Abstract
Metabolites are the small low-molecular-weight biological molecules that are key players in energy conversion and biosynthesis reactions. Both qualitative and quantitative investigation of the entire set of intracellular and extracellular metabolites extracted from growing cells at a specified time of their growth or reproduction cycle is called metabolomics. However, analysis of metabolites is extremely challenging due to factors like metabolites’ reactivity, structural diversity, and broad concentration range. This chapter outlines the past, present, and future development in various extraction and detection protocols of metabolites. These improvements are made majorly in fast sampling as well as quenching of cellular activity, along with quick extraction of the intra- and extracellular metabolites. Furthermore, it will also briefly describe metabolite quantification using modern hyphenated analytical protocols, which chiefly involves use of chromatographic platforms (LC, GC, CE) coupled to mass spectrometry and nuclear magnetic resonance spectroscopy (NMR).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Low molecular weight (>1500 Da) organic and inorganic molecules which are intermediate or end products of enzyme-catalyzed metabolic reactions occurring inside cells during metabolic pathway are known as metabolites (Fiehn 2002). The metabolites do not follow a fixed structural template like DNA, RNA, and proteins, and thus have diverse physical properties (Lu et al. 2017). Indeed, a group of analytical tools are required for studying complete range of metabolites and metabolic networks (Bino et al. 2004). In light of biological dimension, metabolites can be classified as primary or secondary depending upon their role in cell physiological processes, while chemically, water soluble and insoluble (Lu et al. 2017). The solubility is key performer in studying about metabolite’s structure and function, like primary water-soluble metabolites play crucial role in the conversion of nutrients into usable energy, provide the basic building blocks for synthesis of biomolecules and turn over rapidly along with simultaneous execution of the vast majority of metabolic flux (Whitfield et al. 2004).
The field which deals with the identification and characterization of both endogenous and exogenous metabolites is called metabolomics. It is a global metabolite profiling scaffold which involves application of high-resolution analytic techniques along with chemometric statistical tools (Zhang et al. 2012). This will generate an integrated picture of both endogenous and exogenous metabolites including both organic (nucleic acids, peptides, amino acids, vitamins, carbohydrates, polyphenols) and inorganic species. For separation, identification, and accurate measurement of these small molecules, innovational technologies of high resolution are required like nuclear magnetic resonance (NMR), mass spectrometry (MS), high and ultra-performance liquid chromatography (HPLC/UPLC), gas chromatography (GC), CE, liquid chromatography (LC) (Holmes et al. 2008). For representing the functional phenotype of a cell, tissue, and organ, detailed study of small molecules is performed which enables the utilization of biomolecule as biomarkers (Arakaki et al. 2008). Also, for getting more deeper insight into the factors, like dietary and lifestyle, responsible for manipulating specific disease, measurement of metabolites is recommended (Patejko et al. 2017). The major reason for quantification and evaluation of metabolites is its ability to reflect both the normal and pathological biological processes and even pharmacological response to any therapeutic intervention and thus widely accepted in clinical practice for diagnosis (Van der Werf 2003; Villas-Boas et al. 2007; Canelas et al. 2008). Metabolomics has also been useful in defining metabolites which are related to both prognosis and diagnosis of diseases and thus would enhance our understanding to pathophysiology of a disease (Wishart et al. 2016). Hence, the application of set of metabolomics technique provide direction for advancement and integration of robust as well as reliable protocols involved in biomass sampling, cultivation of microbes, isolation/extraction, and quantification of these metabolites.
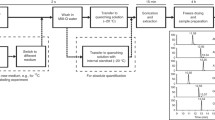
The brief outline of procedure involved in cellular metabolite studies is depicted in Fig. 3.1. Firstly, microbial metabolic activity is rapidly quenched by using different approaches like traditional one where sample’s temperature is changed instantly ranging from < −40 °C to very high >80 °C, or by varying sample pH from highly alkaline to very low acidic. Secondly, the method of filtration or centrifugation is preferred for separating quenched cells from the medium, followed by extraction of intracellular metabolites using organic solvents at high or low temperature from biomass. Later under vacuum, organic solvents are evaporated while the outstanding residue is dissolved in small amount of milli-Q water followed by centrifugation. Finally, the supernatant containing metabolite is stored at low temperature until analyzed by suitable analytical method.
This chapter focuses on the practical challenges that are encountered during analysis of primary metabolites. Initially, we will discuss quenching and extraction steps that are universal in metabolite preparation and critical for accurate measurement. This is followed by in-depth discussion of techniques used for the detection of metabolites which include liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), nuclear magnetic resonance (NMR), and enzyme assays.
3.2 Extraction of Metabolites
Accurate detection and measurement of metabolites inside the cell is quite challenging in metabolomics research, therefore different methods for efficient extraction of metabolites from biological sample is crucial (Mashego et al. 2006). Idyllically, the data obtained from metabolomics shows the detailed image of the metabolic condition of cells at a point of time when cells are harvested. However, sometime many important metabolites quickly get metabolized (<1 mMs−1) in the presence of some external factors like light and temperature (Villas-Boas et al. 2007). As a result, during sample preparation, the amount of these metabolites would get altered thus producing results that differ from the exact one of population’s true metabolic state. This difference in metabolite levels would be minimized or completely eliminated by quenching the cell’s metabolism before or during sampling so as to acquire accurate metabolomics results (Canelas et al. 2008; Da Luz et al. 2014).
The quenching step is often performed while studying the intracellular metabolites as they present inside the cell compartments and thus their extraction is necessary for their quantification. However, sometimes due to factors like temperature, light as well as metabolite interaction, the level and composition of extracellular metabolites in the medium also differ from the real one. Thus, to circumvent such difference in the results of metabolites, the cellular biomass should be quickly separated from growth media and should also be rapidly treated with quenching solution so that the degradation of metabolites could be stopped (Villas-Boas and Bruheim 2007; Patejko et al. 2017). Moreover, the metabolite sample should always be stored at a low temperature of about <20 °C to avoid any damage due to temperature and light (Villas-Boas et al. 2007).
3.2.1 Quenching Methods
As soon as the importance of quickly quenching the cells’ metabolisms and avoiding its leakage is recognized, the accurate measurement of both extracellular as well as intracellular metabolites could be obtained, and a variety of quenching methods have been developed.
3.2.1.1 Perchloric Acid
It is one of the oldest quenching method where sampling is performed by directly adding the acidic solution of perchloric acid into the culture broth (Harrison and Maitra 1968; Strange et al. 1963). Although this approach was efficient but not followed widely because this method provides a mixture of both intra- and extracellular metabolites. Since it damages the cell envelope which results in leakage of intracellular metabolites into the outside medium (Faijes et al. 2007). The quantification of such intracellular metabolites extracted using this approach shows great variability because highly concentrated media components interfere during chemical analysis and degradation of pH-sensitive metabolites (Maharjan and Ferenci 2003).
3.2.1.2 Liquid Nitrogen Method
The problem of previous method is partially addressed by Saez and Lagunas (1976), where they developed a method which quenches the cellular metabolism as well as extracts out the extracellular metabolites separately, but it involves two different steps (Saez and Lagunas 1976). Firstly, using fast filtration biomass is collected followed by quick immersion in liquid nitrogen which enables separation of living cells from culture media. Also, liquid nitrogen decreases the rate of cell metabolism as well as the turnover of metabolites due to its low temperature and thus provides additional time for the extraction of metabolite. Second step involves extraction of intracellular metabolite from biomass using different pH solutions. Although, this is one of the most widely used method but not suitable for the analysis of fast turnover compounds such as NADH, glutamate, pyruvate, ATP, and many others because it takes more than 10 s per sample to actually quench cell metabolism (Saez and Lagunas 1976).
3.2.1.3 Methanol Method
This quenching method was proposed by de Koning and van Dam in 1992, since then this method is considered as the gold standard method of quenching microbial cells (Dekoning and Vandam 1992). In this method, microbial culture is added into methanol solution (60% v/v) or kept at low temperature of about −40 °C. Then, centrifugation is performed to separate cell biomass from the culture medium. This method is advantageous due to its low-temperature strategy which is beneficial in arresting cellular enzymatic activity in less than a second. Next, cells were processed for intracellular metabolite extraction. Even though cold-methanol solution is very efficient and its use continues to be popular these days, there are some issues related to its usage which include little cell leakage and variation in biomolecules content (Wittmann et al. 2004; Villas-Bôas et al. 2005a; Bolten et al. 2007; Villas-Boas and Bruheim 2007; Canelas et al. 2008). For example, it has been found that the concentration of free amino acids was reduced by 90% when cold-methanol solution was used as quenching agent (Bolten et al. 2007).
Additionally, the majority of the quenching methods known till date have been developed as modification of cold-methanol solution along with involvement of liquid nitrogen; however, their success depends largely on the study organism or type of cells.
3.2.2 Extraction of Extracellular Metabolites
The obligatory condition in metabolomics studies is correct estimation of substrate as well as extracellular metabolite concentrations in the culture medium. For quantifying the true values, the time between sampling and quenching of the cell metabolism is considered most crucial because the high turnover rates of metabolites present inside cell in the order of 1–2 s like glucose-6-phosphate and ATP (Dekoning and Vandam 1992). Therefore, for capturing the correct in vivo snapshot of the metabolite pool and cell’s metabolic state ideally, the time gap between collection and quenching of sample should be lesser compared to the turnover rates for such metabolites. The two common methods preferred for biomass separation or sampling are centrifugation and filtration. Further, for quick sampling of biomass, the type of culturing device used is also crucial, i.e., culture flasks or bioreactors (Pinu et al. 2017). Additionally, little modification can be combined with aforementioned separation methods and culturing devices for fast sampling include use of pre-cooled glass tubes which contains 4 mm in diameter cooled glass beads or stainless steel spheres (Theobald et al. 1993). While others prefer to add liquid nitrogen to the medium followed by thawing and centrifugation (Verduyn et al. 1992; Diderich et al. 1999; Van Hoek et al. 1999). Researchers find all methods suitable depending upon the experimental organism, metabolites of interest, and many more.
Although, accurate preparation of metabolites is very essential for getting correct picture of metabolites; however, sample concentration is also a crucial aspect of metabolomics because the metabolites present in sample are diluted. Thus, to improve the analytical tools’ detection limits, the samples are concentrated (Villas-Bôas et al. 2005a; Smart et al. 2010). The most common method applied for concentrating metabolite samples involves removal of water using lyophilizer or freeze dryer. Conversely, organic solvent evaporating systems or vacuum drying methods could also be performed, but care should be taken as metabolites are susceptible to thermal degradation. Indeed, solvent evaporation technique is preferred for concentrating non-aqueous extracellular samples (Pinu et al. 2017).
Even though, the extracted and concentrated samples of metabolites are quenched but need to be stored under appropriate conditions before analysis to ensure the occurrence of no reaction during storage. However, few necessary precautions which have to be taken to avoid any unwanted changes in metabolites during the storage. These storage conditions depend on the types and stability of metabolites. Therefore, the storage of metabolite samples in dark and cool place (−20 °C or −80 °C) is suggested so that metabolite degradation by light and heat could be avoided. This also ensures the possibility that metabolites have retained their original properties. However, still some metabolites lose their characteristics even though they are stored at low temperature by getting oxidized easily which may result in change in their entire features. Thus the most suitable way to protect metabolites from degradation is storing them under vacuum (Villas-Boas et al. 2007). Although, there are few metabolites which remain unaffected by exposure to temperature and are considered as stable which could be stored even at room temperature in dry powder form so that reproducible and high-quality data could be generated.
3.2.3 Extraction of Intracellular Metabolites
The extraction approach for intracellular metabolites differs based on the complexity of structural polymers which are present in microbial cell envelope and also on composition as well as the extent of cross-linking among the polymers and with other cell-wall components. Variety of mechanical and non-mechanical/chemical methods are generally followed for the disruption of cell envelope (Villas-Boas et al. 2007). However, both methods have their own advantage and disadvantage which need to be considered before applying them. Ideally, different approaches together should be used for obtaining optimum performance depending on the cell-wall composition and structure (Villas-Boas et al. 2007).
3.2.3.1 Non-mechanical Lysis
For intracellular metabolite extraction, chemical agents are widely used as non-mechanical method where metabolites extracted in the organic solvents distribute themselves depending on their partitioning coefficients, solvent temperature, solubility between two phases. Use of chemical agents also help in concentrating the metabolites in a single phase which is very crucial for metabolite studies (Villas-Boas et al. 2007). However, care should be taken while selecting the chemical solvent for extraction as the rate of metabolite extraction varies with the nature of chemical. Although, the selection of chemical solvent and extraction method largely depends on the kind of microbe and the set of metabolites of interest. Most extraction methods involving use of chemicals extract only a particular metabolite species (like, amino acids; fatty acids); hence, emphasis has been given to necessitate the use of multiple extraction methods in order to obtain a complete picture of possible intracellular metabolite (Canelas et al. 2009; Duportet et al. 2012; Park et al. 2012; Kim et al. 2013). Usually, organic solvents (both polar and nonpolar) are used for the extraction of intracellular metabolites because they work by attacking cell wall and membrane proteins as well as lipids. This solvent interaction with cell wall and membrane components generates pores and results in the release of metabolites which are present inside the cell (Villas-Bôas et al. 2005a, 2007). In next section, the most common extraction protocols for preparing intracellular samples are given in the following.
3.2.3.1.1 Boiling Ethanol
The method involving the use of boiling buffered ethanol (75% v/v) is widely acceptable, rapid, and simple protocol for the extraction of intracellular metabolite. Briefly, the microbial cells that were quenched earlier are now exposed to the buffered ethanol at 80 °C. This boiling deactivates microbial cell wall enzymes and proteins and thus increases cell disruption, which promote the extraction of intracellular mainly, water-soluble metabolites. Before analysis, the mixture of ethanol and water is evaporated, and the pellets thus obtained are resuspended in water. The main advantage of this method is its excellent reliability (Villas-Boas et al. 2007). However, poor recovery of many metabolites of different classes, like phosphorylated metabolites tricarboxylic acids and nucleotides, is the limitation of this method (Maharjan and Ferenci 2003; Villas-Bôas et al. 2005a). Besides, this metabolite extraction method could not be applied for isolation of thermo-labile metabolites, and the reduced metabolites are susceptible for oxidation (Villas-Boas et al. 2007).
3.2.3.1.2 Cold Methanol
It is simple, fast, and an extensively used method for the extraction of intracellular metabolites from different microbial cells like bacteria (Maharjan and Ferenci 2003; Park et al. 2012), yeasts (Bolten and Wittmann 2008; Kim et al. 2013), and filamentous fungi. This is a very influential method that utilizes single organic solvent for the extraction of metabolite and that too can be effortlessly removed from the extracts by evaporation. Furthermore, the steps involved in the extraction process are usually carried out under very low temperature of approximately lees than −20 °C, and hence it is appropriate for heat-susceptible metabolites. However, the key drawback of this method is its incapability in completely inactivating enzyme and thus there is probability of alteration in intracellular metabolite pools. This method overcome the disadvantages of previous boiling ethanol method as it shows excellent reproducibility and good recovery of metabolites ranging from polar to mid-polar but that is not true for nonpolar metabolites (Villas-Bôas et al. 2005a). Occasionally, for enhancing the cell permeability cold methanol extraction is performed along with freeze–thaw cycles or sonication.
3.2.3.1.3 Buffered Methanol–Chloroform–Water
Folch et al. (1957) by using this buffer first time reported lipid dominating intracellular metabolites extraction from animal tissue. Later, similar buffer was employed for the extraction of polar metabolite from yeast at low temperature ranging from −40 to −20 °C. It is a highly recommended method for the extraction of temperature-sensitive, nonpolar, and polar metabolites from yeasts, bacteria, and filamentous fungi. The involvement of toxic and carcinogenic chloroform assisted in denaturing of complete set of enzymes present in the microbes and inhibited further chemical reactions. Although, it is a painstaking and extremely long method, and the buffers which is used for extraction also may lead to difficulty for different analytical techniques, but it is an excellent method for the recovery of phosphorylated and thermo-labile compounds (Villas-Bôas et al. 2005a, 2007).
3.2.3.1.4 Hot Water
Since 1950s, high-temperature water has been used for bacterial amino acids extraction which later has also been applied for microbial metabolite extraction (Gale 1947; Bagnara and Finch 1972). It is an extremely straightforward and easily executable method along with the ability to quench enzyme activity due to utilization of hot water.
3.2.3.2 Mechanical Lysis of Cell
Number of cell-disruption protocols where cells were broken using mechanical force like microwave, grinding, French press, and ultrasonics are widely followed for the extraction of metabolite from animal and plant cells. Although, these methods are not recommended for the metabolites extraction from microbes; however, two different methods that have been applied for the metabolite extraction from microorganisms include pressurized liquid extraction (PLE) and supercritical fluid extraction (SFE).
3.2.3.2.1 Supercritical Fluid Extraction
In this method, mid-polar to nonpolar metabolites are extracted from microbial cell by using a supercritical fluid such as carbon dioxide (Cocks et al. 1995). For increasing the applicability toward extraction of polar metabolites from microbes, methanol or ethanol is also included along with carbon dioxide as a modifier (Lim et al. 2002). Sometimes, nitrous oxide and xenon could also be applied in place of CO2. The advantage of this method lies in its requirement of small volume of solvent and sample as well as less time requirement.
3.2.3.2.2 Pressurized Liquid Extraction
This method is exclusively performed for the extraction of microbial secondary metabolites. But this method did not find any application in the metabolomics research as the metabolite extracted by this method is very concentrated and hence, not appropriate for high-throughput screening (Gomez-Ariza et al. 2004). On the flip side, only temperature-stable metabolites can be extracted using this method (Villas-Boas et al. 2007).
3.3 Detection of Metabolites
For the quantitative analysis of extracellular and intracellular metabolites, enzyme-based assays were used earlier (Theobald et al. 1993, 1997). But these traditional assays have some drawbacks like large sample volume requirement with detection of very few metabolite per assay, low sensitivity etc. Therefore, for detection of even trace of metabolites more sensitive and reliable analytical methods have been developed which include GC, UPLC/HPLC, and LC (Fig. 3.2). The chromatographic techniques enable partition of metabolites depending on their chemical and physical properties, and then the separated compounds analyzed for mass detection with MS and NMR spectroscopy (Dunn et al. 2005; Dunn and Ellis 2005). The advantages associated with these analytical techniques have increased its applicability and utility which includes high sensitivity of up to picomole, small sample requirement for analysis and the simultaneous quantification of many metabolites belonging to different pathways like tricarboxylic acid cycle, pentose phosphate pathway, and glycolysis (van Dam et al. 2002; Villas-Bôas et al. 2005b).
3.3.1 Mass-Spectrometry (MS)
MS often coupled with chromatography techniques are drawing attention in high-throughput metabolomics. In past few decades, MS has been extensively developed due to which it enjoys the distinguished status in separation science. MS works by analyzing ions produced using ion source by converting sample into ions. These ions generated from sample were then separated based on different trajectories of moving ions due to different mass/charge (m/z) ratios in the presence of electrical and/or magnetic fields and then quantitatively detected. Till date, MS is the most potential technique which is able to measure with high precision, and to produce a wide variety of metabolites during cellular processes. Features that made MS a popular choice for researchers are broad dynamic range, reproducible quantitative analysis, mass accuracy, and the proficiency of evaluating extremely complex biofluids (Lee et al. 2010). MS can analyze biological samples either directly by simply injecting the sample without prior purification or sometime following chromatographic separation. Although, direct-injection MS is extremely fast for metabolic fingerprinting; however, it has some drawbacks like co-suppression and low ionization efficiencies. To overcome the issue, MS is often used as a key hyphenated technique instead alone in metabolomics.
The advantage of using MS in metabolomics is its ability to outlook the effect of nutritional status, time, stress, and environmental perturbation on hundreds of metabolites. Currently, efforts have been made toward MS-based identification of metabolic biomarkers because MS facilitates the finding as well as functional annotation of biomarkers and reconstruction of metabolic networks (Hiller et al. 2010; Weston 2010).
3.3.2 Gas Chromatography-MS
Gas chromatography-mass spectrometry is a versatile technique that can measure a broad spectrum of primarily water-soluble metabolites (Legido-Quigley et al. 2010). Its strength is measurement of low-molecular-weight and volatile analytes, including small species that typically uncharged. For some compound classes, especially for essential oils and volatiles, GC and GC-MS are the only universally applicable analytical methods (Mohler et al. 2007). Besides, for broad metabolic profiling, GC-MS must be chemically derivatized before analysis to increase metabolite stability and volatility. This is typically achieved by the trimethylsilylation derivatization reaction on samples that have been completely dried. The reaction occurs at room temperature in the presence of pyridine as the catalyst (Goodpaster et al. 2011). The extent of derivatization is impacted by solvent and sample cleanliness, and dryness (Gao et al. 2008). In GC, the sample solution is injected into the instrument where it gets mixed with a carrier gas stream (helium or nitrogen) which transports the sample in the form of gas into a separation tube known as the “column.” The various components due to their differential partition in the mobile gas phase and stationary liquid phase get separated in the column. The component which separates in gas comes first to detector while those partitioned into liquid phase comes out later. A standard sample with known concentration is usually injected into the instrument if a sample with an unknown concentration is to be measured where the concentration of an unknown compound is calculated based on the area and retention time of standard sample peak.
GC-MS is the preferred analytical technique for the measurement of many metabolites like very-short-chain fatty acids and alcohols, hydroxy acids, sugars, and monophosphorylated sugars as well as sterols that are hard to measure by other chromatography-MS (An et al. 2010). For general metabolomics, the strengths of GC-MS include outstanding chromatographic peak sharpness and extensive mass spectral libraries for peak identification (Castillo et al. 2011). The effectiveness of GC as a separation method reduces the pressure over MS analysis, leading to cost-effective and robust detection with single quadruple. However, the requirement of hot injection for evaporation of molecules in GC disturbs the quantification of thermolabile compounds, such as di- and triphosphates (including ATP or NADPH) despite derivatization. For example, the guanidinium group of arginine decomposes to yield ornithine (Psychogios et al. 2011). Nevertheless in metabolomics, all signals are recorded, including decomposition products, which can puzzle biochemical interpretations or lead to structures that are absent from chemical libraries such as PubChem (Wu et al. 2010a, b).
GC-MS uses hard electron ionization which complicates the identification of unknown compounds because this type of ionization yields many highly reproducible fragments and rearrangement ions and also responsible for the absence of molecular ions from spectra. Indeed, identification of known compounds by using this ionization source is straightforward as it is based on comparison to mass spectra and retention indices of authenticated standards which have already been deposited in databases. But when chemical ionization approach is applied for GC-MS, molecular ion peaks could easily be seen as it is a softer technique (Legido-Quigley et al. 2010).
3.3.3 Capillary Electrophoresis-MS (CE-MS)
CE-MS is emerged as an influential and promising separation technique which is extremely suitable for the analysis of charged and polar metabolites. In CE, separation of compounds is based on the differences in intrinsic electrophoretic mobilities of compounds, which is dependent on their charge and size. Moreover, as the fundamental mechanism of separation of CE and other chromatographic-based techniques is different, and hence will receive a complementary view of the metabolite composition in a biological sample (Ramautar et al. 2011; Andreas et al. 2015; Kok et al. 2015). In last few years for metabolomics studies, CE-MS is being a choice of study due to its improved concentration sensitivity as a result of advancement in novel interface designs (Zhao et al. 2012; Lindenburg et al. 2015; Ramautar 2016; Zhang et al. 2016). Since the majority of primary metabolites are intrinsically polar, CE-MS serve as a promising adjoining microseparation platform in metabolomics (Britz-McKibbin 2011). In CE, firstly metabolites are separated based on their size and charge, and then selectively detected by using MS which monitor ions over a large range of m/z values.
Although in metabolomics the use of CE–MS is still relatively low as compared to other analytical techniques because of its technical complexity and other constraints such as low sensitivity, method robustness, and migration time variability (Kuehnbaum and Britz-McKibbin 2013; Kohler et al. 2016). Despite, CE-MS possesses a number of crucial advantages over other separation techniques due its ability to perform the reproducible and global profiling of original peptides and metabolites (endogenous) found in a clinical setting (Kami et al. 2013; Pejchinovski et al. 2015; Pontillo et al. 2015; Harada et al. 2016). For biological samples, the first global metabolic profiling using CE-MS was introduced by Soga et al. (2002, 2003). CE-MS has been applied for the analysis of a wide variety of targeted and non-targeted metabolites which include inorganic and organic acids, vitamins, thiols, nucleotides and nucleosides, carbohydrates, and peptides.
3.3.4 Liquid Chromatography-MS (LC-MS)
Diverse analytical methods are available for quantifying extracted metabolites. The most sensitive detection, and thus the broadest metabolome coverage, is achieved by MS-based methods (Theodoridis et al. 2011; Kuehnbaum and Britz-McKibbin 2013; Milne et al. 2013; Junot et al. 2014). Among MS techniques, LC-MS is the most versatile. In LC-MS, analytes are separated on column, ionized at an ion source, separated by a mass analyzer, and detected. Specificity is achieved through the combination of retention time from the column and the MS signature. In metabolomics, to obtain optimal coverage one must employ multiple LC-MS approaches (Patti 2011). Like, reversed-phase chromatography using a C18 column is performed for efficient separation of fatty acids and lipids, but ideally not for the analysis of polar metabolites due to poor retention on column. However, the retention of negatively charged metabolites can be improved by including a cationic ion-pairing agent in the running buffer (Coulier et al. 2006; Luo et al. 2007). These also improve the peak shape for phosphate-containing metabolites (Lu et al. 2008, 2010). But use of ion-pairing agent have a drawback as it takes days to wash out of an LC system, and also suppresses ionization of positively charged metabolites.
Further, the consistent and detailed measurement provided by LC-MS forms the grounds for successive data processing and multivariate data analysis. LC-MS-based large-scale metabolomic technologies are gaining popularity for their application in the diagnosis of human disease by analyzing metabolites in biological samples (Lv et al. 2011). Various biostatistical tools (PLS-DA, PCA) were adopted for classification of metabolites which were separated from different tissue and as a result, approximately 112 hydrophilic metabolites were identified within 8 min of run.
3.3.5 Nuclear Magnetic Resonance
Nuclear magnetic resonance is the most common spectroscopic technique which uniquely identifies and concurrently quantifies a large variety of organic compounds in the micromolar range. The application of this analytical technique in the emerging field of metabolomics provides a “holistic view” of the metabolites under a specific conditions, and hence it is best-fitted as well as beneficial for metabolomic studies (Wu et al. 2010a, b). Conventionally, for metabolomics study of biofluids, NMR is the most widely adopted technique because of its ability to analyze complete biomaterials in the hostile way along with providing the rich structural information. Hence, NMR-based metabolomics allow extensive research of low-molecular-weight metabolites in biological samples with significant improvements. High-resolution NMR is an ideal technique biofluids or tissue extracts’ metabolite profiling and thus, there has been much interest in the establishment of a biomarker for a disease using high-throughput NMR techniques. In drug discovery and development research, NMR could prove to be more beneficial by providing detailed information about the structural transformation of a compound which occurs due to metabolism (Zhang et al. 2010). The features responsible for inclination toward NMR for metabolic studies are its simple and automated operation which is non-selective and non-destructive. Here, the sample that is analyzed in single run could be reused, and provides useful structural information that prove to be an asset in the characterization of each components of complex mixtures (Malet-Martino and Holzgrabe 2011).
Although, for comprehensive metabolite profiling NMR has been found to have relatively low sensitivity; it is currently become a most widely used diagnostic tool due to its specificity. Currently, NMR has also been applied for the study of metabolites associated with a particular disease like those involved in Alzheimer’s disease, prostate cancer, etc. (Jordan and Cheng 2007; Barba et al. 2008). Using NMR-based metabolomics approach, Jung et al. have investigated the link of altered metabolic pattern in plasma and urine of cerebral infarctions patients and also discovered a metabolic biomarkers associated with stroke (Jung et al. 2011). The differential metabolites present in the plasma of stroke patients are characterized by increased amount of lactate, pyruvate, glycolate, and formate, while decreased amount of glutamine and methanol. On the other hand, the metabolite profile of urine of stroke patients shows decrease in citrate, hippurate, and glycine. These detected biomarkers were associated with anaerobic glycolysis and folic acid deficiency. Generally, metabolomics of urine of human being is considered as an important source of information related to the health and therefore it is the most suitable source of studying the status of the global system. This is indicative of the fact that in future magnetic resonance methodologies will be dominating in disease management.
3.4 Conclusion
Although, huge advancement has been made in metabolomics in the last two decades; however, still there is no universally applicable methodology for immediate quenching of cellular metabolic activity, extraction, and analysis of all metabolites of interest. Further, these challenges are exacerbated due to the high degree of chemical diversity such as polar and nonpolar metabolites. Also, the current procedures are so strongly organism-specific that they could not be applied to any other organism without prior optimization. The main problem which remained to be unsolved is leakage of intracellular metabolites into the surrounding medium during quenching and another is the loss of metabolites during extraction which needs to be corrected for increasing the reproducibility of results.
Investigation of the metabolome with analysis of all possible measurable metabolites in the sample could be achieved by metabolic analysis. Based on the type of sample, both targeted and/or non-targeted approaches can be used for supervising hundreds of metabolites at a given time. However, this requires high-throughput and high-end techniques that enable measurement of relative changes in compounds under a wide dynamic range, rather than estimating the absolute concentrations of compounds. The analytical techniques generally useful for these purposes include GC or HPLC/UPLC as separation modules which are coupled with MS for fast and accurate detection of separated metabolites. The cutting-edge analytical technologies enabled the measurement of metabolites and estimation of the changes in metabolite concentrations accurately with precision under defined conditions and thus, have highlighted the effects of perturbations in pathways of interest. Occasionally, researchers also prefer to use more than one analytical method which is complementary to each other, in order to circumvent the pitfalls of one technique and also to avoid improbability to incorrectly measure the metabolites.
The standardization of analytical tools and extraction methods is the prime requisite for metabolite profiling. Therefore, it appears to be more crucial to develop new or modify existing techniques that are dedicated to a particular class of metabolites, i.e., sugar intermediates, organic acids, amino acids, and cofactors. From the above discussions, it is clear that for having the comprehensive metabolite profiles of biological samples, single analytical technique is not sufficient and a combination of different techniques needs to be used for acquiring as much information as possible.
References
An Z, Chen Y, Zhang R, Song Y, Sun J, He J, Bai J, Dong L, Zhan Q, Abliz Z (2010) Integrated ionization approach for RRLC-MS/MS-based metabonomics: finding potential biomarkers for lung cancer. J Proteome Res 9:4071–4081
Andreas NJ, Hyde MJ, Gomez-Romero M, Lopez-Gonzalvez MA, Villasenor A, Wijeyesekera A, Barbas C, Modi N, Holmes E, Garcia-Perez I (2015) Multiplatform characterization of dynamic changes in breast milk during lactation. Electrophoresis 36:2269–2285
Arakaki AK, Skolnick J, McDonald JF (2008) Marker metabolites can be therapeutic targets as well. Nature 456(7221):443
Bagnara AS, Finch LR (1972) Quantitative extraction and estimation of intracellular nucleoside triphosphates of Escherichia coli. Anal Biochem 45:24–34
Barba I, Fernandez-Montesinos R, Garcia-Dorado D, Pozo D (2008) Alzheimer’s disease beyond the genomic era: nuclear magnetic resonance (NMR) spectroscopy-based metabolomics. J Cell Mol Med 12:1477–1485
Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, Trethewey RN, Lange BM, Wurtele ES, Sumner LW (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9:418–425
Bolten CJ, Wittmann C (2008) Appropriate sampling for intracellular amino acid analysis in five phylogenetically different yeasts. Biotechnol Lett 30:1993–2000
Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C (2007) Sampling for metabolome analysis of microorganisms. Anal Chem 79(10):3843–3849
Britz-McKibbin P (2011) Capillary electrophoresis-electrospray ionization-mass spectrometry (CE-ESI-MS)-based metabolomics. Methods Mol Biol 708:229–246
Canelas AB, Ras C, ten Pierick A, van Dam JC, Heijnen JJ, Van Gulik WM (2008) Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics 4:226–239
Canelas AB, ten Pierick A, Ras C, et al (2009) Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal Chem 81(17):7379–7389
Castillo S, Mattila I, Miettinen J, Oresic M, Hyotylainen T (2011) Data analysis tool for comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry. Anal Chem 83:3058–3067
Cocks S, Wrigley SK, Chicarellirobinson MI, Smith RM (1995) High-performance liquid-chromatography comparison of supercritical-fluid extraction and solvent-extraction of microbial fermentation products. J Chromatogr A 697:115–122
Coulier L, Bas R, Jespersen S, Verheij E, van der Werf MJ, Hankemeier T (2006) Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography–electrospray ionization mass spectrometry. Anal Chem 78:6573–6582
Da Luz JA, Hans E, Zeng AP (2014) Automated fast filtration and on-filter quenching improve the intracellular metabolite analysis of microorganisms. Eng Life Sci 14:135–142
Dekoning W, Vandam K (1992) A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral ph. Anal Biochem 204:118–123
Diderich JA, Schepper M, van Hoek P, Luttik MAH, van Dijken JP, Pronk JT, Klaasen P, Boelens MJ, Teixeria de Mattos MJ, van Dam K, Kruckeburg AL (1999) Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J Biol Chem 274:15350–15359
Dunn WB, Ellis DI (2005) Metabolomics: current analytical platforms and methodologies. TrAC Trends Anal Chem 24(4):285–294
Dunn WB, Bailey NJC, Johnson HE (2005) Measuring the metabolome: current analytical technologies. Analyst 130:606–625
Duportet X, Aggio RBM, Carneiro S, Villas-Boas SG (2012) The biological interpretation of metabolomic data can be misled by the extraction method used. Metabolomics 8:410–421
Faijes M, Mars AE, Smid EJ (2007) Comparison of quenching and extraction methodologies for metabolome analysis of lactobacillus plantarum. Microb Cell Factories 6:27
Fiehn O (2002) Metabolomics-the link between genotypes and phenotypes. Plant Mol Biol 48:155–171
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509
Gale EF (1947) The assimilation of amino-acids by bacteria; the passage of certain amino-acids across the cell wall and their concentration in the internal environment of Streptococcus faecalis. J Gen Microbiol 1:53–76
Gao P, Lu C, Zhang F, Sang P, Yang D, Li X, Kong H, Yin P, Tian J, Lu X, Lu A, Xu G (2008) Integrated GC–MS and LC–MS plasma metabonomics analysis of ankylosing spondylitis. Analyst 133:1214–1220
Gomez-Ariza JL, de la Torre MAC, Giraldez I, Morales E (2004) Speciation analysis of selenium compounds in yeasts using pressurised liquid extraction and liquid chromatography-microwave-assisted digestion-hydride generation-atomic fluorescence spectrometry. Anal Chim Acta 524:305–314
Goodpaster AM, Ramadas EH, Kennedy MA (2011) Potential effect of diaper and cotton ball contamination on NMR- and LC/MS-based metabonomics studies of urine from newborn babies. Anal Chem 83:896–902
Harada S, Takebayashi T, Kurihara A, Akiyama M, Suzuki A, Hatakeyama Y, Sugiyama D, Kuwabara K, Takeuchi A, Okamura T, Nishiwaki Y, Tanaka T, Hirayama A, Sugimoto M, Soga T, Tomita M (2016) Metabolomic profiling reveals novel biomarkers of alcohol intake and alcohol-induced liver injury in community-dwelling men. Environ Health Prev Med 21:18–26
Harrison DE, Maitra P (1968) Role of ATP in control of energy metabolism in growing bacteria. J Gen Microbiol 53:S7–S8
Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G (2010) Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem 82:6621–6628
Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P (2008) Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453(7193):396–400
Jordan KW, Cheng LL (2007) NMR-based metabolomics approach to target biomarkers for human prostate cancer. Expert Rev Proteomics 4:389–400
Jung JY, Lee HS, Kang DG, Kim NS, Cha MH, Bang OS, Ryu Do H, Hwang GS (2011) 1H-NMR-based metabolomics study of cerebral infarction. Stroke 42:1282–1288
Junot C, Fenaille F, Colsch B, Becher F (2014) High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom Rev 33:471–500
Kami K, Fujimori T, Sato H, Sato M, Yamamoto H, Ohashi Y, Sugiyama N, Ishihama Y, Onozuka H, Ochiai A, Esumi H, Soga T, Tomita M (2013) Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics 9:444–453
Kim S, Lee DY, Wohlgemuth G, Park HS, Fiehn O, Kim KH (2013) Evaluation and optimization of metabolome sample preparation methods for Saccharomyces cerevisiae. Anal Chem 85:2169–2176
Kohler I, Verhoeven A, Derks RJ, Giera M (2016) Analytical pitfalls and challenges in clinical metabolomics. Bioanalysis 8:1509–1532
Kok MG, Somsen GW, de Jong GJ (2015) Comparison of capillary electrophoresis-mass spectrometry and hydrophilic interaction chromatography-mass spectrometry for anionic metabolic profiling of urine. Talanta 132:1–7
Kuehnbaum NL, Britz-McKibbin P (2013) New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era. Chem Rev 113:2437–2468
Lee Y, Bowen BP, Northen TR (2010) Mass spectrometry-based metabolomics, analysis of metabolite-protein interactions, and imaging. BioTechniques 49:557–565
Legido-Quigley C, Stella C, Perez-Jimenez F, Lopez-Miranda J, Ordovas J, Powell J, van-der Ouderaa F, Ware L, Lindon JC, Nicholson JK, Holmes E (2010) Liquid chromatography-mass spectrometry methods for urinary biomarker detection in metabonomic studies with application to nutritional studies. Biomed Chromatogr 24:737–743
Lim GB, Lee SY, Lee EK, Haam SJ, Kim WS (2002) Separation of astaxanthin from red yeast Phaffia rhodozyma by supercritical carbon dioxide extraction. Biochem Eng J 11:181–187
Lindenburg PW, Haselberg R, Rozing G, Ramautar R (2015) Developments in interfacing designs for CE-MS: towards enabling tools for proteomics and metabolomics. Chromatographia 78:367–377
Lu W, Bennett BD, Rabinowitz JD (2008) Analytical strategies for LC–MS-based targeted metabolomics. J Chromatogr B 871:236–242
Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD (2010) Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a standalone Orbitrap mass spectrometer. Anal Chem 82:3212–3221
Lu W, Su X, Klein MS, Lewis IA, Fiehn O, Rabinowitz JD (2017) Metabolite measurement: pitfalls to avoid and practices to follow. Annu Rev Biochem 86:277–304
Luo B, Groenke K, Takors R, Wandrey C, Oldiges M (2007) Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography–mass spectrometry. J Chromatogr A 1147:153–164
Lv H, Palacios G, Hartil K, Kurland IJ (2011) Advantages of tandem LC-MS for the rapid assessment of tissue-specific metabolic complexity using a pentafluorophenylpropyl stationary phase. J Proteome Res 10:2104–2112
Maharjan RP, Ferenci T (2003) Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem 313:145–154
Malet-Martino M, Holzgrabe U (2011) NMR techniques in biomedical and pharmaceutical analysis. J Pharm Biomed Anal 55:1–15
Mashego MR, van Gulik WM, Vinke JL, Visser D, Heijnen JJ (2006) In vivo kinetics with rapid perturbation experiments in Saccharomyces cerevisiae using a second-generation bioscope. Metab Eng 8:370–383
Milne SB, Mathews TP, MyersDS IPT, Brown HA (2013) Sum of the parts: mass spectrometry–based metabolomics. Biochemistry 52:3829–3840
Mohler RE, Dombek KM, Hoggard JC, Pierce KM, Young ET, Synovec RE (2007) Comprehensive analysis of yeast metabolite GC/GC–TOFMS data: combining discovery-mode and deconvolution chemometric software. Analyst 132:756–767
Park C, Yun S, Lee SY, Park K, Lee J (2012) Metabolic profiling of Klebsiella oxytoca: evaluation of methods for extraction of intracellular metabolites using UPLC/Q-TOF-MS. Appl Biochem Biotechnol 167:425–438
Patejko M, Jacyna J, Markuszewski MJ (2017) Sample preparation procedures utilized in microbial metabolomics: An overview. J Chromatogr 1043:150–157
Patti GJ (2011) Separation strategies for untargeted metabolomics. J Sep Sci 34:3460–3469
Pejchinovski M, Hrnjez D, Ramirez-Torres A, Bitsika V, Mermelekas G, Vlahou A, Zurbig P, Mischak H, Metzger J, Koeck T (2015) Capillary zone electrophoresis on-line coupled to mass spectrometry: a perspective application for clinical proteomics. Proteomics Clin Appl 9:453–468
Pinu FR, Villas-Boas SG, Aggio R (2017) Analysis of intracellular metabolites from microorganisms: quenching and extraction protocols. Meta 7:53
Pontillo C, Filip S, Borras DM, Mullen W, Vlahou A, Mischak H (2015) CE-MS-based proteomics in biomarker discovery and clinical application. Proteomics Clin Appl 9:322–334
Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Zewman JW, Goodfriend T, Wishart DS (2011) The human serum metabolome. PLoS One 6:e16957
Ramautar R (2016) CE-MS in metabolomics: status quo and the way forward. Bioanalysis 8:371–374
Ramautar R, Nevedomskaya E, Mayboroda OA, Deelder AM, Wilson ID, Gika HG, Theodoridis GA, Somsen GW, de Jong GJ (2011) Metabolic profiling of human urine by CE-MS using a positively charged capillary coating and comparison with UPLC-MS. Mol BioSyst 7:194–199
Saez MJ, Lagunas R (1976) Determination of intermediary metabolites in yeast-critical-examination of effect of sampling conditions and recommendations for obtaining true levels. Mol Cell Biochem 13:73–78
Smart KF, Aggio RBM, Van Houtte JR, Villas-Boas SG (2010) Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat Protoc 5:1709–1729
Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T (2002) Simultaneous determination of anionic Internediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 74:2233–2239
Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T (2003) Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res 2:488–494
Strange RE, Wade HE, Dark FA (1963) Effect of starvation on adenosine triphosphate concentration in aerobacter aerogenes. Nature 199:55–58
Theobald U, Mailinger W, Reuss M, Rizzi M (1993) In vivo analysis of glucose-induced fast changes in yeast adenine-nucleotide pool applying a rapid sampling technique. Anal Biochem 214:31–37
Theobald U, Mailinger W, Baltes M, Reuss M, Rizzi M (1997) In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnol Bioeng 55:305–316
Theodoridis G, Gika HG, Wilson ID (2011) Mass spectrometry–based holistic analysis approaches for metabolite profiling in systems biology studies. Mass Spectrom Rev 30:884–906
van Dam JC, Eman MR, Frank J, Lange HC, van Dedem GWK, Heijnen JJ (2002) Analysis of glycolytic metabolites in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionisation with tandem mass spectrometric detection. Anal Chim Acta 460:209–218
Van der Werf MJ (2003) Metabolomics: a revolutionary tool to optimize microbial processes. Abstr Pap Am Chem Soc 226:U84
Van Hoek WPM, Diderich JA, Schepper M, Luttik MAH, van Dijken JP, Pronk JT, Klaasen P, Boelens MJ, Teixeria de Mattos MJ, van Dam K, Kruckeburg AL (1999) Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J Biol Chem 64:4226–4233
Verduyn C, Postma E, Scheffers WA, Van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517
Villas-Boas SG, Bruheim P (2007) Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem 370:87–97
Villas-Bôas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J (2005a) Mass spectrometry in metabolome analysis. Mass Spectrom Rev 24(5):613–646
Villas-Bôas SG, Moxley JF, Akesson M, Stephanopoulos G, Nielsen J (2005b) High-throughput metabolic state analysis: the missing link in integrated functional genomics of yeasts. Biochem J 388:669–677
Villas-Boas SG, Roeseener U, Hansen MAE, Smedsgaard J, Nielsen J (2007) Metabolomics analysis: an introduction. John Wiley & Sons, Hoboken, NJ
Weston DJ (2010) Ambient ionization mass spectrometry: current understanding of mechanistic theory; analytical performance and application areas. Analyst 135:661–668
Whitfield PD, German AJ, Noble PJM (2004) Metabolomics: an emerging post-genomic tool for nutrition. Br J Nutr 92:549–555
Wishart DS, Mandal R, Stanislaus A, Ramirez-Gaona M (2016) Cancer metabolomics and the human metabolome database. Meta 6:1–17
Wittmann C, Kromer JO, Kiefer P, Binz T, Heinzle E (2004) Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal Biochem 327:135–139
Wu J, An Y, Yao J, Wang Y, Tang H (2010a) An optimised sample preparation method for NMR-based faecal metabonomic analysis. Analyst 135:1023–1030
Wu Z, Li M, Zhao C, Zhou J, Chang Y, Li X, Gao P, Lu X, Li Y, Xu G (2010b) Urinary metabonomics study in a rat model in response to protein-energy malnutrition by using gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry. Mol BioSyst 6:2157–2163
Zhang S, Nagana Gowda GA, Ye T, Raftery D (2010) Advances in NMR-based biofluid analysis and metabolite profiling. Analyst 135:1490–1498
Zhang A, Sun H, Wang P, Han Y, Wang X (2012) Modern analytical techniques in metabolomics analysis. Analyst 137:293
Zhang W, Hankemeier T, Ramautar R (2016) Next-generation capillary electrophoresis-mass spectrometry approaches in metabolomics. Curr Opin Biotechnol 43:1–7
Zhao SS, Zhong X, Tie C, Chen DD (2012) Capillary electrophoresis-mass spectrometry for analysis of complex samples. Proteomics 12:2991–3012
Acknowledgments
PG acknowledges Science and Engineering Research Board (SERB), Government of India, for the financial support of NPDF Grant PDF/2016/001670. SG is thankful to INSPIRE, Department of Science and Technology, Govt. of India (IF130678), for their financial assistance.
Competing Interest: There is no competing interest.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Gupta, P., Gupta, S., Pruthi, V. (2020). Techniques for Detection and Extraction of Metabolites. In: Singh, V., Singh, A., Bhargava, P., Joshi, M., Joshi, C. (eds) Engineering of Microbial Biosynthetic Pathways. Springer, Singapore. https://doi.org/10.1007/978-981-15-2604-6_3
Download citation
DOI: https://doi.org/10.1007/978-981-15-2604-6_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-2603-9
Online ISBN: 978-981-15-2604-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)