of mice [99]. Colonic environment supports a dense and abundant bacterial community, predominantly anaerobes having the capacity to use complex carbohydrates that are indigestible in the small intestine. The colon has been reported to be dominated by Lachnospiraceae, Prevotellaceae, and Rikenellaceae [98, 99]. Contrary to the different composition of microbiota within different GI organs, the microbiota of various colorectal mucosal areas in the same organism is conserved structurally in terms of diversity and composition [100, 101]. This property is evident even at the time of localized inflammation [101]. However, fecal/luminal and mucosal composition is significantly different [100, 101]. For instance, Bacteroidetes concentration is reported to be high in fecal/luminal samples than in the mucosal [19, 100]. Conversely, Firmicutes, primarily Clostridium cluster XIVa, are augmented in the mucus layer relative to the lumen [19]. Many experiments in mice colonized with pathogen-free microbiota demonstrated a distinct microbial niche formed by the large intestine’s outer mucus, and the bacterial species existing in the mucus exhibit differential proliferation and resource utilization relative to the same species in intestinal lumen [102].
Inter-individual differences in the arrangement of species and subspecies are suggested to overcome the variations in the organization of community in an individual [100, 103, 104]. The concept of a core microbiota has been projected, suggesting to be a group of the similar abundant species found in all individuals. In the set of microbial genes present between organisms, however, greater comparability can be seen than the taxonomic profile, indicating that the “core microbiota” might be best characterized at a functional rather than organismal level [103]. Individual microbiota arrangements have been recently classified into “community types” that are related with background and can be predictive of one another [105]. Multi-dimensional study of thirty-three samples from various nationalities uncovered the existence of three enterotypes recognizable by differences in the level of one of three genera: Bacteroides (enterotype 1), Prevotella (enterotype 2), and Ruminococcus (enterotype 3) [106]. Nevertheless, there is conflicting data encompassing the presence and development of these enterotypes [21].
2.1.6 Factors Influencing the GI Microbiota
The microbial community’s complexity and richness progress via a number of stages of development spanning from neonatal phase before the apparent stabilization after weaning. In combination with individuality, there are essential inter-linked factors that assume a significant part in forming the microbial composition of human GI. Those factors involve age [107, 108], diet [109, 110], genetics of host [109,110,111], infections, antibiotic usage [108,109,110], physiology of colonization site [69], birth mode [109, 110, 112], feeding type [109, 112], and the birth environment of infants [112].
Technical variation also influences the form of developing microbial composition. For instance, culture-dependent microbe identification procedures are subject to biases that emerge from: (1) sensitivity to oxygen; (2) intractability of some species of bacteria to culturing media; and, (3) competitiveness among fast-growing and slow-growing bacteria. It restricts the existing culture-dependent techniques to be effective for the isolation of only 70% of intestinal microbes in a sample relative to culture-independent methodologies [113].
2.1.6.1 Age
The infant’s microbiota is seeded during childbirth and is at first undifferentiated over the different body habitats. The predominance of aerobic bacteria at time of birth is changed during perinatal and postnatal development. During initial weeks of life, the microbiota diversifies to form a diverse microbial population dominated by anaerobes. This early stage of colonization corresponds with the stimulation of hypothalamic pituitary adrenal (HPA) axis that affects the enteric nervous system thus innervating the GIT 123]. Enteroendocrine cells of gut release a number of metabolically linked peptides, all of which are associated with food consumption, lipid accumulation, energy equilibrium and may be regulated by microbial metabolites, for example, SCFAs. Some investigations have shown that young people have a greater concentration of Bifidobacteria and Clostridia than adults; however, the gut microbiota is more stable during adult life. During old age, a final set of age-related changes in gut microbiota’s composition and function occurs [114]. Aging is related with modified physiological functions, involving function of immune system, which influence the makeup of the gut microbiota. Age-related differences detailed in composition of gut microbiota include rise in the total amount of facultative anaerobes, changes in the proportion of Bacteroidetes to Firmicutes, and a pronounced reduction of Bifidobacteria in humans > 60 years old, during which the immune system begins to weaken. Metabolic shifts that correlate with the development and maturation of gut microbiota can be seen in the excretion profiles of bacterial products of amino acid metabolism and in energy-linked metabolites [115].
2.1.6.2 Diet
Current research indicates that diet affects the gut microbiota enormously [98]. Meta-transcriptomic research has shown the ideal microbiota to be driven by the ability of microbial individuals to metabolize simple sugars, indicating microbiota’s adjustment to the abundance of nutrients in the small intestine [116]. Formation of colonic microbiota depends upon the accessibility of microbiota-accessible carbohydrates (MACs) present in dietary fiber. “Animal-based” or “plant-based” diets result in widespread modifications of human gut microbiota [117]. A crossover study showed the impact of fiber, indicating that otherwise balanced diets high in resistant starch or in non-starch polysaccharide fiber (wheat bran) lead to a powerful and reproducible augmentation of various species of bacteria in the human gut [118].
The role of food-consumed bacteria in gut microbiome had previously been underestimated, potentially as a result of methodological restrictions [119]. Various investigations have indicated that high-calorie diet brings obesity and type-2 diabetes (T2D) both in humans and mice [120,121,122,123,124]. Many evidences propose that the connection among diet and obesity is related to gut microbiota [125,126,127,128,129,130,131]. Changes in diet bring significant and rapid changes in gut microbiome composition, as indicated by various interventional studies [22, 132]. High-fat diet (60% fat) reduces the quantity of bacterial species in the gut microbiome of mice, and the composition of gut microbiome between mice on a high-fat (unpurified) diet and on a regular unpurified diet is totally different. Another study in obese mice having T2D revealed that the abundance of A. muciniphila was reduced and prebiotic feeding of A. muciniphila normalized its abundance, improved metabolic profiles, decreased fat mass, inflammation, and insulin resistance elicited by a high-fat diet [133]. It has been demonstrated that a fiber-rich diet is favorable to health, as it balances the gut microbiome [134]. Studies of 16S rRNA sequencing in humans have categorized the gut microbiota of humans into various enterotypes recognized by the kinds of bacteria present [106]. Enterotypes have been connected with long-term diets, especially those with protein and animal fat. Wu et al. [22] indicated that Bacteroides were related with protein and animal fat, while Prevotella was related to carbohydrates. The authors also examined controlled feeding in ten subjects and discovered that microbiome composition altered within 24 h of starting a low-fat and high-fiber diet or high-fat and low-fiber, and remained stable throughout the 10-d study [22]. The outcomes suggested the strong connection of diet with partitioning of enterotypes. In another study, a plant-based diet rich in legumes, grains, fruits, and vegetables, or an animal-based diet consisting of eggs, meat, and cheese was consumed ad libitum by six male and four female volunteers (aged 21 to 33 years with BMI (in kg/m2) ranging from 19 to 32) for five consecutive days. The subject’s fecal samples were cultured or directly analyzed by 16S rRNA gene sequencing [132]. It was indicated that microbiota changes in the high-fat animal-based diet, and was hypothetically connected to modified fecal bile acid profiles and microorganisms development able of activating inflammatory bowel disease (IBD) [132]. The outcomes demonstrated that a high-fat diet can change the bacteria in the gut and contribute to dysbiosis and eventually disease.
2.1.6.3 Host Genetics
The quantity of different bacteria present in the gut microbiota is affected by the host’s genetic constitution in manners that influence host metabolism and can eventually affect health [135]. It has been found that family members have more comparable microbiota communities than unrelated individuals, and the gut microbiota is more comparable in mono-zygotic than in di-zygotic twins [135]. At present, there are no genome-wide investigations characterizing the specific genes and pathways to determine the gut microbiome composition [136], although some genes of the immune system are related with IBD [137, 138].
The microbiota can also be formed by the immune system of host. This impact is generally constrained to compartmentalization of bacteria in order to prevent opportunistic colonization of host tissue, while species-specific impacts are less likely because of the high levels of functional redundancy in the microbiota [16, 139,140,141,142]. Both anti-microbials collected from the host and administered have a central role in forming the gut microbiota. Paneth cells in GIT produce anti-microbials, for example, angiogenin 4, α-defensins, cathelicidins, collectins, histatins, lipopolysaccharide (LPS)-binding protein, lysozymes, secretory phospholipase A2, and lectins [143]. Such proteins are confined in the mucus layer and are almost absent from the lumen, most likely because of poor mucus dispersion or luminal degradation [144, 145]. Attenuated expression of mucosal α-defensin was observed in ileal Crohn’s disease (CD) patients, featuring the significance of these proteins [146, 147]. Secretory IgA (SIgA), another part of the immune system, co-localizes with gut bacteria in the outer mucus layer and helps with constraining the exposure of epithelial cell surface to bacteria [143, 148]. SIgA is suggested to intercede the shaping of bacterial biofilm by means of binding to SIgA receptors on bacteria [149]. In IgA-deficient individuals, the expression of SIgA receptors by bacteria is reduced [150]. Microbiotic dysbiosis, specifically an over-representation of segmented filamentous bacteria (SFB), arises in mice with IgA deficiency, an impact that might be especially harmful to the host because of the capacity of SFB to firmly bind the epithelium and trigger the immune system [151].
2.1.6.4 Infections
Even though the gut microbiota influences bacterial and viral infections, the opposite is likewise obvious [152,153,154,155,156,157]. One research explored the impact of an enteropathogenic infection caused by Citrobacter rodentium on mice microbiota and discovered that some gut bacterial groups are altered because of C. rodentium infection, including a decrease in the relative abundance of Lactobacillus [158]. A human investigation of Clostridium difficile patients and asymptomatic carriers with the utilization of 16S rRNA gene pyrosequencing revealed that both had decreased microbial richness and diversity relative to healthy individuals [159]. C. difficile infection is characteristic of severe gut microbiota dysbiosis [160, 161]. Transplantation of gut microbiome from healthy donors to infected patients have increased the microbial richness and diversity, and it is, at present, applied clinically [162,163,164,165]. By utilizing a mouse model of hepatitis B virus infection, Chou et al. [152] demonstrated that the clearance of hepatitis B virus infection demands the formation of gut microbiota. It is apparent that the change in gut microbiota of host influences both pathogenesis and clearance of bacterial and viral infections.
2.1.6.5 Antibiotic Usage
Increasing evidence proposes that numerous non-antibiotic drugs including the medications used to treat T2D affect the gut microbiota [166,167,168,169]. The gut microbiota also influences drug efficacy [170, 171]. Antibiotics are ordinarily endorsed drugs that profoundly affect the normal microbiota of gut and their impact is fast, and relentless at times. Broad-spectrum antibiotics decrease the diversity of bacteria while increasing the concentration of certain bacteria that can be utilized by pathogens and reducing the number of beneficial bacteria [172]. The utilization of wide range antibiotics in infants and young children, for example, clindamycin, has been revealed to have the longest-enduring consequences on gut microbiota composition [173,174,175]. Early exposure to antibiotic in neonates can prompt microbial dysbiosis, which might be a predisposing factor for IBD [176]. There is also an association between diet and antibiotic administration. Research in mice and humans has discovered that the utilization of antibiotics early in life can promote obesity later in life, mediated by the modification of gut microbiota [177,178,179]. However, those studies do have limitations. Most of the mice studies on obesity are instigated by a high-fat diet with or without antibiotic treatment utilized by only male mice since they gain more weight than female mice, although no obvious sex bias is observed in human obesity. One study demonstrated that antibiotics modified the gut microbiota of host without altering the host metabolism [180, 181]. Many studies showed that antibiotics lower body weight and improve sensitivity to insulin [182, 183]. Berberine, the primary component of a Chinese herbal extract used for the treatment of bacterial diarrhea, has an anti-diabetic impact by balancing the gut microbiota and reducing glucose and insulin resistance [184, 185].
2.1.6.6 Physical and Biochemical Barriers
Intestinal mucus provides the gut microbiota a source of carbohydrates [186, 187]. The layers of intestinal mucus are made-up around the large, highly glycosylated gel-forming mucin MUC2 (Muc2 in mice), which is secreted by goblet cells [188]. The glycan structures in mucins are different and dependent on four core mucin-type O-glycans including N-acetyl galactosamine, N-acetyl glucosamine, and galactose. O-glycans represent up to 80% of the total molecular mass of Muc2/MUC2 [189]. Mucus is present throughout GIT and is thickest in the colon where it is important to mediate the relationship between host and microbiota [190]. Normalization of layers of host’s intestinal mucus needs long-term microbial colonization [191]. Colonic mucus is separated into two layers comprising of a dense and impermeable internal layer and a loose external coating that is penetrable by bacteria [190]. While the internal layer is almost sterile, the mucin proteins in the external layer, embellished with a rich and diverse collection of O-glycans, provide an energy source and preferential binding sites for commensal bacteria [189, 192, 193]. The type of mucin O-glycosylation depends on the expressed glycosyl transferases and their location in the Golgi apparatus [187], modifications of which influence the composition of microbiota. For example, the presence or absence of H and ABO antigens in GI mucosa, as dictated by the genotype FUT2 (a gene that expresses an α1,2-fucosyl transferase), influences the abundance of numerous bacterial species [194]. Mucus and mucin glycosylation are consequently a key in defining the microbiota and for allowing the selection of most ideal microbial species to mediate host health [195,196,197]. A loss of MACs from mice diet can lead to narrow mucus in the distal colon, increased expression of the inflammatory marker, REGIIIβ, and increased microbe proximity to epithelium [198]. Colonic mucus barrier erosion under dietary fiber deficiency is related with shifting of gut microbiota towards the usage of secreted mucins as a nutrient source [199]. In contrast, administration of A. muciniphila (a mucin degrader) to mice avoids the development of high-fat diet-induced obesity and strengthens metabolic endotoxemia-induced inflammation by restoring the gut barrier [133, 200]. The protective function of A. muciniphila could be recapitulated by utilizing its purified membrane protein or the pasteurized bacterium [201]. It has been recently shown that supplementation of A. muciniphila reduces fat mass and alleviates body weight gain in chow diet-fed mice by mitigating metabolic inflammation [202]. The capability of A. muciniphila was therefore suggested as an alternative therapy to target human obesity and related disorders.
The ability of gut bacteria to use dietary or mucin glycans is directed by the collection of polysaccharide lyases (PLs) and glycoside hydrolases (GHs) encoded by their genomes [187]. Many species serve as generalists capable of degrading many polysaccharides, while others are specialists in targeting specific glycans [203]. Bacteroidetes encode a lot more glycan-cleaving enzymes than members of Firmicutes [204]. The genome of Bacteroides thetaiotaomicron contains 260 GHs, relative to 97 hydrolases encoded by humans [205]. The most represented family in the gut microbiota is GH13 family, which includes enzymes associated with the starch breakdown [204]. The biochemical and structural characterization of extensive degrading assembly of prominent gut species like B. thetaiotaomicron or Bacteroides ovatus uncovered that the identification and breakdown of complex carbohydrates by the human gut microbiota is considerably more complex than previously recommended [206,207,208,209,210,211]. Firmicutes members also show some unique and complex highlights, such as the recent discovery of amylosomes in the resistant starch using Ruminococcus bromii bacterium [212].
Mutations and lateral gene transfer can lead to diversification of microbial population [213, 214]. New bacterial functions encourage niche variation, making it a positive feedback loop where more diversification can occur [215, 216]. Additionally, interaction between gut microbes permits colonization by a diverse set of microorganisms, shaping the gut microbiota community. One mechanism proposed to intervene this impact is microbial cross-feeding. Several products of carbohydrate fermentation, including succinate, lactate, and 1,2-propanediol, do not generally aggregate to higher levels in the healthy adult human’s colon, because they can act as substrates for other bacteria, including propionate and butyrate producers [217]. For instance, acetate produced by R. bromii (fermentation of resistant starch) [218] or lactate produced by lactic acid bacteria (Lactobacilli and Bifidobacteria) provides substrate for other microbiota members such as Eubacterium hallii and Anaerostipes caccae which convert it into butyrate [219, 220]. B. ovatus has recently been shown to conduct extra-cellular insulin digestion at its own expense, but to the benefit of other species that provide reciprocal advantages [221]. Such association is especially obvious in the outer mucus layer where mucin-degrading bacteria give mono- or oligo-saccharides to bacteria lacking specialized mucolytic ability [102]. For instance, the limit of cleaving sialic acid off mucins is confined to bacterial groups encoding GH33 sialidases. Numerous bacteria, including pathogens, for example, Salmonella typhimurium or C. difficile, lack a sialidase but harbor a “nan cluster” dedicated to the metabolism of sialic acid, and hence depend on other members of gut microbiota to supply them with this carbon source [222]. Intramolecular trans-sialidase, new class of sialidases is recently recognized in strains of Ruminococcus gnavus that can help the gut commensal bacteria to adapt to the niche of mucosa [186, 223, 224]. This action may give such bacteria a competitive nutritional advantage over other species in the gut mucosal environment, particularly in IBD which are rich in short, sialylated mucin glycans [186, 225]. Accessibility of sulfated compounds in the colon, either organic (host mucins and dietary amino acids) or inorganic (sulfites and sulfates), may impact specific bacterial groups like sulfate-reducing bacteria, which are gut microbiota occupants involved in the etiology of intestinal disorders, for example, IBS, IBD, or colorectal cancer [226].
As extensively reviewed, the bile acids distribution in small and large intestine can influence the dynamics of bacterial community within the gut [227, 228]. Essential bile acids, like taurocholate, can give homing signals to gut bacteria and encourage spore germination, as well as alleviate microbiota recovery after antibiotics or toxin-induced dysbiosis [113]. In addition, decreased concentration of bile acid in gut can play a significant part in permitting pro-inflammatory microbial taxa to expand [229].
2.1.6.7 Mode of Birth
Birth mode determines the microbial population to which babies are exposed at time of birth. For example, vaginal birth exposes infants to the microbes that are presently colonizing the birth canal of mother. Infants born via vaginal delivery have a comparative microbiota to that of their own mother as compared to other mothers [77, 230]. On the other hand, no substantial difference has been found between the microbiota of mothers and children delivered by C-section [77, 230]. Environmental factors (air, delivery and surgical equipment, other infants and health care workers) seem to affect the infant’s microbiome delivered by C-section [69, 231]. Recent results for C-section-delivered infants showed that a time of labor before surgery was related to infants with a microbiota that looked like that of vaginally delivered infants, while infants born without any duration of labor had a microbiota that resembled that of the skin of mother [232]. C-section is recommended to be a reason for microbial disruption at early stages of life and this disturbance in microbial colonization influences host-microbial interaction that can prompt long-term metabolic results in the host [233,234,235]. Furthermore, C-section infants have higher chances of developing atopic diseases in the initial two years after birth, when compared to vaginally born infants based on data collected from 2500 full-term healthy newborns in LISA-Study [236].
The birth mode effect on acquiring Lactobacillus in infant’s GIT is a good example of birth mode impact on the gut microbiota. In the maternal vagina, Lactobacillus is exceptionally common with an IndVal index of 0.922 [232]. Infants delivered through the mother’s birth canal contain Lactobacillus as part of their microbiome profile, but those delivered by C-section do not [234]. One more study detected less Lactobacillus genus in the infant’s microbiome profile delivered by C-section (n = 17, detection rate = 6%) versus vaginal (n = 134, detection rate = 37%) [237]. This variation in Lactobacilli detection rates, however, disappeared by the age of three [237].
The level of bacteria within an individual’s microbiota in the genera of Bacteroides and Clostridium (Bacteroides fragilis and Clostridium difficile) is also connected with birth mode [77, 230, 231, 238,239,240,241]. In the Netherlands study of KOALA Birth Cohort (n = 1032), diverse bacterial species from stool samples obtained at one month of age were identified by real-time quantitative PCR assays [238]. Infants delivered by unassisted vaginal mode (n = 826) had reduced quantity of C. difficile and relatively high quantity of B. fragilis in comparison to C-section infants [238]. On the other hand, the inverse relationship was indicated by stool samples of infants delivered by C-section (n = 108) [238]. Identification of C. difficile on the hands and in the stools from healthy hospital personnel could be connected to ecological factors rather than with the mother [238, 242]. C. difficile was regarded a microorganism that only exists in hospitals [243] and was absent in women’s vaginal swabs before delivery [244, 245]. This could clarify the C. difficile levels in the infants born in hospital and by C-section [238]. A study of 24 infants has further indicated the low abundance of Bacteroidetes (p = 0.002) in C-section-delivered infants (n = 9) in comparison to vaginally delivered infants [77]. Remarkably, this decrease in Bacteroidetes abundance continued for the first two years following birth [77]. The above studies are consistent with earlier studies that illustrate deferred formation of Bacteroides in first six months [231] and one year of life [246] of C-section infants.
Not all investigations have discovered a relationship between birth mode, the development and inheritance of GI microbiota. For instance, an investigation of 21 infants discovered that birth mode did not influence population of microbes in premature babies during the initial three months after birth [247, 248]. Studies have shown that infants delivered via C-section appear to have: less quantity of anaerobes; less diverse microbiota [77, 231, 249]; slower colonization of microbial population [239]; and, they develop atopic diseases [249] and metabolic disorders [235] more often than infants delivered by unassisted vaginal mode.
2.1.6.8 Type of Feeding
Methods of feeding may also influence the concentration of certain bacterial groups in infant’s gut microbiota. The primary food, added into GIT postpartum is milk and its composition is known to have a direct influence on the development of early GI microbiota [250, 251]. This effect can occur by providing: fundamental nutrients for proliferation of bacteria [250]; immuno-modulatory molecules [252]; and, microbes able to colonizing the infant [253]. The form of feeding contributes towards the early post-natal growth of GI flora which is confirmed by a reported closeness between microbial composition in colostrum and the meconium of infants that were breast-fed from the first hour after birth [254]. Shared bacterial DNA has been found in human breast milk and infant’s fecal samples [255]. This association is increasingly articulated between infants, their mother’s milk, and areolar skin as compared to a random mother (p < 0.001) [256]. Such outcomes, together, are associated with the vertical movement of microbial species to the infant’s gut, mediated by breast milk [256].
Methodologies focused on culture have detected more assorted microbiomes in formula-fed infants as opposed to breast-fed infants [246]. This finding has been confirmed by culture-independent studies [257, 258]. For instance, Lee et al. [257] described the impact of feeding type on the microbiota of 20 vaginally born Korean infants. Fecal samples from 10 predominantly breast-fed and 10 formula-fed babies were collected at age of four weeks. Relatively limited quantities of formula supplementation (once every 24 h in the first week after birth) to breast-fed infants changed the microbial profile to motif close to that found for formula-fed infants exclusively [259]. Some formula-fed infants were fed a diet containing 70 to 100% of formula milk, and they were also exposed to breast milk [257]. In this analysis, five bacterial species were found to be present in the fecal samples of all infants (both formula- and breast-fed groups contained Bifidobacterium longum, Streptococcus lactarius, Streptococcus salivarius, Lactobacillus gasseri, and Streptococcus pseudopneumoniae). Lee et al. [257] argued that the existence of these bacterial species in these babies’ intestines must be independent of the feeding type, and therefore these species represent specific commensal bacteria found in 4 week-old Korean infants. The higher abundance of B. longum, L. gasseri, and S. pseudopneumoniae, and lesser abundance of S. lactarius, and S. salivarius were observed in breast-fed babies as compared to formula-fed babies. These outcomes are consistent with the predictions that disclosure to varied feeding types, breast or formula milk changes the relative abundance of certain commensal bacteria.
On the whole, formula-fed infants have more stable and diverse GI microbial populations with high levels of facultative and strict anaerobes as compared to breast-fed infants [257, 260,261,262]. Fecal samples of breast-fed infants are less complex, have high quantity of aerobic bacteria, and have shown more changes in the microbial composition in the first year following birth [257, 261, 262]. Studies recommend that once the introduction of solid foods into the diet begins, the distinctions in microbial population among breast and formula-fed infants are lost and microbial communities migrate towards an intricate adult microbiome [69, 250].
2.1.6.9 Birth Environment of the Infants
Disclosure of multiple extra-uterine disorders at time of early development of gut adds to the colonization and development of infant’s GI microbiota. It is known that infants delivered by C-section are more vulnerable to ecological factors [263, 264]. It is especially valid for premature infants having high possibility of developing a flora that reflects NICU (Neonatal Intensive Care Unit), owing to the immatureness of their GIs and extended vulnerability to the environment [251].
The path of microbial transmission from surroundings to neonates is difficult to confirm yet investigations have demonstrated that microbes from the surroundings can be separated from fecal samples of neonates [265, 266]. However, cross-transference among patients and spread of a multi-drug resistant (MDR) strain, Acinetobacter baumannii additionally prompted an outburst in a Tunis NICU. 31 infants (26–41 weeks gestational age) got pneumonia induced by MDR A. baumannii and 10 deaths occurred because of infection after the transfer of MDR A. baumannii from an infant to another hospital’s epidemic-associated surgical ward [266]. Such outcomes are agreeable with reviews that infants belonging to different geographical regions/hospitals harbor diverse microbial communities [261, 265]. Despite the fact, the PiPS experiment, a double-blind randomized placebo-controlled trial of probiotic treatment with Bifidobacterium breve was conducted to prevent sepsis and necrotizing enterocolitis in 1310 premature babies (born in the range of 23–30 weeks period of gestation) from 24 hospitals. The probiotic strain of B. breve was reported to be recognized in the feces of 37% of infants in the placebo arm, in comparison to 85% of the intervention arm, showing that ecological-associated parameters lead to cross-colonization of B. breve in infants [267]. Interestingly, this PiPS trial indicated no distinction in the microbial diversity of babies microbiome in two arms of the study [268].
The environment of hospital, handling, feeding, and treatment mechanisms can improve microbial transference to infants [265]. Nonetheless, information of transmission mechanisms, dominating microbial communities in the environment of hospitals and the strains of bacteria with high probability of effectively colonizing the infant’s GI remain subtle and are worth investigating in further studies.
2.1.6.10 Other Factors
Various ecological parameters have been involved in forming the microbiota that involves surgery, geographic location, depression, smoking, and living arrangements (rural/urban) [73, 269,270,271].
2.2 Gut Microbiota Balance and Health
Bacteria are colonized in the human GIT from the time of human birth. The species and quantity of the flora are dynamically changing with conditions such as life, diet, and environment until a stable adult microbiota is established. Total number of bacteria in the intestinal tract of normal people is as many as 1014 [272]. This bacterial community is mainly composed of obligate anaerobic bacteria, aerobic bacteria, and facultative anaerobic bacteria. Among them, anaerobic bacteria are more prevalent than aerobic bacteria, and 60% of anaerobic bacteria are thick-walled bacteria, more than 20% Bacteroides [273]. The intestinal flora of healthy people can be roughly divided into three categories: (1) Intestinal dominant bacteria, mainly obligate anaerobic bacteria that are symbiotic with the host, including Bifidobacterium, Bacteroides, Lactobacillus, Clostridium genus, with nutrition, immune regulation and metabolism; (2) pathogenic bacteria coexisting with the host, mainly facultative anaerobic bacteria, when the intestinal flora is disordered, can cause disease; (3) Pathogens, such as Proteus and Pseudomonas, due to the small number of bacteria and long-term colonization opportunities, once the body’s immunity is low, the number is beyond the normal range, causing disease. Due to the bactericidal action of gastric acid and intestinal peristalsis, the number of bacteria in the stomach is very small, the small intestine acts as a transition zone, the jejunum is dominated by a small amount of aerobic bacteria, and the number of ileal bacteria is large, mainly gram-negative anaerobic bacteria in the colon. The number and type are obviously increased, the concentration can reach 1012 cfu/mL, mainly composed of anaerobic bacteria such as Bifidobacterium, Lactobacillus, Bacteroides, and Clostridia [274]. The terminal colon is very different and is regulated by pathophysiological conditions. In healthy individuals, the host maintains a steady state symbiotic relationship with the microbe, the host provides a nutritious and stable environment, and the microbes participate in the protective barrier of the intestinal mucosa. The gut microbiota provides a broad, anaerobic or hypoxic, constant temperature environment, which helps the host to improve the decomposition efficiency of nutrients, increase the absorption of beneficial substances, synthesize nutrients and essential vitamins needed by the body, and maintain the nervous system. Stability promote the immune system. In an unbalanced state, dysbacteriosis affects host growth, development, health and disease, and can also affect drug treatment [275].
2.2.1 Gut Microbiota and Gut Barrier
A layer of polarized columnar epithelial cells and epithelial area, including lamina propria, enteric nervous system, connective tissue, and muscular layer, are present in intestinal mucosa. There are four types of intestinal barriers: mechanical barriers, immune barriers, chemical barriers, and biological barriers. First is the mechanical barrier which is tightly connected. The intestinal mucosa is not only an anatomical physical structure, but more importantly it is an intestinal barrier. The energy through the intestinal epithelial cells is primarily through an extra-cellular pathway, with specific membrane channels and pumps, as well as a para-cellular pathway that is regulated by tight junctions. Under the microscope, they look like discrete contacts of a series of adjacent cells. Eventually a complex tight junction is formed that maintains the normal structure of the intestinal mucosal cells. Next to the immune barrier, this barrier helps the intestinal cells to secrete IgA normally. The third barrier is a chemical barrier in which microorganisms and antigens in the gut are degraded in a non-specific manner through the gastric acid environment, pancreatic fluid, and biliary secretions. Digestive enzymes are mainly proteases, lipases, amylases, and nucleases that kill microorganisms by destroying the cell walls of bacteria [276]. A large amount of digestive juice produced by the intestine can adulterate the toxin and clean out the intestinal lumen, making it hard for potential pathogenic bacteria to bind to the intestinal epithelium, thereby shortening the presence of potentially toxic or pathogenic substances in the intestinal lumen. It can stimulate the secretion of gastric acid protease. Finally, the biological barrier, the intestinal flora is located in the outermost layer of the mucus, is an important part of the metabolism, proliferation, and maintenance of the intestinal barrier of the epithelial barrier [4]. However, the interaction between microorganisms and intestinal epithelial cells is twofold. Some are considered pathogens, while others are considered symbiotic. The symbiotic flora limits the colonization of pathogens by competing for nutrients and niches, changing pH, releasing antibacterial substances that allow exchanges between species, and optimizing the number of beneficial microorganisms. Of course, the gut flora also provides other important functions for the host. The results indicate that the native bacteria can regulate gene expression involved in a variety of crucial intestinal functions that includes absorption of nutrients, mucosal barrier enhancement, angiogenesis, xenogeneic metabolism, and postnatal intestinal maturation [277]. The intestinal barrier plays a significant role in maintenance of human health. The destruction of intestinal barrier can cause dysfunction of the body and lead to a variety of disorders.
2.2.2 Gut Microbiota in Metabolism
The mixed oxygen in the food is consumed by aerobic and facultative bacteria in the upper part of the intestine, and the closure of the intestinal wall makes the large intestine meet the anaerobic environment required by the obligate or facultative anaerobic bacteria fermentation. In the large intestine, crude fibers and non-starch polysaccharides (NSP), which cannot be decomposed and used by the host, become the raw materials for its fermentation and eventually produce volatile fatty acids, thus providing energy for the host. At the same time, volatile fatty acids can also promote the growth of intestinal epithelial cells, accelerate the repair of intestinal damaged mucosa, and even regulate the gene expression of epithelial cells, inhibit the occurrence of enteritis and colon cancer, thereby promoting the health of the host. In addition to producing beneficial substances, intestinal microbial fermentation in the body also produces metabolites that inhibit host growth. Intestinal microorganisms degrade tyrosine and tryptophan into highly toxic phenol and aromatic compounds in the intestinal tract and expel them from the urine, but these phenol compounds are not found in the urine of sterile mice. Ammonia is another toxic waste produced by microbial urease fermentation of amino acids in the intestinal tract. However, urea hydrolysis in sterile animals cannot take place, and the concentration of ammonia in the colon of normal animals is several times the concentration required for cell damage, which inhibits the growth of the host. Therefore, the main mechanism of using antibiotics to promote growth may be to reduce the inhibiting effect of toxic and harmful substances produced by intestinal microbial fermentation on the growth of animals [278].
2.2.2.1 Lipid Metabolism
Fiaf is an endocrine signal expressed in intestinal epithelium, liver, and adipose tissue that activates the Tie2 receptor and initiates intracellular signal transduction to inhibit lipoprotein lipase (LPL) activity and reduce triglyceride deposition in adipose cells. Backhed et al. found that the total body fat content, weight of epididymal fat pad, and LPL activity of aseptic fed Fiaf+/+ mice and conventionally fed Fiaf−/− and Fiaf+/+ mice were higher than those of aseptic fed Fiaf+/+ mice. The inhibition of intestinal microorganisms on the expression of Fiaf and the deletion of the mutation of Fiaf gene will lead to the decrease of the expression of Fiaf in intestinal epithelial cells, weakening the inhibition of Fiaf on IPL activity and promoting the storage of triglycerides in fat cells. The fat precipitation effect caused by Fiaf gene deletion is consistent with the effect of microbial inhibition of Fiaf. Srebp-1 and ChREBP are transcription factors mediating the lipid response of liver cells to insulin and glucose. Acetyl CoA carboxylase (Acc) and fatty acid synthase (Fas) genes are the target sequences of srebp-1 and ChREBP, which can promote the synthesis and storage of fat. Studies have proved that ChREBP mRNA in liver of conventionally fed mice was significantly increased (p < 0.01), and srebp-1 mRNA was also significantly increased (p < 0.05).
2.2.2.2 Protein Metabolism
The proteins ingested by the host are mainly broken down into amino acids that can be absorbed and utilized by protease and peptidase. Studies have shown that although only a few bacteria contain protease, almost all bacteria have peptidase. As a result, intestinal microbes are able to independently break down the proteins taken by the host to meet their own needs. The proteins degraded and utilized by intestinal microorganisms cannot be utilized by the host. Amino acids that are broken down by gut microbes but not used can be used by the host to help digest proteins. Intestinal microbes can not only break down proteins but also use ammonia in the intestine to synthesize bacterial proteins. Microorganisms in the rumen of cattle are able to synthesize bacterial proteins from ammonia and provide proteins to the host. In the case of protein deficiency, ammonia formed by the degradation of amino acids by intestinal microorganisms can enter the host and recycle to synthesize amino acids, which makes up for the deficiency of protein and is beneficial to the growth of the host.
2.2.2.3 SCFAs Production
At least four different pathways allow the SCFAs to signal to the host. First, SCFAs, particularly butyrate, serve as an energy substrate for colonocytes [67, 68], and in retaliation to decreased availability of energy, germ-free mice slow down the transportation through small intestine to permit more time for nutrient absorption [69]. Second, propionate act as a substrate for gluconeogenesis and can stimulate intestinal gluconeogenesis, by signaling through the central nervous system (CNS) to defend the host from diet-induced obesity and glucose intolerance [64]. Third, acetate and butyrate, can act as inhibitors of histone deacetylase [70, 71]. Fourth, SCFAs signal through G-protein-coupled receptors like GPR41 and GPR43, and thus affecting various crucial processes including inflammation [72] and enteroendocrine regulation [73]. SCFAs generation is, however, just one feature of microbial metabolism in the gut [279].
2.2.2.4 Bile Acid Conversion
Bile acids are generated in the liver, stored in the gall bladder, and secreted into the duodenum after consumption. Bile acids have long been known as single emulsifiers for absorption of lipids, and have also been found to be effective signaling molecules regulating other metabolic pathways. Intestinal flora is a significant controller of bile acid metabolism. Intestinal flora can not only regulate the synthesis of bile acid but also promote it to produce secondary metabolites. Therefore, the diversity of bile acids in germ-free mice is much less than that in colonized mice [280].
Bile acids can bind to cell receptors like farnesoid X receptors (NR1H4) and G-protein-coupled receptor (GPCRs, TRG5), and are involved in regulating lipid metabolism and maintaining homeostasis of the body’s internal environment. Activation of FXR has a crucial role in modulating bile acid equilibrium in the body. Studies have shown that the activation of the ileum FXR receptor can promote the increase of the expression level of the growth factor (FGF)19 gene in fibroblasts and the homologous FGF15 gene in mice, thereby inhibiting the synthesis of bile acid. In addition, the activation of FXR receptor also promotes the expression of small heterodimer (SHP) genes, the transcription level of ileum bile acid-binding protein (IBABP) gene, and the expression level of organic solute transporter-ost beta gene, thereby regulating the absorption and transport of bile acid in the terminal ileum. Activation of TRG5 receptor induces glp-1 secretion by intestinal L cells, which improves liver and pancreas role and enhances glucose tolerance in mice suffering from obesity. Studies have shown that TGR5, which activates brown fat tissue and muscle, enhances expenditure of energy and prevents diet-induced obesity. The intestinal flora, therefore, can be used to regulate the metabolism of bile acid pool of FXR and TGR5 receptors to adjust and control signal, and regulate the body fat metabolism and sugar metabolism, and finally play a decisive role for diabetes and obesity. In addition, the study of Baghdasaryan et al. on the mouse model of bile duct sclerosis showed that inhibiting the absorption of intestinal bile acid can effectively improve the cholestatic liver and bile duct injury in mice. Molecular concatenates (anti-apoptotic protein Bcl2, long non-coding rna-hi9, and nuclear receptor Shp) can maintain normal liver function by regulating the balance of bile acids in the body. Therefore, maintaining bile acid homeostasis is an important prerequisite for improving body health [281].
2.2.3 Gut Microbiota and Host Immunity
Firmicutes and Bacteroides are the most important intestinal bacteria in animals. Firmicutes are mainly gram-positive bacteria, such as Clostridium, Streptococcus, and Lactobacillus. Bacteroidetes are mainly gram-negative bacteria, including Bacteroidetes multiformis and ovalis. An important role of intestinal bacteria is to improve the host’s digestion and utilization efficiency of nutrients. However, in the process of co-evolution with the host, animal intestinal microbes have evolved more functions. For example, intestinal microbes can regulate intestinal development, angiogenesis, and lymphocyte development as signal molecules. In addition, intestinal bacteria also has an extremely crucial role in protecting the host from pathogens. By competing with bacterial pathogens for dietary nutrients, intestinal bacteria limit the rapid colonization of pathogens in the intestinal tract. Gut microbes can also stimulate host immune responses. However, the association between microbes in the intestinal tract and the host is not always mutually beneficial. For example, Enterococcus faecalis, one of the most important flora in the human intestinal tract, can invade mucosal tissues and increase the incidence of bacteremia and infectious endocarditis in humans.
Intestinal microorganisms are rebooting and regulating factors of host innate immunity and adaptive immunity. When the body is exposed to pathogenic factors, the body will activate related receptors, for example, Toll-like receptors (TLRs) and nod-1ike receptors (NLRs), to activate inflammatory response and kill pathogenic factors. Therefore, it is important to comprehend the association of gut microbes and immune system [282].
2.2.4 Gut Microbiota and Innate
Intestinal microbes are known as “superorganisms” that encode genes for breaking down dietary fiber, amino acids, and drugs. Intestinal microbes can promote the formation of immune function and influence the composition of T-cell subsets. Wu et al. established a sterile chicken model, indicating that intestinal microorganisms can promote the development of spleen and improve immunity. Gut microbes can adjust the immune function of the immune system, for example, Bifidobacterium stimulating immune cells to secrete IL-6, IL-1, that promote differentiation of mature B-lymphocytes and T-lymphocyte proliferation, enhance the killing ability of NK cells. In addition to this, some strains of Bifidobacterium having anti-inflammatory activity increase the secretion of intestinal IgA, and induction of mature dendritic cells [283].
Modulation of immune system is not only affected by microbial flora, but also the reaction in the microbial flora of immune system played a key function in shaping gut microbes group. SIg-A the secretion of intestinal lamina propria of gram-negative bacteria have special affinity, can pack by bacteria, inhibit bacteria and intestinal epithelial cells, specific binding to prevent bacteria in intestinal epithelial cell adhesion, shifting to avoid bacteria through intestinal epithelium [284].
2.2.5 Diet-Mediated Production of Beneficial or Detrimental Metabolites by the Gut Microbiome
Microbial metabolites are produced by microorganism–microorganism and host–microorganism interactions.
2.2.5.1 Polyamines
Putrescine, spermine, and spermidine are polycationic molecules present in all living cells and are essential to many biological functions that includes gene transcription and translation, growth of cell, and death. The intestinal tract comprises a large amount of polyamines, derived from diet and de novo by host and microbial cells. Polyamines are accountable for increasing the integrity of intestinal epithelial cells (IECs) barrier [285]. Polyamines, as demonstrated by the in vitro studies, can promote the generation of inter-cellular junction proteins, which are essential for controlling para-cellular permeability and reinforcing epithelial barrier function.
2.2.5.2 SCFAs
Bacterial fermentation in the colon produces SCFAs (acetic acid, butyric acid, and propionic acid) as their main metabolic end products by using undigested complex carbohydrates as substrates. SCFA concentrations in the gut [31] are dependent upon microbiota composition, intestinal transit time, microbiota-host metabolic flux of SCFAs, and fiber content of host diet [286]. These microbiota-generated metabolites are crucial sources of energy for gut microbiota and IECs. Apart from acting as substrates for energy production, SCFAs have various regulatory functions, and their impact on physiology and immunity of host is still apparent.
2.2.5.3 Formyl Peptides
Formyl peptide receptors (FPRs) can recognize conserved N-formyl peptide motifs that are present in bacteria, and their closely associated motifs present in mitochondria. Non-formylated endogenous ligands are also detected by FPRs, which includes serum amyloid A, protein annexin, cathelicidin anti-microbial peptide. Instigation of FPRs results in enlisting the leukocytes and generation of pro-inflammatory cytokines, super oxides, and enzymes to fight infections. FPRs are stated by innate immune cells, endothelial cells, epithelial cells, neural cells, and muscle cells, and many studies suggested the instigation of FPRs on non-phagocytic cells to be necessary to achieve tissue homeostasis after infection or injury [287].
2.3 Gut Microbiota Dysbiosis and Disease
Stability of the intestinal micro-ecology is an indispensable part of human health. The imbalance of intestinal micro-ecology may induce a series of diseases, such as T2D, autoimmune diseases, senile dementia, obesity, IBD, depression, IBS, Alzheimer’s disease, cancer, etc. According to “China’s adult diabetes prevalence and control status,” the prevalence of diabetes in adults aged 18 and over in China has reached 11.6%. Diabetes has become one of the most important and difficult public health problems in China.
From the birth of the baby, the bacteria settle into the intestines. Under the influence of dietary intake and environmental conditions, the ratio of various intestinal microbes tends to be stable. Therefore, each individual’s gut microbiota is unique in the genus and species level, but has a strong universality at the door level, such as Bacteroides and thick-walled bacteria. The microbiota colonizes for a long time and forms a gut micro-ecology with its living environment. These intestinal flora participate in the regulation of human health through various ways such as absorption of energy, alteration of intestinal permeability, production of SCFAs, choline metabolism, bile acid metabolism, and brain–gut axis. Therefore, the intestinal flora is closely related to the metabolism and immunity of the human body. In addition, the normal intestinal flora prevents the invasion of foreign pathogenic microorganisms by establishing mechanical, biological, and immune barriers, and maintains the stability and micro-ecological equilibrium of intestinal environment. Probiotics colonize the intestinal mucosa to create a biological barrier, reducing the infection and colonization of pathogenic microorganisms. Certain probiotics produce anti-bacterial substances that suppress the growth and reproduction of noxious bacteria [288].
When the internal or external environment causes imbalance of intestinal micro-ecology, it will lead to disease. In Gordon’s study, the intestinal flora of obese mice was transplanted into sterile mice, which showed a significant increase in body weight [289]. Taiwanese scholars have found that WEGL can alleviate metabolic disorders caused by intestinal flora imbalance and obesity [290]. AIEC bacteria in the gut of CD patients can adhere to and invade IECs. AIEC releases macrophages and releases IFN-γ and TNF-α, which enhances its own value and aggravates inflammation [291]. A study by the Tokyo University of Science and the University of Tokyo pointed out that laminarin in seaweed can prevent the occurrence of IBD by increasing the number of Lactobacilli in the intestine [292].
Investigations have shown that gut microbiota diversity is the key to gut health. Some treatments can reduce the diversity of intestinal microbes, so the patient relapses after stopping the drug. Microbiota may also promote the resistance of pathogenic species to drugs, or lead to the expansion of disease-causing populations and enhance virulence [293]. Research on the gut microbiota has become the key to treating these diseases.
2.3.1 Gut Microbiota and Metabolic Disorders
The human’s gut microbiome as a part of the digestive system, can participate in the body’s digestion of nutrients, and can affect the body’s own metabolic activities [294]. Among them, Bacteroides bacteria can degrade a large group of plant polysaccharides (such as cellulose, hemicellulose, pectin, resistant starch, etc.) that cannot be digested in the human body, thus providing additional energy to the host. For the extra energy provided by bacteria (mainly in the form of carbohydrates), the body combines it into fat storage in adipose tissue, making the effect Bacteroides on the body’s sugar metabolism a major cause of obesity [295]. Similarly, Phylum Firmicutes bacteria that degrade non-degradable polysaccharides in the body’s digestive tract are also likely to be a major contributor to obesity.
In 2004, a study by Backhed et al. [296] found that gut microbes may affect the body’s energy storage, suggesting that obesity may be associated with it. Studies have shown that gut microbes use the body’s undigested polysaccharide metabolism to produce small molecular compounds that can be used by the body to increase their energy, and in mouse models, gut microbes can increase the host’s metabolic rate, increase its ineffective circulation, and store excess energy in fat form. Intestinal microorganisms increase the density of capillaries under the intestinal microflu, which contributes to the absorption of nutrients; the intestinal microbe inhibits the expression of the intestinal epithelial to Fiaf and may promote the synthesis of fat in the liver.
Imbalances in the gut microbiome can lead to metabolic disorders, such as insulin resistance due to steady state imbalances [297], which cause abnormalities in the sugar metabolism of the TMA/FMO3/TMAO pathway regulation. The use of sugar-reducing lipid-adjusting side intervention after 3 months can significantly reduce blood sugar lipid levels in patients with combined hyperlipidemia in obese T2D, improve insulin resistance, and be equal to metformin, while regulating the patient’s intestinal flora, increasing the beneficial bacteria represented by Blautia and Faecalibacterium. Changes in the structure of the flora were significantly related to an improvement in blood sugar lipid levels [298].
A new study has seen [299] a change in the composition of the fecal microbiome in postmenopausal obese women with low-calorie diet interventions, preserving the core microbiome and changing the structure of some functional microbiomes. At the same time, the concentration of fecal bile acid decreased significantly, which was related to the metabolic pathways of amino acids, radon, and lipids in plasma. Intestinal flora can also produce SCFAs by fermenting soluble dietary fiber [300, 301], and SCFAs can reduce serum triglyceride and cholesterol levels by inhibiting the activity of liver lip-creation enzymes, promoting the production of cholesterol oxidase that accelerates the degradation of cholesterol, improves liver utilization, and increases bile acid synthesis [29], lower serum cholesterol. Intestinal flora regulates fat cytokines, component binding proteins, and other genes and enzymes to regulate blood lipids [30,31,32]. There have been a large number of experiments and clinical studies which showed that the disorder of intestinal flora structure is related to metabolic syndrome.
2.3.2 Gut Microbiota and Hepatic Disorders (e.g. NAFLD and ALD)
Recent reports have indicated that gut microbiota is closely associated with alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD). ALD is a series of liver lesions due to long-term heavy drinking. According to pathological features, it is divided into mild alcoholic liver disease, alcoholic hepatitis, alcoholic fatty liver, alcoholic liver fibrosis, and alcoholic cirrhosis. One of its pathogenesis is the damage of the intestinal barrier. The damage of intestinal barrier results in intestinal micro-ecological disorders, enhanced permeability of intestinal mucosa, displacement of a large number of bacteria and endotoxin (LPS) in the intestinal tract, and excessive production of inflammatory factors, thereby accelerating the occurrence and development of the disease [302]. Inokuchi et al. found that alcohol favors the development of gram-negative bacteria such as Proteobacteria in intestine, thereby reducing the number of anaerobic bacteria such as Bifidobacteria. Since the Proteobacteria are considered to be important bacteria that initiate the innate immune system, an increase in the number of Proteobacteria can result in activation of immune system, which will promote the development of chronic inflammation of the liver [303]. Bull-Otterson et al. found that alcohol intake can cause damage to the local immune defense system of the GI tract, promote the growth of intestinal bacterial overgrowth (SIBO), and significantly reduce the number of thick-walled bacteria and Bacteroides in the intestine. Gram-positive (Actinomycetes) and gram-negative (Proteobacteria and Prevotella) increased in number, and LPS in the intestine was released in large quantities, causing liver damage [303]. NAFLD has become a reason of chronic liver disease (CLD), and its occurrence is the result of a combination of genetics, environment, and lifestyle. A growing number of reports have indicated that the imbalance of intestinal microecology is involved in the evolution and progression of NAFLD, mainly through the function of enteric axis, and elevated levels of bacterial lipopolysaccharide (LPS) in the systemic or portal or circulation in various CLDs [303]. The study found that there was a rise in the amount of SIBO and inflammatory factor, tumor necrosis factor alpha in NASH patients [303]. In summary, the relationship between microbial populations and NAFLD and ALD can be represented by the following figure:

Regulating the intestinal flora becomes a new direction for the treatment of ALD and NAFLD. Use of probiotics and prebiotics can regulate the intestinal flora to prevent or treat NAFLD. Kirpich et al. found that ALD patients were supplemented with Bifidobacterium and germ lactic acid bacteria to maintain the integrity of the intestinal barrier, rebuild the balance of intestinal microbes, and prevent intestinal microbial translocation and harmful inflammatory reactions [304].
2.3.3 Gut Microbiota and Autoimmune Diseases: Inflammatory Bowel Diseases
The dysregulation of gut flora may lead to a variety of autoimmune diseases, including IBD. Autoimmune refers to the phenomenon that the body’s immune system produces antibodies and sensitized lymphocytes against its own tissue components, causing an immune response. When autoimmunity causes dysfunction of its own tissues and organs and clinical symptoms appear, it is called autoimmune disease (AID). At present, there are more than 30 kinds of autoimmune diseases, most of which are primary and a few are secondary. The cause of primary autoimmune disease is unknown, closely related to genetic factors, and is divided into organ-specific and non-organ-specific. Target antigens and lesions of organ-specific AIDs are often restricted to a specific organ. Target antigens and lesions of non-organ-specific AIDs are often systemic or systemic, and secondary refers to other diseases or treatments. The dysregulation of intestinal microecology may lead to a variety of autoimmune-related diseases. Intestinal microorganisms can directly affect the body’s innate immune system through TLRs and other related immune receptors, and have a significant function in the pathogenesis of a variety of auto-immune and inflammatory diseases [305]. Recent studies have shown that a variety of auto-immune diseases, for example IBD, metabolic syndrome, multiple sclerosis, rheumatoid arthritis, etc., are associated with abnormal changes in intestinal microecology [305]. Many studies have shown that small molecules secreted by intestinal bacteria can enter the cell through transporters or endocytosis on the surface of intestinal mucosal cells, and activate a series of signal pathways related to cell survival. It was found that patients with IBD have different degrees of intestinal microbial abnormalities, the most common is the reduction of thick-walled bacteria and the increase of Proteobacteria. Some people have suggested through clinical analysis that patients with active IBD have lower abundance of Clostridium sphaeroides, Clostridium sp., Bifidobacteria, in the active period and remission period of ulcerative colitis. The abundance of E. coli and Lactobacilli did not differ between the active phase of IBD and the remission period [305]. By altering the population or community of microorganisms, reshaping the structure and function of intestinal microbes, and then regulating immunity, it is expected to provide new possibilities for the treatment of autoimmune diseases.
2.3.4 Gut Microbiota and Cardiovascular Disease
Community structure modifications in the gut microbiota are closely associated with cardiovascular disease (CVD). CVD is considered to be one of the major causes of death in contemporary human diseases, with the most common diseases including hypertension, coronary atherosclerosis, and heart failure. Trimethylamine N-oxide (TMAO), a metabolic derivative formed by the intestinal flora, can increase atherosclerosis and promote the risk of cardiovascular diseases such as chronic heart failure [306]. Yang et al. found that the abundance of intestinal flora in the hypertension group decreased significantly from both clinical observation and animal experiments. The main reason was the decrease in the number of probiotics such as Bifidobacteria. Some scholars believe that the intestinal flora metabolites may regulate blood pressure through the buffer system of SCFAs receptor-olfactory receptor 78 and G-protein coupled receptor orphan [306]. In recent years, gene sequencing has found that the intestinal flora of patients with coronary heart disease is disordered, and the content of E. coli, Helicobacter pylori, and Streptococcus is increased, and Bifidobacteria, Lactobacillus content is reduced [306]. The metabolite TMAO of the intestinal flora is also associated with atherosclerosis. Experiments have shown that plasma levels of TMAO are positively correlated to mouse atherosclerotic plaque load [306]. Patients with heart failure are often accompanied by gastrointestinal congestion, prone to loss of appetite, abdominal distension and other symptoms, decreased gastrointestinal motility leads to accumulation of gastrointestinal contents, a large number of bacteria can easily destroy intestinal homeostasis, causing dysbacteriosis. Further research found that the pathogenic bacteria such as Salmonella and Shigella in the intestinal flora of the patients increased significantly [306]. A study has shown that the severity of heart failure is also related to TMAO. Therefore, changing the intestinal ecology through probiotics will be a new entry point for the prevention and cure of cardiovascular diseases.
2.3.5 Intestinal Microflora and Nervous System Diseases
2.3.5.1 Microbiota–Gut–Brain Axis
At present, many mental diseases (autism, Parkinson’s disease, and Alzheimer’s disease) are highly related to intestinal flora, and our joys and sorrows may also be regulated by flora. Many of our desires and preferences may also be affected by intestinal flora, including appetite food preferences, and even sexual orientation. These connections involve an important chain of relationships: the bacteria–intestine–brain axis. Although our brain and intestines are located in two separate parts of our body, there is a very strong relationship between them. In fact, there may be three channels in the bacteria–intestine–brain axis. One is the nerve channel, the second is the blood channel, and the third is the immune channel. Some intestinal substances may pass through the intestinal barrier, through the blood, pass over the brain–blood barrier, thus affecting the brain. Some of the cells involved in the intestinal immune response may repeat the same immune response in the brain.
2.3.5.2 Intestinal Microbiology Group Is Closely Related to Neurological Diseases
One study found that many patients with Parkinson’s disease suffer from severe constipation for a long time before they are diagnosed. Bacteria in the human gut decompose undigested proteins into toxic substances such as ammonia, mercaptan, indole, hydrogen sulfide, and histamine. These toxic substances can be excreted from the body through the stool. However, the intestinal function of the elderly is declining, especially in elderly patients with constipation. It is very difficult for elderly patients to rule out these toxic substances. Over time, toxic substances will accumulate in large quantities. When toxic substances accumulate to a certain extent, they will slowly enter the brain with blood circulation. Damage to the CNS can lead to Alzheimer’s disease. For Parkinson’s disease, higher the enterobacteriaceae in the intestinal tract of patients, more serious the symptoms will often be, and the pathogenic protein in the brain, α-synaptic nucleoprotein, is also closely related to the pathological changes of the enteric nervous system. [307].
The researchers first bred two groups of mice that produced too much α-synaptic nucleoprotein, which is thought to be one of the “culprits” of Parkinson’s disease. The only difference between the two groups was that one group had a complete intestinal microflora and the other group was sterile. The results showed that aseptic mice not only did not show the symptoms of Parkinson’s disease but also performed much better in running, pole climbing, and other motor performance tests. The researchers then fed some aseptic mice with SCFAs formed by the decomposition of food fiber by intestinal flora and transplants intestinal flora obtained from the feces of patients with Parkinson’s disease. As a result, all of the mice developed symptoms of Parkinson’s disease and it is concluded that intestinal microbiome is an important promoter of this disease. Changes in the composition of intestinal flora or intestinal bacteria themselves may contribute to or even lead to deterioration of motor function, which is the main symptom of Parkinson’s disease.
In this framework, antibiotics, probiotics, diet, fecal bacteria transplants, and meditation, which may regulate flora, may be ideal tools and the best way to treat neurological or mental illness.
2.3.6 Intestinal Microflora and Cancer
2.3.6.1 Importance of Microorganisms in Human Cancer
Cancer is the number one killer of human health, but the complex relationship between the mechanism of cancer and environmental microorganisms has been difficult to prove. Since the partial success of William Coley’s attempt to treat sarcomas with local injection of bacteria (Coley’s toxin) in the late nineteenth century, the relationship between cancer and pathogens such as bacteria, viruses, and fungi has attracted worldwide attention [308]. Especially after the first discovery of microbial membrane on the surface of cancer cell mucosa by Christine et al., the study of the interaction between human microorganisms, especially intestinal microorganisms and cancer has become a hot topic.
2.3.6.2 Progress of Intestinal Microbiome in Cancer Research
Intestinal microflora is not only related to the formation of the immune system, but also to the interaction between the immune system. Under normal homeostasis conditions, intestinal symbiotic bacteria are recognized by TLRs and has a crucial role in maintaining the homeostasis of intestinal epithelial cells. In the experiment of chemical induction of intestinal epithelial cell injury in mice, Rakoff-Nahoum found that mice lacked a key connector molecule in the microbial ligand or linker protein pathway produced by pathogenic microorganisms and intestinal symbiotic bacteria which will aggravate the damage to the cells [309]. It can be seen that the health of the body and disease state is the outcome of interaction between pathogenic bacteria and intestinal flora. Upadhyay et al. demonstrated that the intestinal microbe group interacts with the immune response and forms the related lipid metabolism by affecting obesity. Russell et al. found that if Candida albicans mutates in intestinal flora, the specific chemicals produced will affect the immune response and make the immune system oversensitive and produce allergic diseases.
For example, related studies have shown that Clostridium nucleatum is a common bacteria living in human large intestine, and it is also considered to be a key leader in colon cancer. In addition, intestinal Clostridium and Bacteroides are also one of the pathogenic bacteria of colon cancer. The researchers have found that a group of probiotic bacteria in the intestinal tract can stimulate intestinal cells to activate the Nrf2 signaling pathway, which has a protective effect on small intestinal cells [310]. This finding is of great significance for the use of bacteria to treat intestinal diseases and to reduce the intestinal damage caused by cancer radiotherapy.
2.3.6.3 Achievements of Intestinal Microbiome in Cancer Prevention and Control
French scientist Sophie Viaud used a cyclophosphamide anticancer drug to change the composition of the intestinal microbial population, driving gram-positive bacteria into the secondary lymphatic system, triggering a special helper T cell attack on the tumor. In order to achieve the therapeutic effect of killing tumor, Chen et al. found that the intestinal microflora of individuals is dominated by bacteria that use different fibers, such as Plumeria and Bacteroides to ferment the fiber in food into SCFAs. Butyric acid, as the preferred energy source of colon cells, can promote intestinal barrier function and reduce inflammation. Therefore, feeding fiber can optimize the structure and function of intestinal flora, which is very important for the early prevention and control of the disease.
Researchers at Xin Zhou University and Tokyo Pharmaceutical University in Japan have used transgenic technology to develop a Bifidobacterium whose life activities can cut off the nutritional supply of cancer cells, thereby inhibiting the growth of tumor tissue, a technology that can be used to treat cancer. Bifidobacterium is a common bacteria in human intestinal tract, which is easy to survive in anoxic environment, and the interior of breast and chest cancer tissue belongs to anoxic state.
2.3.6.4 Research Prospect of Intestinal Microbiome
Intestinal flora plays a significant part in regulating anxiety, emotional disorders and other neurological diseases, and chronic diseases such as IBD, type I diabetes, obesity, cardiovascular disease, and cancer [311]. It is worth noting that, intestinal microbiome can maintain homeostasis in the human body, and may also produce potential carcinogenic toxins and metabolites through bacteria to have a negative impact on cancer prevention. Therefore, in the future, anti-tumor therapy can be carried out through the combination of intestinal microbiome and its metabolites with immunotherapy, or it can also be combined with the traditional method of directly targeting malignant cells for anti-tumor therapy. Based on the immune response induced by intestinal microorganism group and the mechanism of cancer induction, high efficient anticancer strains were screened to develop new and efficient anticancer agents.
2.3.7 Renal Diseases
Although intestinal flora lives in the gut, its role is not limited to the digestive system. The effect of intestinal flora on human body is systemic through its influence on human metabolism and immune function. The kidney is the main organ of excretion of metabolites in the body and also the important site of deposition of immune complex. Therefore, intestinal flora has a crucial role in the development and treatment of renal diseases. For example, Vaziri et al. found that the quantity of Firmicutes and Bacteroidetes in the intestinal tract of chronic renal failure rats was lower, especially that of Lactobacillus and Prevotellaceae. Wong et al. found that in patients having end-stage renal dirty disease, the abundance of bacteria producing ammonia, indole, cresol, and other harmful metabolites increased, while the abundance of bacteria producing SCFAs (including Lactobacillus and Prevotellaceae) decreased. IS, PCS, and PAG can be detected in the early stages of renal dysfunction. Meanwhile, kidney stone disease is closely related to changes in intestinal flora. The main pathological change of kidney stone disease is crystal formation in the kidney, and its incidence rate is increasing day by day. Stern et al. used 16sRNA test to find that intestinal Bacteroides in patients with kidney stones had a higher abundance, while Prevotella was lower. Eubacterium and E. coli were negatively correlated with urinary oxalic acid and citric acid content at 24 h, respectively. Calcium oxalate stone is a common type of KSD. Gnanandarajah et al. suggested that the lack of bacterial colonization in the intestine was a risk factor for calcium oxalate urolithiasis. Sadaf et al. found that oxalate Bacillus and Lactobacillus prevent stone deposition and formation in the kidney by producing enzymes conducive to oxalate degradation. Xiaoying et al. found that Enterobacteriaceae was significantly elevated in kidney stone disease. Recently, it was found that the fecal microbial diversity of patients with recurrent idiopathic calcium calculi was low, and the expression of oxalate degradation related bacteria gene was significantly reduced, which was negatively correlated with oxalate excretion. At the same time, it is also believed that kidney stone disease is not caused by the lack of oxalate formate bacteria or one kind of bacteria, but is related to the extensive changes of intestinal flora. IgAN is deposited in the glomeruli by a polyimmune complex containing IgA, causing kidney damage. DeAngelis et al. discovered that the composition of intestinal flora in IgAN patients changed, mainly manifested by the increase of Streptococcus, Enterobacter and the decrease of Bifidobacteria [312].
Ley et al. sequenced 16S ribosomal RNA genes in fat and lean mice and found that the number of Bacteroides in fat mice was relatively high. For obese and non-obese people, human trials also showed the same changes in bacteria as animal studies. T2D patients are also often associated with differences compared with the normal population. Larsen et al. compared the degree of abnormality in the types and quantities of intestinal flora of T2D group and non-T2D group, and it was found that E. coli, Salmonella, and Vibrio cholerae belong to proteobacteria are present in the intestines of T2D patients, and the proportion of bacterial flora change related to blood glucose concentration. Qin et al. found that T2D patients were accompanied by moderate-intensity bowel. The proportion of trace bacteria was unbalance, which was reflected by the benefit of producing butyric acid of Hoffmann-La Roche Inc. A large number of bacteria were lost, while the number of harmful bacteria such as Clostridium was increased. The diabetic patients were supplemented with probiotics, prebiotics, and other microecological preparations to make intestinal flora; after being regulated and reaching steady state, its blood glucose level will also improve. Intestinal flora structural changes (e.g. reduction of Bacteroidetes/Firmicutes ratio, butyric acid production, salt bacteria, etc.) is closely related to T2D and may pass through those involved in SCFAs, LPS, fence-induced fat factors and bile acids in vivo synthesis, induces the body to produce a variety of mechanisms (such as chronic inflammatory response, generation Endotoxemia, etc.), which then leads to the destruction of islet beta cells [313]. T2D reduces the body’s sensitivity to insulin, and ultimate leads to death. Therefore, intestinal flora and T2D were actively studied to make full use of intestinal flora for better control of T2D patients’ blood sugar.
References
Sender R, Fuchs S, Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. https://doi.org/10.1371/journal.pbio.1002533
Bengmark S (1998) Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut 42:2–7. https://doi.org/10.1136/gut.42.1.2
Backhed F (2005) Host-bacterial mutualism in the human intestine. Science 307:1915–1920. https://doi.org/10.1126/science.1104816
Neish AS (2009) Microbes in gastrointestinal health and disease. Gastroenterology 136:65–80. https://doi.org/10.1053/j.gastro.2008.10.080
Hooper LV, Gordon JI (2001) Commensal host-bacterial relationships in the gut. Science 292:1115–1118
Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. https://doi.org/10.1126/science.1124234
Luckey TD (1972) Introduction to intestinal microecology. Am J Clin Nutr 25:1292–1294
Natividad JMM, Verdu EF (2013) Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res 69:42–51. https://doi.org/10.1016/j.phrs.2012.10.007
den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. https://doi.org/10.1194/jlr.R036012
Bäumler AJ, Sperandio V (2016) Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. https://doi.org/10.1038/nature18849
Gensollen T, Iyer SS, Kasper DL, Blumberg RS (2016) How colonization by microbiota in early life shapes the immune system. Science 352:539–544. https://doi.org/10.1126/science.aad9378
Chang C, Lin H (2016) Dysbiosis in gastrointestinal disorders. Best Pract Res Clin Gastroenterol 30:3–15. https://doi.org/10.1016/j.bpg.2016.02.001
Schroeder BO, Bäckhed F (2016) Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 22:1079–1089. https://doi.org/10.1038/nm.4185
Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host-bacterial mutualism in the human intestine. Science 307:1915–1920
Qin J, Li R, Raes J, Arumugam M et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Ley RE, Peterson DA, Gordon JI (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848
Eckburg PB, Bik EM, Bernstein CN, Purdom E et al (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Tannock GW (2007) What immunologists should know about bacterial communities of the human bowel. Semin Immunol 19:94–105
Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M et al (2013) Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7:949–961
Arumugam M, Raes J, Pelletier E, Le Paslier D et al (2011) Enterotypes of the human gut microbiome. Nature 12:174–180
Jeffery IB, Claesson MJ, O’Toole PW, Shanahan F (2012) Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol 10:591–592
Wu GD, Chen J, Hoffmann C, Bittinger K et al (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108
Huse SM, Ye Y, Zhou Y, Fodor AA (2012) A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242
Wu GD, Chen J, Hoffmann C, Bittinger K et al (2012) NIH public access. Science 334:105–108
Knights D, Ward TL, McKinlay CE, Miller H et al (2014) Rethinking enterotypes. Cell Host Microbe 16:433–437
Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D et al (2014) Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. https://doi.org/10.1371/journal.pone.0093827
Mizrahi-Man O, Davenport ER, Gilad Y, White BA (2013) Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One 8:e53608. https://doi.org/10.1371/journal.pone.0053608
Suau A et al (1999) Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65:4799–4807
Hugon P, Dufour JC, Colson P, Fournier PE et al (2015) A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis 15:1211–1219. https://doi.org/10.1016/S1473-3099(15)00293-5
Li J, Jia H, Cai X, Zhong H et al (2014) An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. https://doi.org/10.1038/nbt.2942
Donohoe DR, Garge N, Zhang X, Sun W et al (2011) The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13:517–526. https://doi.org/10.1016/j.cmet.2011.02.018
Kobyliak N, Virchenko O, Falalyeyeva T (2016) Pathophysiological role of host microbiota in the development of obesity. Nutr J 15:1–12. https://doi.org/10.1186/s12937-016-0166-9
Guarner F, Malagelada JR (2003) Gut flora in health and disease. Lancet 361:512–519. https://doi.org/10.1016/S01406736(03)12489-0
Borre YE, O’Keeffe GW, Clarke G, Stanton C et al (2014) Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med 20:509–518. https://doi.org/10.1016/j.molmed.2014.05.002
Morrison DJ, Preston T (2016) Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200. https://doi.org/10.1080/19490976.2015.1134082
den Besten G, van Eunen K, Groen AK, Venema K et al (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. https://doi.org/10.1194/jlr.R036012
Macfarlane GT, Gibson GR, Cummings JH (1992) Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72:57–64. https://doi.org/10.1111/j.1365-2672.1992.tb04882.x
LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG et al (2017) Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Factories 16:1–10. https://doi.org/10.1186/s12934-017-0691-z
Chakraborti CK (2015) New-found link between microbiota and obesity. World J Gastrointest Pathophysiol 6:110–119. https://doi.org/10.4291/wjgp.v6.i4.110
Li X, Shimizu Y, Kimura I (2017) Gut microbial metabolite short chain fatty acids and obesity. Biosci Micro, Food Heal 36:135–140. https://doi.org/10.12938/bmfh.17-010
Pingitore A, Chambers ES, Hill T, Maldonado IR et al (2017) The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab 19:257–265. https://doi.org/10.1111/dom.12811
Schönfeld P, Wojtczak L (2016) Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res 57:943–954. https://doi.org/10.1194/jlr.R067629
Cooper DN, Martin RJ, Keim NL (2015) Does whole grain consumption alter gut microbiota and satiety? Healthc (Basel, Switz) 3:364–392. https://doi.org/10.3390/healthcare3020364
Everard A, Cani PD (2014) Gut microbiota and GLP-1. Rev Endocr Metab Disord 15:189–196
Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B et al (2014) The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 5:3611. https://doi.org/10.1038/ncomms4611
Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF (2003) Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 52:1442–1447
Cousin FJ, Jouan-Lanhouet S, Theret N, Brenner C et al (2016) The probiotic Propionibacterium freudenreichii as a new adjuvant for TRAIL-based therapy in colorectal cancer. Oncotarget 7:7161–7178. https://doi.org/10.18632/oncotarget.6881
Ni J, Wu GD, Albenberg L, Tomov VT (2017) Gut microbiota and IBD: Causation or correlation? Nat Rev Gastroenterol Hepatol 14:573–584
Pascale A, Marchesi N, Marelli C, Coppola A et al (2018) Microbiota and metabolic diseases. Endocrine 61:357–371. https://doi.org/10.1007/s12020-018-1605-5
Flint HJ, Bayer EA (2008) Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann New Y Acad Sci 1125:280–288
van den Abbeele P, Gérard P, Rabot S, Bruneau A et al (2011) Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol 13:2667–2680. https://doi.org/10.1111/j.1462-2920.2011.02533.x
McOrist AL, Miller RB, Bird AR, Keogh JB et al (2011) Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J Nutr 141:883–889. https://doi.org/10.3945/jn.110.128504
Grootaert C, Van Den Abbeele P, Marzorati M, Broekaert WF et al (2009) Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 69:231–242. https://doi.org/10.1111/j.1574-6941.2009.00712.x
Begley M, Hill C, Gahan CGM (2006) Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72:1729–1738. https://doi.org/10.1128/AEM.72.3.1729-1738.2006
Ley R, Turnbaugh P, Klein S, Gordon J (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. https://doi.org/10.1038/nature4441021a
Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA et al (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. https://doi.org/10.1073/pnas.0504978102
Komaroff AL (2017) The microbiome and risk for obesity and diabetes. JAMA 317:355. https://doi.org/10.1001/jama.2016.20099
Mazmanian SK, Round JL, Kasper DL (2008) A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. https://doi.org/10.1038/nature07008
Wen L, Ley RE, Volchkov PY, Stranges PB et al (2008) Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455:1109–1113. https://doi.org/10.1038/nature07336
Kim YS, Milner JA (2007) Dietary modulation of colon cancer risk. J Nutr 137:2576S–2579S
Lau SKP, Woo PCY, Woo GKS, Fung AMY et al (2004) Eggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagn Microbiol Infect Dis 49:255–263. https://doi.org/10.1016/j.diagmicrobio.2004.04.012
Kraatz M, Wallace RJ, Svensson L (2011) Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int J Syst Evol Microbiol 61:795–803. https://doi.org/10.1099/ijs.0.022954-0
Lau SK, Woo PC, Fung AM, Chan K et al (2004) Anaerobic, non-sporulating, gram-positive bacilli bacteraemia characterized by 16S rRNA gene sequencing. J Med Microbiol 53:1247–1253. https://doi.org/10.1099/jmm.0.45803-0
Finamore A, Palmery M, Bensehaila S, Peluso I (2017) Antioxidant, immunomodulating, and microbial-modulating activities of the sustainable and ecofriendly Spirulina. Oxidative Med Cell Longev 2017:3247528. https://doi.org/10.1155/2017/3247528
Shin NR, Lee JC, Lee HY, Kim MS et al (2014) An increase in the Akkermansia spp. Population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63:727–735. https://doi.org/10.1136/gutjnl-2012-303839
Hampson DJ, La T, Phillips ND (2015) Emergence of Brachyspira species and strains: reinforcing the need for surveillance. Porc Heal Manag 1:8. https://doi.org/10.1186/s40813-0150002-1
Galperin MY (2008) New feel for new phyla. Environ Microbiol 10:1927–1933. https://doi.org/10.1111/j.1462-2920.2008.01699.x
Yamauchi M, Lochhead P, Morikawa T, Huttenhower C et al (2012) Colorectal cancer: a tale of two sides or a continuum? Gut 61:794–797. https://doi.org/10.1136/gutjnl-2012-302014
Mackie RI, Sghir A, Gaskins HR (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69:1035S–1045S
Trahair J (2001) Digestive system. In: Harding R, Bocking AD (eds) . Cambridge University Press, Cambridge, pp 137–153. ISBN 0521645433
Trahair JF, Harding R (1994) Development of the gastrointestinal tract. In textbook of fetal physiology; Thorburn, G.D., Harding, R., Eds. Oxford University Press, New York, NY.. ISBN 0198577486
Aagaard K, Ma J, Antony KM, Ganu R et al (2014) The placenta harbors a unique microbiome. Sci Transl Med 6:237ra65. https://doi.org/10.1126/scitranslmed.3008599
Rodriguez JM et al (2015) The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26:26050
Koenig JE, Spor A, Scalfone N, Fricker AD et al (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108:4578–4585. https://doi.org/10.1073/pnas.1000081107
Avershina E, Storrø O, Øien T, Johnsen R et al (2014) Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol 87:280–290. https://doi.org/10.1111/1574-6941.12223
Aagaard K, Riehle K, Ma J, Segata N et al (2012) A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One 7:e36466. https://doi.org/10.1371/journal.pone.0036466
Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K et al (2014) Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. https://doi.org/10.1136/gutjnl-2012-303249
Salminen S (2004) Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53:1388–1389. https://doi.org/10.1136/gut.2004.041640
Backhed F, Roswall J, Peng Y, Feng Q et al (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. https://doi.org/10.1016/j.chom.2015.05.012
Bäckhed F (2011) Programming of host metabolism by the gut microbiota. Ann Nutr Metab 58:44–52. https://doi.org/10.1159/000328042
Palmer C, Bik EM, DiGiulio DB, Relman DA et al (2007) Development of the human infant intestinal microbiota. PLoS Biol 5:e177. https://doi.org/10.1371/journal.pbio.0050177
Dethlefsen L, Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108:4554–4561. https://doi.org/10.1073/pnas.1000087107
Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R et al (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108:4586–4591. https://doi.org/10.1073/pnas.1000097107
Biagi E, Nylund L, Candela M, Ostan R et al (2010) Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5:e10667. https://doi.org/10.1371/journal.pone.0010667
Claesson MJ et al (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178
Woodmansey EJ, McMurdo MET, Macfarlane GT, Macfarlane S (2004) Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol 70:6113–6122. https://doi.org/10.1128/AEM.70.10.6113-6122.2004
Biagi E, Candela M, Turroni S, Garagnani P et al (2013) Ageing and gut microbes: perspectives for health maintenance and longevity. Pharmacol Res 69:11–20. https://doi.org/10.1016/j.phrs.2012.10.005
Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R et al (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. PNAS 108:458691
Adlerberth I, Wold AE (2009) Establishment of the gut microbiota in western infants. Acta Paediatr 98:22938
Marques TM, Wall R, Ross RP, Fitzgerald G et al (2010) Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol 21:14956.12
Eckburg PB, Bik EM, Bernstein CN, Purdom E et al (2005) Diversity of the human intestinal microbial flora. Science 308:16358
Qin J, Li R, Raes J, Arumugam M et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:5965
Backhed F (2011) Programming of host metabolism by the gut microbiota. Ann Nutr Metab 58:4452
Yatsunenko T, Rey FE, Manary MJ, Trehan I et al (2012) Human gut microbiome viewed across age and geography. Nature 486:2227
Borre YE, Moloney RD, Clarke G, Dinan TG et al (2014) The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol 817:373403
Murphy JMRK, Stanton C, Ross RP, Kober OI et al (2015) The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26:26050. https://doi.org/10.3402/mehd.v26.26050
Macpherson AJ, McCoy KD (2013) Stratification and compartmentalisation of immunoglobulin responses to commensal intestinal microbes. Semin Immunol 25:358–363. https://doi.org/10.1016/j.smim.2013.09.004
Donaldson GP, Lee SM, Mazmanian SK (2015) Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. https://doi.org/10.1038/nrmicro3552
Gu S, Chen D, Zhang JN, Lv X et al (2013) Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8:e74957. https://doi.org/10.1371/journal.pone.0074957
Eckburg PB (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638. https://doi.org/10.1126/science.1110591
Lavelle A, Lennon G, O’Sullivan O et al (2015) Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 64(10):1553–1561
Li H, Limenitakis JP, Fuhrer T, Geuking MB et al (2015) The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. https://doi.org/10.1038/ncomms9292
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL et al (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484. https://doi.org/10.1038/nature07540
Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M et al (2010) Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5. https://doi.org/10.1371/journal.pone.0009836
Ding T, Schloss PD (2014) Dynamics and associations of microbial community types across the human body. Nature 509:357–360. https://doi.org/10.1038/nature13178
Arumugam M, Raes J, Pelletier E, Le Paslier D et al (2011) Enterotypes of the human gut microbiome. Nature 473:174–180. https://doi.org/10.1038/nature09944
Odamaki T, Kato K, Sugahara H, Hashikura N et al (2016) Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16:90
Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H et al (2015) Role of the normal gut microbiota. World J Gastroenterol 21:8787–8803
Wen LL, Duffy A (2017) Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J Nutr 147:1468S–1475S
Voreades N, Kozil A, Weir TL (2014) Diet and the development of the human intestinal microbiome. Front Microbiol 5:494
Spor A, Koren O, Ley R (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290
Nagpal R, Tsuji H, Takahashi T, Nomoto K et al (2017) Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird’s-eye view. Front Microbiol 8:1388
Browne HP, Forster SC, Anonye BO, Kumar N et al (2016) Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533:543–546
Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G et al (2008) Fecal microflora of Greek healthy neonates. Anaerobe 14(2):94–101
Samuel BS, Shaito A, Motoike T, Rey FE et al (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci 105(43):16767–16772
Zoetendal EG, Raes J, van den Bogert B, Arumugam M et al (2012) The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6:1415–1426. https://doi.org/10.1038/ismej.2011.212
David LA, Maurice CF, Carmody RN, Gootenberg DB et al (2013) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. https://doi.org/10.1038/nature12820
Walker AW, Ince J, Duncan SH, Webster LM et al (2011) Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. https://doi.org/10.1038/ismej.2010.118
Veiga P, Pons N, Agrawal A, Oozeer R et al (2014) Changes of the human gut microbiome induced by a fermented milk product. Sci Rep 4:6328
Field AE, Willett WC, Lissner L, Colditz GA (2007) Dietary fat and weight gain among women in the Nurses Health Study. Obesity (Silver Spring) 15(967–76)
Shai I, Schwarzfuchs D, Henkin Y, Shahar DR et al (2008) Weight loss with a low carbohydrate, Mediterranean, or low-fat diet. N Engl J Med 359:29–41
Sacks FM, Bray GA, Carey VJ, Smith SR et al (2009) Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 360:859–873
Mozaffarian D, Hao T, Rimm EB, Willett WC et al (2011) Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med 364:2392–2404
Winzell MS, Ahren B (2004) The high-fat diet-fed mouse: a model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes 53:S215–S219
Musso G, Gambino R, Cassader M (2010) Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care 33:2277–2284
DiBaise KK, Frank DN, Mathur R (2012) Impact of the gut microbiota on the development of obesity: current concepts. Am J Gastroenterol Suppl 1:22–27
Cani PD, Bibiloni R, Knauf C, Waget A et al (2008) Changes in gut microbiota control metabolic endotoxemia induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M et al (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696
Scott KP, Gratz SW, Sheridan PO, Flint HJ et al (2013) The influence of diet on the gut microbiota. Pharmacol Res 69:52–60
Graf D, Di Cagno R, Fak F, Flint HJ et al (2015) Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis 26:26164
Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK et al (2016) Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215
David LA, Maurice CF, Carmody RN, Gootenberg DB et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563
Everard A, Belzer C, Geurts L, Ouwerkerk JP et al (2013) Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071
Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345
Goodrich JK, Waters JL, Poole AC, Sutter JL et al (2014) Human genetics shape the gut microbiome. Cell 159:789–799
Blekhman R, Goodrich JK, Huang K, Sun Q et al (2015) Host genetic variation impacts microbiome composition across human body sites. Genome Biol 16:191
Thompson-Chagoyàn OC, Maldonado J, Gil A (2005) Aetiology of inflammatory bowel disease (IBD): role of intestinal microbiota and gut-associated lymphoid tissue immune response. Clin Nutr 24:339–352
Rehman A, Sina C, Gavrilova O, Hasler R et al (2011) Nod2 is essential for temporal development of intestinal microbial communities. Gut 60:1354–1362
Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336:1268–1273. https://doi.org/10.1126/science.1223490
Macpherson AJ (2000) A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288:2222–2226. https://doi.org/10.1126/science.288.5474.2222
Macpherson AJ, Uhr T (2004) Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303:1662–1665. https://doi.org/10.1126/science.1091334
Cash HL (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130. https://doi.org/10.1126/science.1127119
McGuckin MA, Lindén SK, Sutton P, Florin TH (2011) Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. https://doi.org/10.1038/nrmicro2538
Hooper LV, Macpherson AJ (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10:159–169. https://doi.org/10.1038/nri2710
Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG et al (2008) Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771. https://doi.org/10.1136/gut.2007.141481
Wehkamp J (2004) NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut 53:1658–1664. https://doi.org/10.1136/gut.2003.032805
Wehkamp J, Salzman NH, Porter E, Nuding S et al (2005) Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A 102:18129–18134. https://doi.org/10.1073/pnas.0505256102
Rogier EW, Frantz A, Bruno M, Kaetzel C (2014) Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens 3:390–403. https://doi.org/10.3390/pathogens3020390
Bollinger RR, Everett ML, Palestrant D, Love SD et al (2003) Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109:580–587. https://doi.org/10.1046/j.1365-2567.2003.01700.x
Friman V et al (1996) Decreased expression of mannose-specific adhesins by Escherichia coli in the colonic microflora of immunoglobulin A-deficient individuals. Infect Immun 64:2794–2798
Suzuki K, Meek B, Doi Y, Muramatsu M et al (2004) Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A 101:1981–1986. https://doi.org/10.1073/pnas.0307317101
Chou HH, Chien WH, Wu LL, Cheng CH et al (2015) Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc Natl Acad Sci U S A 112:2175–2180
Singh P, Teal TK, Marsh TL, Tiedje JM et al (2015) Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3:45
Qin N, Zheng B, Yao J, Guo L et al (2015) Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci Rep 5:14771
Zaiss MM, Rapin A, Lebon L, Dubey LK et al (2015) The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43:998–1010
Yang L, Poles MA, Fisch GS, Ma Y et al (2016) HIV induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS 30:19–29
Zilberman-Schapira G, Zmora N, Itav S, Bashiardes S et al (2016) The gut microbiome in human immunodeficiency virus infection. BMC Med 14:83
Hoffmann C, Hill DA, Minkah N, Kirn T et al (2009) Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun 77:4668–4678
Zhang L, Dong D, Jiang C, Li Z et al (2015) Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 34:1–7
Seekatz AM, Young VB (2014) Clostridium difficile and the microbiota. J Clin Invest 124:4182–4189
Blanchi J, Goret J, Megraud F (2016) Clostridium difficile infection: a model for disruption of the gut microbiota equilibrium. Dig Dis 34:217–220
Youngster I, Russell GH, Pindar C, Ziv-Baran T et al (2014) Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 312:1772–1778
Kelly CR, Ihunnah C, Fischer M, Khoruts A et al (2014) Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol 109:1065–1071
Youngster I, Sauk J, Pindar C, Wilson RG et al (2014) Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clin Infect Dis 58:1515–1522
Bashan A, Gibson TE, Friedman J, Carey VJ et al (2016) Universality of human microbial dynamics. Nature 534:259–262
Xu X, Zhang X (2015) Effects of cyclophosphamide on immune system and gut microbiota in mice. Microbiol Res 171:97–106
Imhann F, Bonder MJ, Vich Vila A, Fu J et al (2016) Proton pump inhibitors affect the gut microbiome. Gut 65:740–748
Devkota S (2016) Microbiome. Prescription drugs obscure microbiome analyses. Science 351:452–453
Forslund K, Hildebrand F, Nielsen T, Falony G et al (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266
Kang MJ, Kim HG, Kim JS, Oh DG et al (2013) The effect of gut microbiota on drug metabolism. Expert Opin Drug Metab Toxicol 9:1295–1308
Yoo DH, Kim IS, Van Le TK, Jung IH et al (2014) Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos 42:1508–1513
Modi SR, Collins JJ, Relman DA (2014) Antibiotics and the gut microbiota. J Clin Invest 124:4212–4218
Cho I, Yamanishi S, Cox L, Methe BA et al (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626
Gough EK, Moodie EE, Prendergast AJ, Johnson SM et al (2014) The impact of antibiotics on growth in children in low and middle income countries: systematic review and meta-analysis of randomised controlled trials. BMJ 348:g2267
Kozyrskyj AL, Ernst P, Becker AB (2007) Increased risk of childhood asthma from antibiotic use in early life. Chest 131:1753–1759
Shaw SY, Blanchard JF, Bernstein CN (2010) Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol 105:2687–2692
Trasande L, Blustein J, Liu M, Corwin E et al (2013) Infant antibiotic exposures and early-life body mass. Int J Obes 37(16–23)
Cox LM, Yamanishi S, Sohn J, Alekseyenko AV et al (2014) Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721
Mahana D, Trent CM, Kurtz ZD, Bokulich NA et al (2016) Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med 8:48
Fujisaka S, Ussar S, Clish C, Devkota S et al (2016) Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest 126:4430–4443
Reijnders D, Goossens GH, Hermes GD, Neis EP et al (2016) Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab 24:63–74
Membrez M, Blancher F, Jaquet M, Bibiloni R et al (2008) Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J 22:2416–2426
Chou CJ, Membrez M, Blancher F (2008) Gut decontamination with norfloxacin and ampicillin enhances insulin sensitivity in mice. Nestle Nutr Workshop Ser Pediatr Program 62:127–137
Han J, Lin H, Huang W (2011) Modulating gut microbiota as an antidiabetic mechanism of berberine. Med Sci Monit 17:RA164–RA167
Chang W, Chen L, Hatch GM (2015) Berberine as a therapy for type 2 diabetes and its complications: from mechanism of action to clinical studies. Biochem Cell Biol 93:479–486
Tailford LE, Owen CD, Walshaw J, Crost EH et al (2015) Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun 6:7624. https://doi.org/10.1038/ncomms8624
Arike L, Hansson GC (2016) The densely O-glycosylated MUC2 mucin protects the intestine and provides food for the commensal bacteria. J Mol Biol
Ouwerkerk JP, de Vos WM, Belzer B (2013) Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol 27:25–38. https://doi.org/10.1016/j.bpg.2013.03.001
Johansson MEV, Larsson JMH, Hansson GC (2011) The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108:4659–4665. https://doi.org/10.1073/pnas.1006451107
Gustafsson JK, Ermund A, Johansson MEV, Schutte A et al (2012) An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol 302:G430–G438. https://doi.org/10.1152/ajpgi.00405.2011
Johansson ME, Jakobsson HE, Holmén-Larsson J, Schütte A et al (2015) Normalization of host intestinal mucus layers requires long-Term microbial colonization. Cell Host Microbe 18:582–592. https://doi.org/10.1016/j.chom.2015.10.007
Juge N (2012) Microbial adhesins to gastrointestinal mucus. Trends Microbiol 20:30–39. https://doi.org/10.1016/j.tim.2011.10.001
Tailford LE, Crost EH, Kavanaugh D, Juge N (2015) Mucin glycan foraging in the human gut microbiome. Front Genet 6:131. https://doi.org/10.3389/fgene.2015.00081
Rausch P, Rehman A, Kunzel S, Hasler R et al (2011) Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A 108:19030–19035. https://doi.org/10.1073/pnas.1106408108
Arpaia N, Campbell C, Fan X, Dikiy S et al (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. https://doi.org/10.1038/nature12726
Furusawa Y, Obata Y, Fukuda S, Endo TA et al (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. https://doi.org/10.1038/nature12721
Zarepour M, Bhullar K, Montero M, Ma C et al (2013) The mucin MUC2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun 81:3672–3683. https://doi.org/10.1128/IAI.00854-13
Earle KA, Billings G, Sigal M, Lichtman JS et al (2015) Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18:478–488. https://doi.org/10.1016/j.chom.2015.09.002
Desai MS, Seekatz AM, Koropatkin NM, Kamada N et al (2016) A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. https://doi.org/10.1016/j.cell.2016.10.043
Li J, Lin S, Vanhoutte PM, Woo CW et al (2016) Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoemice. Circulation 133:2434–2446. https://doi.org/10.1161/CIRCULATIONAHA.115.019645
Plovier H, Everard A, Druart C, Depommier C et al (2016) A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. https://doi.org/10.1038/nm.4236
Zhao S, Liu W, Wang J, Shi J et al (2017) Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol 58:1–14. https://doi.org/10.1530/JME-16-0054
Cockburn DW, Koropatkin NM (2016) Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 428:3230–3252. https://doi.org/10.1016/j.jmb.2016.06.021
El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. https://doi.org/10.1038/nrmicro3050
Cantarel BL, Lombard V, Henrissat B, Appanna VD (2012) Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. https://doi.org/10.1371/journal.pone.0028742
Larsbrink J, Rogers TE, Hemsworth GR, McKee LS et al (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut bacteroidetes. Nature 506:498–502. https://doi.org/10.1038/nature12907
Rogowski A, Briggs JA, Mortimer JC, Tryfona T et al (2015) Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. https://doi.org/10.1038/ncomms8481
Cuskin F, Lowe EC, Temple MJ, Zhu Y et al (2015) Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. https://doi.org/10.1038/nature13995
Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ et al (2016) Molecular dissection of xyloglucan recognition in a prominent human Gut symbiont. MBio 7:e02134–e02115. https://doi.org/10.1128/mBio.02134-15
Foley MH, Cockburn DW, Koropatkin NM (2016) The Sus operon: a model system for starch uptake by the human gut bacteroidetes. Cell Mol Life Sci 73:2603–2617. https://doi.org/10.1007/s00018-016-2242-x
Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS et al (2017) Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541:407–411. https://doi.org/10.1038/nature20828
Ze X et al (2015) Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. MBio 6:e01058–e01015
Bjedov I (2003) Stress-induced mutagenesis in bacteria. Science 300:1404–1409. https://doi.org/10.1126/science.1082240
Xu J et al (2007) Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol 5:1574–1586
Svanback R, Bolnick DI (2007) Intraspecific competition drives increased resource use diversity within a natural population. Proc R Soc B-Biol Sci 274:839–844. https://doi.org/10.1098/rspb.2006.0198
Emerson BC, Kolm N (2005) Species diversity can drive speciation. Nature 434:1015–1017. https://doi.org/10.1038/nature03450
Louis P, Flint HJ (2016) Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19(1):29–41
Ze X, Duncan SH, Louis P, Flint HJ (2012) Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. https://doi.org/10.1038/ismej.2012.4
Louis P, Scott KP, Duncan SH, Flint HJ (2007) Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 102:1197–1208. https://doi.org/10.1111/j.1365-2672.2007.03322.x
Duncan SH, Louis P, Flint HJ (2004) Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. https://doi.org/10.1128/AEM.70.10.5810-5817.2004
Rakoff-Nahoum S, Foster KR, Comstock LE (2016) The evolution of cooperation within the gut microbiota. Nature 533:255–259. https://doi.org/10.1038/nature17626
Juge N, Tailford L, Owen CD (2016) Sialidases from gut bacteria: a mini-review. Biochem Soc Trans 44:166–175. https://doi.org/10.1042/BST20150226
Crost EH, Tailford LE, Le Gall G, Fons M et al (2013) Utilisation of mucin glycans by the human Gut symbiont Ruminococcus gnavus Is strain-Dependent. PLoS One 8:e76341. https://doi.org/10.1371/journal.pone.0076341
Crost EH et al (2016) The mucin-degradation strategy of Ruminococcus gnavus: the importance of intramolecular trans-sialidases. Gut Microbes 7(4):302–312
Larsson JMH, Karlsson H, Crespo JG, Johansson MEV et al (2011) Altered o-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis 17:2299–2307. https://doi.org/10.1002/ibd.21625
Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR (2012) Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 3:448. https://doi.org/10.3389/fphys.2012.00448
Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS (2014) Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. https://doi.org/10.1097/MOG.0000000000000057
Staley C, Weingarden AR, Khoruts A, Sadowsky MJ (2017) Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol 101:47–64. https://doi.org/10.1007/s00253-016-8006-6
Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB et al (2013) Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58:949–955. https://doi.org/10.1016/j.jhep.2013.01.003
Bäckhed F, Roswall J, Peng Y, Feng Q et al (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703
Martin R, Makino H, Yavuz AC, Ben-Amor K et al (2016) Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498
Chu DM, Ma J, Prince AL, Antony KM et al (2017) Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 23:314–326
Cox LM, Yamanishi S, Sohn J, Alekseyenko AV et al (2014) The intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721
Dominguez-Bello MG, Costello EK, Contreras M, Magris M et al (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975
Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM et al (2016) Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 22:250–253
Negele K, Heinrich J, Borte M, Von Berg A et al (2004) Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr Allergy Immunol 15:48–54
Nagpal R, Tsuji H, Takahashi T, Kawashima K et al (2016) Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol 7:1997
Penders J, Thijs C, Vink C, Stelma FF et al (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521
Wampach L, Heintz-Buschart A, Hogan A, Muller EEL et al (2017) Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol 8:738
Fallani M, Young D, Scott J, Norin E et al (2010) Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 51:77–84
Biasucci G, Rubini M, Riboni S, Morelli L et al (2010) Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86:13–15
Kim KH, Fekety R, Batts DH, Brown D et al (1981) Isolation of clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis 143:42–50
Rousseau C, Poilane I, De Pontual L, Maherault AC et al (2012) Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 55:1209–1215
Gabriel I, Olejek A, Stencel-Gabriel K et al (2017) The influence of maternal vaginal flora on the intestinal colonization in newborns and 3-month-old infants[J]. J Matern Fetal Neonatal Med 31(11):1448–1453
Al Jumaili IJ, Shibley M, Lishman AH, Record CO (1984) Incidence and origin of Clostridium difficile in neonates. J Clin Microbiol 19:77–78
Adlerberth I, Lindberg E, Åberg N, Hesselmar B et al (2006) Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res 59:96–101
Arboleya S, Binetti A, Salazar N, Fernández N et al (2012) Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79:763–772
Stewart CJ, Embleton ND, Clements E, Luna PN et al (2017) Cesarean or vaginal birth does not impact the longitudinal development of the gut microbiome in a cohort of exclusively preterm infants. Front Microbiol 8:1008
Biasucci G, Benenati B, Morelli L, Bessi E et al (2008) Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr 138:1796S–1800S
Guaraldi F, Salvatori G (2012) Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2:94
Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL et al (2014) Development of the preterm infant gut microbiome: a research priority. Microbiome 2:38
Li C, Liu Y, Jiang Y, Xu N et al (2017) Immunomodulatory constituents of human breast milk and immunity from bronchiolitis. Ital J Pediatr 43:8
Williams JE, Price WJ, Shafii B, Yahvah KM et al (2017) Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk. J Hum Lact 33:540–551
Collado MC, Rautava S, Aakko J, Isolauri E et al (2016) Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep Sep 6:23129
Perez PF, Doré J, Leclerc M, Levenez F et al (2007) A bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724–e732
Pannaraj PS, Li F, Cerini C, Bender JM et al (2017) Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 171:647
Lee SA, Lim JY, Kim BS, Cho SJ et al (2015) Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract 9:242–248
Bezirtzoglou E, Tsiotsias A, Welling GW (2011) Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17:478–482
Bullen CL, Tearle PV, Stewart MG (1977) The effect of ‘humanised’ milks and supplemented breast feeding on the faecal flora of infants. J Med Microbiol 10:403–413
Le Huërou-Luron I, Blat S, Boudry G (2010) Breast-v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 23:23–36
Stark PL, Lee A (1982) The microbial ecology of the large bowel of breastfed and formula-fed infants during the first year of life. J Med Microbiol 15:189–203
Cresci GA, Bawden E (2015) Gut microbiome: what we do and don’t know. Nutr Clin Pract 30:734–746
Fanaro S, Chierici R, Guerrini P, Vigi V (2003) Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 92:48–55
Makino H, Kushiro A, Ishikawa E, Kubota H et al (2013) Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One 8:e78331
Brooks B, Firek BA, Miller CS, Sharon I et al (2014) Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2:1
Touati A, Achour W, Cherif A, Hmida HB et al (2009) Outbreak of Acinetobacter baumannii in a neonatal intensive care unit: antimicrobial susceptibility and genotyping analysis. Ann Epidemiol 19:372–378
Costeloe K, Bowler U, Brocklehurst P, Hardy P et al (2016) A randomised controlled trial of the probiotic Bifidobacterium breve BBG-001 in preterm babies to prevent sepsis, necrotising enterocolitis and death: the probiotics in preterm infantS (PiPS) trial. Health Technol Assess 20:1–194
Millar M, Seale JJ, Greenland M, Hardy P et al (2017) The microbiome of infants recruited to a randomised placebo-controlled probiotic trial (PiPS Trial). EBioMedicine 20:255–262
Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E et al (2013) Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 8:e59260. https://doi.org/10.1371/journal.pone.0059260
Jiang H, Ling Z, Zhang Y, Mao H et al (2015) Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48:186–194. https://doi.org/10.1016/j.bbi.2015.03.016
Tyakht AV, Kostryukova ES, Popenko AS, Belenikin MS et al (2013) Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun 4:2469. https://doi.org/10.1038/ncomms3469
Francisco G (2003) Malagelada Juan-RGut flora in health and disease[J]. Lancet 361(9356):512–519
Sghir A, Gramet G, Suau A et al (2000) Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization[J]. Appl Environ Microbiol 66(5):2263–2266
Mm QE (2011) Gut microbiota and the role of probiotics in therapy[J]. Curr Opin Pharmacol 11(6):593–603
Sandra C (2018) Intestinal barriers protect against disease[J]. Science 359(6380):1097–1098
Sarker S, Gyr K (1992) Non-immunological defence mechanisms of the gut. Gut 33:987–993
Szymanowska-Powa owska D, Orczyk D, Leja K (2014) Biotechnological potential of Clostridium butyricum bacteria. Braz J Microbiol 45:892–901
Nijiang Y (2012) Research progress on the relationship between intestinal microorganisms and host metabolism[J]. Feed Expo 7:9–12
Rooks MG, Garrett WS (2016) Gut microbiota, metabolites and host immunity[J]. Nat Rev Immunol 16(6):341–352
Huangxiaoyan W (2012) The molecular mechanism of intestinal microorganisms regulating lipid metabolism[J]. Feed Industry 33(18):59–62
Pew G (2017) Zhuweiyun. Advances in the study of the metabolic axis of intestinal microorganisms in animal hosts[J]. J Microbiol 2:161–169
Jinlei, W (2018) Advances in the study of the relationship between intestinal microorganisms and host immunity[J]. Modern Animal Husbandry, 358(09):57–63.
Peterson DA, McNulty NP, Guruge JL, Gordon JI (2007) IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2:328–339
Wu JJ, Lai SM, Pan K et al (2015) Effects of intestinal flora on intestinal development, mucosal morphology and immune organs development of chicks. Chin J Anim Nutr 27(4):1101–1109
Liu L et al (2009) Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am J Phys Cell Phys 296:C801–C810
Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT (1987) Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227
Lewandowski T et al (2013) Staphylococcus aureus formylmethionyl transferase mutants demonstrate reduced virulence factor production and pathogenicity. Antimicrob Agents Chemother 57:2929–2936
Liuruixue LZ (2016) Research progress in microecological balance and human health of intestinal flora[J]. Food Indus Technol 37(06):383–387+391
Turnbaugh PJ, Ley RE, Mahowald MA et al (2006) An obesity-associated gut microbiome with increased capacity for energy harvest[J]. Nature 444(7122):1027–1031
Chang CJ, Lin CS, Lu CC et al (2015) Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota[J]. Nat Commun 6(7489):1
Barnich N, Carvalho FA, Glasser AL et al (2007) CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease[J]. J Clin Invest 117:1566–1574
Tang C, Kamiya T, Liu Y et al (2015) Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine[J]. Cell Host Microbe 18:183–197
Bäumler Andreas J (2016) Sperandio Vanessa. Interactions between the microbiota and pathogenic bacteria in the gut.[J]. Nature 535(7610):85–93
Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host—bacterial mutualism in the human intestine[J]. Science 307:1915–1920
Chenheng, Yesheng, Zuo Yanwen, Pei Xiaofang (2008) Research progress in the relationship between intestinal microorganisms and obesity[J]. Modern Prev Med 035(004):608–609,616
Gerba CP, Gramos DM, Nwachuku N (2002) Comparative inactivation of Enteroviruses and adenovirus 2 by UV light[J]. Appl Environ Microbiol 68(10):5167–5169
Jialianqun, S, Lvmeijun, S, Chenlijuan, CS, Zhang, L, Yu, YG. Discussion on the relationship between intestinal microbiological homeostasis and glycolipids metabolism based on the theory of “temper astigmatism”[J/OL]. Liaoning J Tradit Chin Med: 1–8[2019-07-15].
Tong X, Xu J, Lian F et al (2018) Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. MBio 9(3):e02392–e02317
Aleman JO, Bokulich NA, Swann IR et al (2018) Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women[J]. J Transl Med 16(1):244
Eckburg PB, Bik EM, Bernstein CN et al (2005) Diversity of the human intestinal microbial flora[J]. Science 308(5728):1635–1638
Ewaschuk JB, Diaz H, Meddings L et al (2008) Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function[J]. Am J Physiol Gastrointest Liver Physiol 295(5):G1025–G1034
Gaorunping LY (2019) Microbial intervention as a new target for advanced treatment of non-alcoholic fatty liver disease[J]. Journal of Clinical Hepatitis (02):35, 333
Qian L, Hachengyong Z, Yubin Z (2018) Research progress on treatment of alcoholic liver disease based on liver and intestine axis[J]. Pharm Biotechnol 25(04):368–371
Zhangdong (2017) Effects of intestinal flora disorders and probiotics on alcoholic liver disease and its mechanism[D]. Qingdao University
Lizhao X, Wangziqian N, Limengtao ZX (2017) The role of intestinal microorganisms in autoimmune diseases[J]. Chin J Clin Immunol Allergy 11(01):61–68
Wang Meng, Wang Dengjielin, Mengguannan, Yu Li Radium, Jiang Hong (2018) Advances in the study of intestinal microorganisms and cardiovascular diseases[J]. Medical Overview 24(18):3543–3547+3553
Lederberg J (2000) Infectious history. Science 288:287–293
Marchesi JR, Ravel J (2015) The vocabulary of microbiome research: a proposal. Microbiome 3:31
Pfeiffer JK, Sonnenburg JL (2011) The intestinal microbiota and viral susceptibility. Front Microbiol 2:1–6
Zhao L, Shen J (2010) Whole-body systems approaches for gut microbiota-targeted, preventive healthcare. J Biotechnol 149:183–190
Dishaw LJ, Cannon JP, Litman GW, Parker W (2014) Immune-directed support of rich microbial communities in the gut has ancient roots. Dev Comp Immunol 47:36–51
Panyutong P, Jun Z (2019) Research progress on the correlation between intestinal flora and kidney disease[J]. Chin J Microecol 31(06):729–733
Genglin YC (2019) Research progress on the correlation between intestinal flora and type 2 diabetes[J]. Medical Overview 25(10):2034–2038
Sender R, Fuchs S, Milo R (2016) Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164:337–340
Jiménez E, Marín ML, Martín R et al (2008) Is meconium from healthy newborns actually sterile?[J] Res Microbiol 159(3):0–193
Horinaka M, Yoshida T, Kishi A et al (2010) Lactobacillus strains induce TRAIL production and facilitate natural killer activity against cancer cells secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function[J]. FEBS Lett 584(3):577–582
Bengmark S , Cocco PD , Clemente K et al (2011) Bio-ecological control of chronic liver disease and encephalopathy[J]. Minerva Med 102(4):309–319
Lee KJ, Kim YB, Kim JH et al (2008) The alteration of enterochromaffin cell, mast cell, and lamina propria T lymphocyte numbers in irritable bowel syndrome and its relationship with psychological factors[J]. J Gastroenterol Hepatol 23(11):1689–1694
Saleh M, Trinchieri G (2010) Innate immune mechanisms of colitis and colitis-associated colorectal cancer[J]. Nat Rev Immunol 11(1):9–20
Progress of Wang Lin, Li Bing, and Zhujian et al (2016) Chin Agri Bull 32(5):10–15
Chenxiaolin RH (2014) Relationship between intestinal microbial groups and intestinal immunity[J]. Gastroenterol Hepatol Impur 23(11):1245–1248
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Zhang, C., Virk, A.K., Khan, I., Qin, H. (2020). Gut Microbiota and Health. In: Li, X., Liu, P. (eds) Gut Remediation of Environmental Pollutants. Springer, Singapore. https://doi.org/10.1007/978-981-15-4759-1_2
Download citation
DOI: https://doi.org/10.1007/978-981-15-4759-1_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4758-4
Online ISBN: 978-981-15-4759-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)