Abstract
Soil acts as an natural abode for plants as well as for diverse micro/macroflora and fauna, and thus provides the rhizosphere environment, the fraction of soil surrounding the root system, where plants interact with their root microbiomes. Plants maintain a continuum of interactions with associated microbes that have effects on their cellular physiology resulting in changes in development and function, which can have both positive and negative outcomes. Plant-microbial or microbial-microbial interactions that occur at the plant root-soil interface can also have dynamic effects on the rhizosphere microbiome that greatly affects the overall health and vigor of plants, a key metric in agricultural productivity. This chapter reviews the current understanding of the range of interactions happening in the rhizosphere and the recent advancements that next generation sequencing technologies have had on the ability to identify and classify the rhizosphere microbiome.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
“We know more about the movement of celestial bodies than about the soil underfoot.”
This remark from Leonardo da Vinci can still be used to describe the current knowledge gaps in the understanding of soil microbiomes. Although scientific perspectives and approaches have accelerated our understanding in recent generations, we have just begun to break the shackles in this field of science as characterizing and understanding the diversity underlying the biology and ecology of the soil microbiome are still one of the great challenges in science. Soils act as the natural biological support with a complex group of micro- and macrofauna that represents one of the most important and richest microbial communities on earth. This complex ecosystem that harbors vast genetic diversity, which has only just begun to be detected, let alone harnessed, is hypothesized to contain new forms of antibiotics, catalysts, and metabolites that could be utilized for yet to be imagined purposes in human medicine, agriculture, and beyond (Gans et al. 2005; Wagg et al. 2014).
As the home of many forms of life, mediating diverse and multiphyletic interactions, the soil acts as the factory of life. Soil organisms consist of harmful, beneficial, and neutral living components in the perspective of plant health and productivity. The rhizosphere, defined as all soil under plant root influence, acts as a hotspot between soil inhabiting organisms and plant roots, thus providing the temporal and special cues that drive the dynamic interactions that occur in it (Hinsinger and Marschner 2006; Raaijmakers et al. 2009). The rhizosphere communities are largely made up of organisms such as archaea, bacteria, oomycetes, fungi, algae, arthropods, and their associated viruses as well as other classes of living organisms (Buée et al. 2009; Mendes et al. 2013).
Many biotic and abiotic factors including but not limited to soil type, soil properties, prevailing environmental conditions, inhabiting flora and fauna, and agricultural management practices have strong influence on the structural and functional diversity of rhizosphere communities (Berg and Smalla 2009). Also, the importance of plant species and how their root properties play a large role in the function of soil-phyto-microbial interactions cannot be understated. Plants in their natural environment are surrounded by a remarkable diversity of plant associated microbes, termed as “the plant microbiota.” Establishment of microbiota in and around the plant depends on multiple factors that facilitate niche colonization, a term used for establishment of a population of species in a community, taking account of totality of biological and environmental factors affecting the species and utilization of these factors in the species establishment (Bulgarelli et al. 2013; Vandermeer 1972). Clearly, the majority of bacterial diversity in the soil is dominated by a few bacterial phyla such as Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, and Proteobacteria (Fierer et al. 2009). However, it has been shown that the presence of the host plant drives the diversity in the rhizosphere microbiome filtering few classes of bacteria and selecting for Actinobacteria and Proteobacteria classes. Understanding this complex biophysicochemical interplay and its components is imperative to use soil biodiversity for protective agriculture. Next generation omics such as metagenomics and metabolomics approaches provides a remarkable tool to understand the complex plant microbiota and their intertwined complex interactions.
To understand microbial population, the use of next generation sequencing (NGS) provides an excellent tool for detection of genetic diversity represented by 16S rRNA coding gene sequencing, shotgun metagenome (whole genome) or transcriptome profiling. Classification of microbial groups can be achieved by analyzing similarity of sequenced reads to a known reference which has predetermined taxonomical units for phylotyping, or by similarity of sequenced reads within a microbial community and assigning operational taxonomic units (OTUs) (Chen et al. 2013; Schloss and Westcott 2011). Here we will attempt to sketch the rhizosphere microbial interaction in detail and the genomics approaches utilized to classify the microbial community involved in such interactions.
5.1.1 Friends and Foes: A Side to Pick
Rhizosphere acts as a battlefield between plants and microbes, full of microbial warriors, few acting as foe, few as friends, and the rest not directly taking part in this battle but just being part of the dynamic microbiome interactions with microbiome structure and community, deciding the fate of such interactions around the plant roots. Many bacterial and fungal communities help plants to derive micro- and macronutrients essential for plant growth and development. Plant reciprocates the favor by providing habitat and carbon sources to the bacterial and fungal communities in its rhizosphere. Such mutualistic relationships benefit both interacting partners and are also utilized in agricultural practices. However, pathogenic microbes within the microbiome are greedy and seek out opportunities to utilize the plant carbon sources without reciprocating any energy. This invasion and infection of the plant system to use its nutrient source for its establishment, colonization, and reproduction are the foundation of a host-pathogen relationship and once established as a specialized pathogen begin a molecular arms race where both constantly evolve to gain the upper hand in the interaction.
5.1.2 Microbiome Communities as a Friend for Plant
Root microbiomes provide a remarkable pool of friends for plants, helping them in the nutrient cycling and maintenance of rhizosphere activity essential for proper plant health and vigor. Many bacterial communities majorly belonging to Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria inhabit a soil zone that is in close proximity to the plant root interface, making up an active site of the soil biome. Few of these bacterial species help in N2 fixation symbiotically or nonsymbiotically. Legumes fix atmospheric N2 symbiotically with a specific group of bacteria in the genus Rhizobium (Phillips 1980). Rhizobia, a group of gram-positive, spore-forming bacteria, belong to the α-subclass of Proteobacteria (Rhizobiaceae) and nodulate legumes (Hong et al. 2012; Peters et al. 1995). The Proteobacteria have been classified into six different genera, namely, Rhizobium, Bradyrhizobium, Mesorhizobium, Allorhizobium, Azorhizobium, and Sinorhizobium, which are phylogenetically separated from each other (De Lajudie et al. 1998). The Rhizobia reduce atmospheric N2 inside specialized organs, “nodules,” which are formed as an outcome of a multistep process initiated with infection of plant roots by Rhizobia due to positive chemotaxis to root exudates (Caetano-Anollés et al. 1992). The Rhizobia bacteria are attracted to amino acids, the dicarboxylic acids present in the root exudates, and even to very low concentrations of excreted flavonoids, which are exudates from the secretory part of rhizodeposition, and the bacteria once in contact subsequently attach to the plant root surface. The infection proceeds with specific attachment of bacterial polysaccharides (lipochitooligosaccharides) to specific lectin proteins of host plants, followed by root hair curling, deforming, and branching (Bohlool and Schmidt 1974; Van Rhijn and Vanderleyden 1995; Yuan et al. 2017). Infection threads are formed, cortical cells divide to form nodules, and Rhizobia are released in the plant cell cytoplasm through the infection thread (Wood and Newcomb 1989) and colonize and initiate biological nitrogen fixation, a symbiotic interaction between the plant and the transformed bacteriod. Biological nitrogen fixation (BNF) is an important input of nitrogen (N) in the global agricultural systems. BNF is catalyzed by a complex metalloenzyme, nitrogenase, which is composed of two main components: a heterotetrameric core and a homodimeric dinitrogenase reductase subunit (Kim and Rees 1994). The dinitrogenase reductase is an iron (Fe) protein containing 4 Fe atoms (Fe4S4), and the heterotetramer is a molybdenum-iron (MoFe) protein containing 2 Mo and 30 Fe atoms (Peters et al. 1995). This enzyme system helps the reduction of dinitrogen (N2) that involves reduction of Fe binding protein by different electron carriers (like ferredoxin), followed by transfer of one electron from Fe protein to Mo-Fe protein, and subsequently from Mo-Fe protein to the N2 (the substrate) in the last step of reduction (Kim and Rees 1994). This conversion (reduction) of atmospheric N2 to ammonia (NH3) can only be carried out by a small group of microorganisms like Rhizobia, Frankia and Azospirillum, known as “diazotrophs” (Burns and Hardy 2012).
The first discovered Rhizobium genes related to nitrogen fixation were nif (fixation) and nod (nodulation) genes, and it was found that the nod genes and genes related to host specificity were tightly clustered with the nif genes(Long 2001). Dinitrogenase reductase subunit of the nitrogenase complex is encoded by the nifH gene, while the heterotetrameric core is encoded by nifD and nifK (Yates et al. 1992). The nifH gene is a universally accepted biomarker for BNF as it is found to be highly conserved among N2-fixing organisms in different natural environments (such as marine, terrestrial, and hydrothermal sites) and is widely used to study the ecological and evolutionary aspects of N2-fixing bacteria (Izquierdo and Nüsslein 2006; Mehta et al. 2003; Wartiainen et al. 2008). A previous study by Hennecke et al. (1985) reported that the phylogeny of the nifH genes is closely related to the 16S rRNA gene and the nifH genes and might have evolved with the bacterial evolution. This finding is in contrast with the convention of “lateral gene transfer” among microorganisms. However, a comprehensive study focused on sequence analysis of a large number of nifH genes was carried out and found that the phylogenetic trees of nifH genes were inconsistent with the phylogenetic tree of 16S rRNA; instead, a phylogenetic similarity was found with nodA genes (Haukka et al. 1998). The nifH genes were mostly studied by culture-independent approaches because these provide better understanding of the bacterial community than the culture-dependent method microbiome community studies (Poly et al. 2001). Along with PCR amplification followed by denaturing gradient gel electrophoresis (DGGE), other approaches like DNA hybridization analysis, restriction fragment analysis, and cloning and sequencing approaches were used to study the diversity and abundance of nifH genes (Freitag and Prosser 2003; Hamelin et al. 2002; Neufeld et al. 2001; Stres et al. 2004). Composition of nifH gene pools were also studied in various environments using various techniques, such as polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP), PCR cloning, DGGE, and fluorescently labeled terminal (FLT)-RFLP (Chelius and Lepo 1999; Piceno and Lovell 2000; Zehr et al. 1995).
Different soil physicochemical properties like texture, total carbon (C), and total N influence the variation in nifH gene pools, and the nifH genetic structures are the outcomes of the adaptation of the changing or constant soil environmental conditions (Poly et al. 2001). In a gene expression study on Azotobacter vinelandii, it was revealed that the nifH gene expression is positively correlated with N2-fixation (Bürgmann et al. 2003). Again, different dry bean cultivars showed differential nifH gene expressions, depending on their symbiotic efficiencies with the rhizobia strains, correlated with their N2-fixation potentials (Sanyal et al. 2020, Akter et al. 2013).
The diversity and abundance of nifH genes have been associated with N2-fixation rates and thus depend on the structure and dynamics of bacterial communities (Hsu and Buckley 2009; Reed et al. 2010). PCR and qPCR are considered as powerful tools to study specific gene expression and soil microbiome diversity (Bürgmann et al. 2003; Zehr and McReynolds 1989). Zehr and McReynolds (1989) designed degenerate universal PCR primers to target a wide range of variable nifH gene sequences in diverse bacterial communities and for the first time were able to study community composition. Later, many universal primers were designed to target the nifH gene in the most conserved portions of amino acid sequences (Bürgmann et al. 2004). In an “in silico” analysis, 51 universal and 35 group specific nifH primers were assayed using more than 20,000 nifH gene sequences, and 15 universal markers were identified that successfully targeted more than 90% of the N2-fixers (Gaby and Buckley 2017). However, this study also revealed that many of these primers targeted genes that are not related to N2-fixation, and were often found to miss significant variants of nifH genes (Gaby and Buckley 2017).
5.1.3 Microbial Foes: A Concern for Plant Health
Soil-borne pathogens are major yield limiting factor in most agriculture crops worldwide. Soil-borne pathogens can survive in the bulk soil as a part of the soil microbiome for several years. When a suitable host plant is available, the pathogens identify the host plant by perceiving cues from the rhizosphere and initiate infection to cause disease. The rhizosphere acts as a playground to the complex microbial community, which can also affect the host pathogen interaction influencing the outcome of the pathogen infection process, resulting in compatible (susceptibility and disease) or incompatible (resistant) interactions. Bacterial pathogens such as Erwinia and Pectobacterium species cause disease in plants and reduce their health and vigor and are important soilborne pathogens. Fungi and oomycetes make up one of the most important groups of plant pathogens accounting for more than 70% of all the major crop diseases (Agrios 2005). Fungi are eukaryotic, filamentous, multicellular, and heterotrophic organisms. They produce a network of hyphae called the mycelium and absorb nutrients from their host (Alexopoulos et al. 1996). Most of host-pathogen associations are very specific with a specialized host range. The host-limiting fungi are termed as biotrophs (feed from living host cells without killing them) or necrotrophs (kill the host tissues to derive nutrients as part of the colonization process). Some of the fungal pathogens overwinter in soil and may survive for years in soil even in the absence of their host. Nevertheless, controlling root fungal pathogens is a major challenge compared to foliar pathogens that attack the aboveground portions of the plant (Bruehl 1987). Fusarium root rot is a prominent root disease and is caused by a complex of several Fusarium species. For example, four species of the Fusarium (F. graminearum, F. culmorum, F. poae, and F. sporotrichioides) are mainly associated with the foot and crown rot disease in wheat (Kuzdraliński et al. 2014). Another devastating disease of wheat and barley is Fusarium head blight (FHB), which also causes grain contamination by secreting harmful mycotoxins, leaving them useless for consumption as well as planting material (McMullen et al. 2012). To date, at least 15 known species of the F. graminearum species complex have been reported as the causal agent of FHB (O’Donnell et al. 2008; Wang and Cheng 2017). Specific species were more prevalent than others in FHB disease complex to wheat, and such variations are related to the ecological preference of the pathogens. One of the most common species, F. oxysporum, causes vascular wilt disease in a wide variety of economically important crops (Beckman 1987). Sudden death syndrome, a major yield-limiting soybean disease, is caused by F. virguliforme (previously named Fusaium solani f. sp. glycines).
Signaling processes are the first and most critical step in defining either success or failure in establishing disease. Fungal and oomycete pathogens depend on several different signaling molecules from the host plant to germinate, grow, and persist in the rhizosphere. Many cues such as amino acids, organic acids, flavonols, glucosinolates, indole compounds, fatty acids, and polysaccharides are secreted from microbial community and plant roots in the rhizosphere to establish communication. This wide range of secreted compounds is collectively known as rhizodeposition (Dakora and Phillips 2002; De-la-Peña and Loyola-Vargas 2014). The type and composition of root secretion are dependent on plant species and cultivars, growth stage, and stress factors, which all greatly influence the rhizosphere microbiome communities. Root exudates can affect the microbial community in the soil favoring beneficial microbes while preventing the growth of harmful microbes (Huang et al. 2014; Szoboszlay et al. 2016). Chemical analysis of vegetable root exudates revealed the presence of major organic acids such as citric, succinic, and malic acid and major sugars such as fructose and glucose (Badri et al. 2012; Kamilova et al. 2006). Low concentrations of phenolic compounds like p-hydroxybenzoic, gallic, coumaric, cinnamic, ferulic, salicylic, and sinamic acids in root exudates stimulated conidial germination of F. oxysporum f. sp. niveum, while inhibitory effects were observed at higher concentrations (Wu et al. 2008a, b) indicating roles of rhizodeposition in the species dynamics of rhizosphere microbiota. A study by Wasmann and VanEtten (1996) showed that transformation-mediated chromosome loss and pisatin demethylase gene (PDA1) disruption can decrease the virulence of the pea pathogen Nectria haematococca (anamorph F. solani f. sp. pisi). Pisatin (a phytoalexin) can be detoxified by cytochrome P-450-mediated demethylation. Recently, a cluster of five pea pathogenicity (PEP) genes, which are expressed during N. haematococca infection, were identified in close proximity to PDA1 on a supernumerary chromosome (Han et al. 2001). The PEP gene cluster have differences in GC-content and codon bias compared to genes on essential chromosomes indicating that horizontal transfer occurred for these pathogenicity gene clusters.
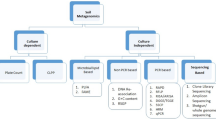
5.2 Metagenomics Approaches for Taxonomical and Functional Classification of Microbiomes
To understand and unravel the components of microbiomes, it is required to adopt a comprehensive approach rather than studying each component separately. Use of molecular techniques such as PCR, qPCR, and DGGE provides valuable tools to study individual elements of the microbiome, but they are insufficient to undertake large-scale analyses to understand the system as a whole (Sharpton 2014). The advent of next generation sequencing techniques and recent high-throughput sequencing technologies (HTST) coupled with state-of-the-art bioinformatics pipelines to assemble sequencing reads provided a boom for soil microbiome studies to explore the diverse taxonomical inhabitants in the rhizosphere. Reduced sequencing cost of per mega base of DNA sequence (Fig. 5.1) further made technology available to all, and genome sequencing became more common specially for small genomes such as fungal, bacterial, and viral whole genome sequencing and pan-genome sequencing (Wetterstrand 2016). Today, many approaches are available for microbiome species identification and classification in conjunction with NGS platforms.
An estimation of raw cost of per megabase (Mb) of DNA sequence over a period of time (reproduced from the National Institutes of Health (Wetterstrand 2016))
5.2.1 PhyloChip and Amplicon-Based Classification of Microbiomes
Identification and phylogenetic classification of fungal species largely rely on internal transcribed spacer 1 (ITS1) and ITS2 region sequencing of the ribosomal RNA (rRNA) cistron as a universal DNA barcode (Schoch et al. 2012; Stielow et al. 2015). However, ITS2 is considered as a region of choice for fungal classification due to better representation in databases and less variability in length (Nilsson et al. 2009; Op De Beeck et al. 2014). For fungal and oomycete genomics and functional data analysis, FungiDB provides an excellent database for the design of experimental primers to carry out phylogenetic analysis (Stajich et al. 2012). Nonetheless, the major portion of the soil microbiome is encompassed by prokaryotes such as bacterial species; thus, we will limit our discussion to the perspective of bacterial microbiome. Nuclear 16S rRNA (ribosomal RNA) gene sequencing has been successfully utilized to identify prokaryotic microbial diversity in natural and agricultural ecosystems (Fierer et al. 2012; Pace et al. 1986). The ubiquitous prokaryotic 16S rRNA cistron is comprised of nine V1-V9 hypervariable regions flanked by conserved DNA sequences suitable for universal primer binding and amplification. Thus, the diversity in variable region is utilized to classify the organisms based on sequence dissimilarities (Chakravorty et al. 2007; Ree et al. 1989). The 16S rRNA gene sequences for bacterial microbiome studies are available from the National Center for Biotechnology Information (NCBI) or other secondary datasets such as EzBioCloud (Yoon et al. 2017). The recent interest of the scientific community to study the soil microbiome in agricultural and non-agricultural ecosystems has led to the development and utilization of the high-density 16S rRNA gene oligonucleotide microarray popularly known as the PhyloChip (Brodie et al. 2007). In the study by Weinert et al. (2011), a high-density 16S rRNA probe-based PhyloChip array capable of detecting 8741 known OTUs was utilized to study and characterize the bacterial microbiota of two different field sites with different soil properties from the rhizosphere of three different potato cultivars. Some 2432 OTUs were detected and classified into 43 phyla in which only one phyla comprised of less than 10 OTUs. Interestingly, differences in soil between two sites influence root bacterial microbiota describing 28% divergence in detected bacterial OTUs, whereas only 9% of the OTUs showed cultivar dependency. Approximately 4% of the OTUs were dependent on both cultivar and site, belonging to Proteobacteria, Bacteroidetes, Firmicutes, Chloroflexi, and Streptomycetaceae phyla. Thus, these phyla dominated the bacterial community in each site irrespective of cultivars grown. This study concludes that historical profiling of soil plays a major role in determining the potato rhizosphere bacterial microbiota composition rather than the effect of the cultivar genotype itself. However, such probe-based studies relied on dominant ribotypes in the sample profile and were limited to the number of known probe OTUs on the PhyloChip. The latest generation of PhyloChip arrays includes 60,000 OTUs capable of classifying147 phyla and 1123 classes of bacterial and archaeal domains, thus enormously increasing its capacity (Hazen et al. 2010). Using the high density PhyloChip, Mendes et al. (2011) identified 33,346 bacterial and archaeal OTUs in the rhizosphere of sugar beet plants grown in disease suppressive soil for Rhizoctonia solani, a fungal pathogen (Mendes et al. 2011). Interestingly, Proteobacteria, Firmicutes, and Actinobacteria were found associated with disease suppression in the study samples (Mendes et al. 2011). Thus, this study implicates a role of natural modification in microbial communities to provide disease suppression, a remarkable finding useful for agricultural crop settings. A study carried out on wild and domesticated barley (Bulgarelli et al. 2015) to understand associated bacterial root microbiota indicated a large share of microbiota is comprised of Actinobacteria, Bacteroidetes, and Proteobacteria using the 16S rRNA survey with an NGS approach. Interesting inferences were made upon annotation and functional classification of sample sequences acquired from bulk soil and host inhabited rhizosphere and root samples. Root and rhizosphere bacterial taxas were significantly enriched for the 12 functional categories of proteins considered to be important for adaptation of microbes to interact with the host root along with protein families for sugar transport and iron mobilization. Thus, the presence of the host barley plants drives the diversity of the rhizosphere for protein functional categories considered to be important for host-microbe interactions either pathogenic or mutualistic (Bulgarelli et al. 2015).
5.2.2 Shotgun Metagenomics for Microbiome Studies
Despite the majority of studies utilizing DNA barcodes and 16S rRNA amplicon-based pyrosequencing, shotgun sequencing approaches for metagenome analyses provide a remarkable upgrade to classify and understand the structure and function of rhizosphere microbiome interactions. PCR-based sequencing approaches can introduce amplification bias (Hong et al. 2009) and require availability of known taxonomically informative genetic markers and thus make analysis difficult for novel or highly diversified bacterial taxa. Further, horizontal gene transfer between distantly related taxons, especially for 16S rRNA genetic locus, can result in overrepresentation of diversity (Acinas et al. 2004). Thus, shotgun approaches enables researchers to study uncultured microbiota and also overcome the majority of the aforementioned concerns for PCR-based sequencing approaches. The shotgun sequencing approach was successfully utilized in a study conducted by Castañeda and Barbosa (2017) to investigate the taxonomic diversity of bacterial and fungal species and their metabolic function in the forest and vineyard soils in Chile. They found bacterial OTU abundance in both habitats, i.e., forest and vineyard, and Proteobacteria accounted for the majority of bacterial phyla and most of the fungal communities were assigned to the Ascomycota. Thus, using shotgun approaches enabled researchers to study the soil from both the bacterial and fungal perspective using a single DNA library, whereas in amplicon-based sequencing, it is required to make fungal and bacterial amplicon libraries separately based on ITS and 16S rRNA markers, respectively.
5.2.3 Approaches to Acquire and Analyze Metagenomics Data
Various large-scale coordinated efforts were initiated to investigate and characterize the soil microbial taxonomy and functional diversity such as the Earth Microbiome Project (Gilbert et al. 2014), ECOFINDERS (http://projects.au.dk/ecofinders/), and TerraGenome (Vogel et al. 2009). These initiatives provide a good resource of information for any interested novice laboratories that decide to make the leap into soil microbiome studies. However, many practical roadblocks still prevail before carrying out a microbiome analyses. Since the majority of environmental microbes are unculturable, this is a major hurdle to acquiring and studying representative natural environment samples. For example, using soil samples from a single experimental site, Delmont et al. found that the DNA extraction methods can produce larger variations in the metagenomic composition of the microbiome than either the season of sampling or the soil depth (Delmont et al. 2012). Thus, just DNA extraction methods used can have a profound effect on confounding the data for analysis of metagenomic variation. Since the requirement of a high-quality DNA sample is the first step and depending on the sequencing platform, DNA amplification steps may be required for low DNA yielding soil samples (permafrost soil), these PCR steps further add bias to the samples (Mackelprang et al. 2011). Thus, to adopt a common efficient method although unlikely to develop is required for a true representation of sample.
One of the important steps to exploring the complexity of soil microbiomes is the analysis of large amount of sequencing data after the NGS run. Many bioinformatics pipelines and methods have been described. The recent availability of many online analysis tools such as Galaxy and KBase that can perform sequence quality analysis and assembly has made this task a bit less complex and has provided an excellent alternative for setting up a computing facility and expertise in “big data” analysis. Open-source pipelines such as Quantitative Insights Into Microbial Ecology (QIIME2) (Caporaso et al. 2010) or MEGAN6 (Huson et al. 2007, 2016) are very useful tools, which can be installed locally to analyze data offline. EBI metagenomics provides a cloud-based automated pipeline for analysis and archiving of metagenomic data with tools to determine the phylogenetic diversity as well as functional and metabolic analysis of submitted datasets (Mitchell et al. 2016) (Fig. 5.2). Many informative web resources and literature are also available to walk through the process of data analysis, yet researchers must pave their own paths when utilizing these publicly available tools (Table 5.1) (Clooney et al. 2016; Kunin et al. 2008; Oulas et al. 2015; Quince et al. 2017; Thomas et al. 2012).
An illustration and schematic overview describing the rhizosphere microbiome and its interactions. PCR amplicon-based or shotgun metagenomics approaches can be adopted using next generation sequencing (NGS) approaches. Various bioinformatics pipelines can be adopted for sequence assembly and alignment for microbiome identification, taxonomical classification, and functional annotation
5.3 Concluding Remarks
Biodiversity loss is a major concern affecting ecosystem homeostasis and stability among its inhabitants. Soil biodiversity comprised of micro- and macrofauna is still largely understudied; thus, we have insufficient information on the effects that climate change and environmental pollution are having on the soil ecosystems worldwide. The majority of these soil inhabitants or the microbiome components play a crucial role in soil health, nutrient cycle, rhizosphere interactions, and ecosystem function; thus, mass extinction and loss of ecosystem homeostasis could be happening under foot, and we may never be aware of it or its consequences until it is too late.
On the rhizosphere frontline between plant and microbes, many microbial friends, foes, and spectators stand tall in the arena presenting numerous possibilities for interaction. The microbiome structure and community decide the fate of such interactions in the rhizosphere. Many bacterial and fungal pathogen communities prevail as a part of soil microbiome, yet introduction of the agricultural systems sufficiently modify the microbiome structure of particular soils leading to the establishment of a unique microbial community. In a study conducted by Costa et al. (2007) comparing uncultured Pseudomonas in bulk and rhizosphere soil, it was observed that strawberries infected with Verticillium dahliae attract fluorescent pseudomonads having close relatedness to P. fluorescens strain F113, a biocontrol agent. This strain produces DAPG (2,4-diacetylphloroglucinol), a phenolic compound having antiphytopathogenic properties (Bangera and Thomashow 1999; Costa et al. 2007). Thus, such examples indicate the summoning of beneficial bacteria from microbial pools by plant for their own benefits. On the other hand, pathogenic microbes act as foes continuously invading plants as a host to gain nutrients required for their pathogenic lifestyle. Many fungal pathogens such as Sclerotinia sclerotiorum, Fusarium oxysporum, and Pythium spp. cause root disease in a wide plant host range adversely affecting the plant health as previously discussed. Symbiotic microbes live in a mutualistic relationship with plants and thus work as a friend in the soil microbiome fraction. The use of Bacillus and Trichoderma strains (Benítez et al. 2004; Chowdhury et al. 2015) is common as a biocontrol agent to augment the soil microbial population to keep check on pathogenic fungal strains, and they are extensively studied. Pseudomonas-based formulations are also popular for soil augmentation and biocontrol (Weller 2007). Interaction among microbial communities, plant roots, and rhizodeposition to establish rhizosphere microbiota is a complex mechanism and needed to be understood for use for improved agricultural practices and soil health.
The integration of modern genetic tools is allowing for the dissection of the complex structure and makeup of the rhizosphere microbiome in relation to plant-microbe and microbe-microbe interactions and for the study of their intertwined molecular signaling pathways. Next generation sequencing approaches for metagenomics and metabolomics analysis coupled with conventional phenotype-dependent approaches to study the structure and function of microbial life in the plant rhizosphere illuminated and enhanced our limited understanding of these interactions and its components. This information will help us to make better use of the currently underutilized and vastly untapped resources of the soil microbiome. Shotgun-based NGS approaches will not only enable us to classify different known soil microbes but will also help to identify rare species prevailing in certain ecosystem important for its maintenance. It is important to establish a larger comprehensive catalog of microbial communities in different soil types for future comparative studies and taxonomical and functional classification. Characterizing the specific inhabitants of soil microbiome communities will allow us to pinpoint their contributions and cooperation in natural ecosystems and bring them under the purview of biotechnological manipulations to work as microbial factories. The microbiome acts as a dynamic entity and requires its largest to smallest community fractions to work properly as a system of ‘fellowship or disunity’ for its maintenance. Individual microbial contribution to its microbiome entity can be sketched by the famous remark quoted by the fictional character Galadriel in J.R.R Tolkien’s novel Lord of the Rings—“Even the smallest person can change the course of the future.”
References
Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF (2004) Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186:2629–2635
Agrios G (2005) Plant pathology, 5th edn. Academic Press, San Diego, CA
Akter Z, Pageni B, Lupwayi N, Balasubramanian P (2013) Biological nitrogen fixation and nif H gene expression in dry beans (Phaseolus vulgaris L.). Can J Plant Sci 94:203–212
Alexopoulos CJ, Mims C, Blackwell M (1996) Introductory mycology. John Wiley and Sons, New York, NY
Badri DV, Chaparro JM, Manter DK, Martinoia E, Vivanco JM (2012) Influence of ATP-binding cassette transporters in root exudation of Phytoalexins, signals, and in disease resistance. Front Plant Sci 3:149
Bangera MG, Thomashow LS (1999) Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J Bacteriol 181:3155–3163
Beckman, C. (1987). The nature of wilt diseases of plants (APS press)
Benítez T, Rincón AM, Limón MC, Codón AC (2004) Biocontrol mechanisms of Trichoderma strains. Int Microbiol 7:249–260
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bohlool BB, Schmidt EL (1974) Lectins: a possible basis for specificity in the rhizobium–legume root nodule symbiosis. Science 185:269–271
Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL (2007) Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A 104:299–304
Bruehl, G. (1987). Soilborne plant pathogens (Macmillan publishing company)
Buée M, De Boer W, Martin F, van Overbeek L, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17:392–403
Bürgmann H, Widmer F, Sigler WV, Zeyer J (2003) mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl Environ Microbiol 69:1928–1935
Bürgmann H, Widmer F, Von Sigler W, Zeyer J (2004) New molecular screening tools for analysis of free-living diazotrophs in soil. Appl Environ Microbiol 70:240–247
Burns R, Hardy R (2012) Nitrogen fixation in bacteria and higher plants. Springer, Berlin
Caetano-Anollés G, Wrobel-Boerner E, Bauer WD (1992) Growth and movement of spot inoculated rhizobium meliloti on the root surface of alfalfa. Plant Physiol 98:1181–1189
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Castañeda LE, Barbosa O (2017) Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. Peer J 5:e3098
Chakravorty S, Helb D, Burday M, Connell N, Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69:330–339
Chelius M, Lepo J (1999) Restriction fragment length polymorphism analysis of PCR-amplified nifH sequences from wetland plant rhizosphere communities. Environ Technol 20:883–889
Chen W, Zhang CK, Cheng Y, Zhang S, Zhao H (2013) A comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS One 8:e70837
Chowdhury SP, Hartmann A, Gao X, Borriss R (2015) Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front Microbiol 6:780
Clooney AG, Fouhy F, Sleator RD, O’ Driscoll A, Stanton C, Cotter PD, Claesson MJ (2016) Comparing apples and oranges?: next generation sequencing and its impact on microbiome analysis. PLoS One e0148028:11
Costa R, Gomes NCM, Krögerrecklenfort E, Opelt K, Berg G, Smalla K (2007) Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ Microbiol 9:2260–2273
Dakora FD, Phillips DA (2002) Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245:35–47
De Lajudie P, Willems A, Nick G, Moreira F, Molouba F, Hoste B, Torck U, Neyra M, Collins MD, Lindström K et al (1998) Characterization of tropical tree rhizobia and description of Mesorhizobium plurifarium sp. nov. Int J Syst Bacteriol 48(Pt 2):369–382
De-la-Peña C, Loyola-Vargas VM (2014) Biotic interactions in the rhizosphere: a diverse cooperative enterprise for plant productivity. Plant Physiol 166:701–719
Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA et al (2012) Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6:1677–1687
Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC (2009) Global patterns in belowground communities. Ecol Lett 12:1238–1249
Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R (2012) Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017
Freitag TE, Prosser JI (2003) Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl Environ Microbiol 69:1359–1371
Gaby JC, Buckley DH (2017) The use of degenerate primers in qPCR analysis of functional genes can cause dramatic quantification bias as revealed by investigation of nifH primer performance. Microb Ecol 74:701
Gans J, Wolinsky M, Dunbar J (2005) Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390
Gilbert JA, Jansson JK, Knight R (2014) The earth microbiome project: successes and aspirations. BMC Biol 12:69
Hamelin J, Fromin N, Tarnawski S, Teyssier-Cuvelle S, Aragno M (2002) nifH gene diversity in the bacterial community associated with the rhizosphere of Molinia coerulea, an oligonitrophilic perennial grass. Environ Microbiol 4:477–481
Han Y, Liu X, Benny U, Kistler HC, VanEtten HD (2001) Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J 25:305–314
Haukka K, Lindström K, Young JP (1998) Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl Environ Microbiol 64:419–426
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL et al (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208
Hennecke H, Kaluza K, Thöny B, Fuhrmann M, Ludwig W, Stackebrandt E (1985) Concurrent evolution of nitrogenase genes and 16S rRNA in rhizobium species and other nitrogen fixing bacteria. Arch Microbiol 142:342–348
Hinsinger P, Marschner P (2006) Rhizosphere—perspectives and challenges—a tribute to lorenz hiltner 12–17 September 2004—Munich, Germany. Plant Soil 283:vii–viii
Hong S, Bunge J, Leslin C, Jeon S, Epstein SS (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3:1365–1373
Hong Y, Ma Y, Wu L, Maki M, Qin W, Chen S (2012) Characterization and analysis of nifH genes from Paenibacillus sabinae T27. Microbiol Res 167:596–601
Hsu S-F, Buckley DH (2009) Evidence for the functional significance of diazotroph community structure in soil. ISME J 3:124–136
Huang X, Chaparro J, Reardon K, Zhang R, Shen Q, Vivanco J (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:267–275
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, Ruscheweyh H-J, Tappu R (2016) MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957
Izquierdo JA, Nüsslein K (2006) Distribution of extensive nifH gene diversity across physical soil microenvironments. Microb Ecol 51:441–452
Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B (2006) Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant-Microbe Interact 19:250–256
Kim J, Rees DC (1994) Nitrogenase and biological nitrogen fixation. Biochemistry 33:389–397
Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P (2008) A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev 72:557–578, Table of Contents
Kuzdraliński A, Szczerba H, Tofil K, Filipiak A, Garbarczyk E, Dziadko P, Muszyńska M, Solarska E (2014) Early PCR-based detection of Fusarium culmorum, F. graminearum, F. sporotrichioides and F. poae on stem bases of winter wheat throughout Poland. Eur J Plant Pathol 140:491–502
Long SR (2001) Genes and signals in the rhizobium-legume symbiosis. Plant Physiol 125:69–72
Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK (2011) Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371
McMullen M, Bergstrom G, Wolf E, Dill-Macky R, Hershman D, Shaner G, Sanford D (2012) A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis 96:1712–1728
Mehta MP, Butterfield DA, Baross JA (2003) Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl Environ Microbiol 69:960–970
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663
Mitchell A, Bucchini F, Cochrane G, Denise H, ten Hoopen P, Fraser M, Pesseat S, Potter S, Scheremetjew M, Sterk P et al (2016) EBI metagenomics in 2016—an expanding and evolving resource for the analysis and archiving of metagenomic data. Nucleic Acids Res 44:D595–D603
Neufeld JD, Driscoll BT, Knowles R, Archibald FS (2001) Quantifying functional gene populations: comparing gene abundance and corresponding enzymatic activity using denitrification and nitrogen fixation in pulp and paper mill effluent treatment systems. Can J Microbiol 47:925–934
Nilsson RH, Ryberg M, Abarenkov K, Sjökvist E, Kristiansson E (2009) The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol Lett 296:97–101
O’Donnell K, Ward TJ, Aberra D, Kistler HC, Aoki T, Orwig N, Kimura M, Bjørnstad S, Klemsdal SS (2008) Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet Biol 45:1514–1522
Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV (2014) Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629
Oulas A, Pavloudi C, Polymenakou P, Pavlopoulos GA, Papanikolaou N, Kotoulas G, Arvanitidis C, Iliopoulos I (2015) Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol Insights 9:75–88
Pace NR, Stahl DA, Lane DJ, Olsen GJ (1986) The analysis of natural microbial populations by ribosomal RNA sequences. In: Marshall KC (ed) Advances in microbial ecology. Springer, Boston, MA, pp 1–55
Peters JW, Fisher K, Dean DR (1995) Nitrogenase structure and function: a biochemical-genetic perspective. Annu Rev Microbiol 49:335–366
Phillips D (1980) Efficiency of symbiotic nitrogen fixation in legumes. Annu Rev Plant Physiol 31:29–49
Piceno YM, Lovell CR (2000) Stability in natural bacterial communities: I. nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb Ecol 39:32–40
Poly F, Ranjard L, Nazaret S, Gourbière F, Monrozier LJ (2001) Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262
Quince C, Walker AW, Simpson JT, Loman NJ, Segata N (2017) Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 35:833–844
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Ree HK, Cao KM, Thurlow DL, Zimmermann RA (1989) The structure and organization of the 16S ribosomal RNA gene from the archaebacterium Thermoplasma acidophilum. Can J Microbiol 35:124–133
Reed SC, Townsend AR, Cleveland CC, Nemergut DR (2010) Microbial community shifts influence patterns in tropical forest nitrogen fixation. Oecologia 164:521–531
Sanyal D, Solanki S, Ameen G, Brueggeman RS, Chatterjee A (2020) Understanding the expression dynamics of symbiont rhizobial nifH and nitrogen assimilatory NR and GS genes in dry bean (Phaseolus vulgaris L.) genotypes at various growth stages. Legum Sci. https://doi.org/10.1002/leg3.26
Schloss PD, Westcott SL (2011) Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium, and Fungal Barcoding Consortium Author List (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246
Sharpton TJ (2014) An introduction to the analysis of shotgun metagenomic data. Front Plant Sci 5:209
Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF et al (2012) FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–D681
Stielow JB, Lévesque CA, Seifert KA, Meyer W, Iriny L, Smits D, Renfurm R, Verkley GJM, Groenewald M, Chaduli D et al (2015) One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35:242–263
Stres B, Mahne I, Avgustin G, Tiedje JM (2004) Nitrous oxide reductase (nosZ) gene fragments differ between native and cultivated Michigan soils. Appl Environ Microbiol 70:301–309
Szoboszlay M, White-Monsant A, Moe LA (2016) The effect of root exudate 7,4′-dihydroxyflavone and Naringenin on soil bacterial community structure. PLoS One 11:e0146555
Thomas T, Gilbert J, Meyer F (2012) Metagenomics - a guide from sampling to data analysis. Microb Inform Exp 2:3
Van Rhijn P, Vanderleyden J (1995) The rhizobium-plant symbiosis. Microbiol Rev 59:124–142
Vandermeer JH (1972) Niche theory. Annu Rev Ecol Syst 3:107–132
Vogel T, Simonet P, Nalin R, Philippot L (2009) TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol 7:252
Wagg C, Bender SF, Widmer F, van der Heijden MGA (2014) Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci U S A 111:5266–5270
Wang C-L, Cheng Y-H (2017) Identification and trichothecene genotypes of Fusarium graminearum species complex from wheat in Taiwan. Bot Stud (Taipei, Taiwan) 58:4
Wartiainen I, Eriksson T, Zheng W, Rasmussen U (2008) Variation in the active diazotrophic community in rice paddy—nifH PCR-DGGE analysis of rhizosphere and bulk soil. Appl Soil Ecol 39:65–75
Wasmann C, VanEtten H (1996) Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Molecular Plant-microbe Interactions: MPMI (USA)
Weinert N, Piceno Y, Ding G-C, Meincke R, Heuer H, Berg G, Schloter M, Andersen G, Smalla K (2011) PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75:497–506
Weller DM (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97:250–256
Wetterstrand K (2016) DNA sequencing costs: data from the NHGRI genome sequencing program (GSP)
Wood S, Newcomb W (1989) Nodule morphogenesis: the early infection of alfalfa (Medicago sativa) root hairs by rhizobium meliloti. Can J Bot 67:3108–3122
Wu H, Liu D, Ning-Ling, Bao W, Ying R, Ou Y, Huo Z, Li Y, Shen Q (2008a) Effects of vanillic acid on the growth and development of Fusarium oxysporum f. sp. niveum. Allelopath J 22:111–121
Wu H-S, Raza W, Fan J-Q, Sun Y-G, Bao W, Liu D-Y, Huang Q-W, Mao Z-S, Shen Q-R, Miao W-G (2008b) Antibiotic effect of exogenously applied salicylic acid on in vitro soilborne pathogen, Fusarium oxysporum f.sp.niveum. Chemosphere 74:45–50
Yates M, Stacey G, Burris R, Evans H (1992) Biological nitrogen fixation. 685–735
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yuan Z, Zhang Z, Wang X, Li L, Cai K, Han H (2017) Novel impacts of functionalized multi-walled carbon nanotubes in plants: promotion of nodulation and nitrogenase activity in the rhizobium-legume system. Nanoscale 9:9921–9937
Zehr JP, McReynolds LA (1989) Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55:2522–2526
Zehr JP, Mellon M, Braun S, Litaker W, Steppe T, Paerl HW (1995) Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol 61:2527–2532
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Solanki, S. et al. (2020). Friends and Foes: Phyto-Microbial Interactions in Molecular Perspective. In: Kumar, M., Kumar, V., Prasad, R. (eds) Phyto-Microbiome in Stress Regulation. Environmental and Microbial Biotechnology. Springer, Singapore. https://doi.org/10.1007/978-981-15-2576-6_5
Download citation
DOI: https://doi.org/10.1007/978-981-15-2576-6_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-2575-9
Online ISBN: 978-981-15-2576-6
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)