Abstract
Viruses are known for their utilization of the host machinery to aid their replication within the infected cells. The viral nucleic acid, their intermittent forms contain pathogen-associated molecular patterns (PAMP) which enable the pattern recognition receptors (PRR) to distinguish them from self and mount a response against them. Highly robust targets for the PRRs are the cell entry points and phases of viral replication. RNA-sensing PRR can be classified as endosomal PRR and cytosolic PRR. Both enveloped and non-enveloped viruses utilize endosomal compartments to undergo proteolytic cleavage or pH-dependent conformational changes to ensure membrane fusion. Thus, monitoring the endosomal compartments in order to restrict the virus from penetrating into the cytoplasm is a key antiviral strategy. Endosomal compartments are guarded by Toll-like receptors (TLRs) which are the earliest discovered PRRs. TLR3, 7, and 8 specifically cater to viral RNA sensing. They can detect dsRNA and stable stem structures of ssRNA. Cytosolic PRRs include RIG-I like receptors (RLR) and nucleotide-binding oligomerization domain-containing (NOD)—like receptors (NLR). RLRs are cytosolic helicases which detect viral RNA, ds RNA, short 5′ppp RNA, and RNase L cleaved RNA. Retinoic acid-inducible gene I product (RIG-I), melanoma differentiation-associated antigen 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicases belonging to the SF2 superfamily. Other RNA helicases like SNRNP200 which belongs to the Ski-2 superfamily, or DDX60 belonging to the Ski-2-like helicase family are also involved in viral RNA sensing. NLRs like NLRP3 and NOD2 are cytosolic RNA-sensing PRRs. In addition to these, RNA-binding proteins like RNase L, protein kinase R (PKR), and interferon-inducible transmembrane protein (IFIT) are also present which play pertinent roles to enhance the antiviral immunity during a viral infection. Recognition of PAMPs or DAMPs by PRRs activate various signaling cascades which ultimately triggers the transcription of type I/III IFN, production of various pro-inflammatory cytokines, and various other genes that can ensure an intracellular antiviral state which will aid in containing the viral infection. Furthermore, these interferons can lead to the expression of multiple interferon-stimulated genes (ISGs) which induce antiviral activities controlling the life cycle of the virus, restricting its replication, and transmission to the surrounding cells. This chapter will elaborate on these RNA-sensing PRRs, their structures, agonists, mechanism of activation, and response mounted against viral infection.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Viruses are known for their utilization of the host machinery to aid their replication within the infected cells. The viral nucleic acid, its intermittent forms contain pathogen-associated molecular patterns (PAMP) which enable the pattern recognition receptors (PRR) to distinguish them from self and mount a response against them. Highly robust targets for the PRRs are the cell entry points and phases of viral replication. RNA-sensing PRR can be classified as endosomal PRR and cytosolic PRR. Both enveloped and non-enveloped viruses utilize endosomal compartments to undergo proteolytic cleavage or pH-dependent conformational changes to ensure membrane fusion. Thus, monitoring the endosomal compartments in order to restrict the virus from penetrating into the cytoplasm is a key antiviral strategy. Endosomal compartments are guarded by Toll-like receptors (TLRs) which are the earliest discovered PRRs. TLR3, 7, and 8 specifically cater to viral RNA sensing. They can detect dsRNA and stable stem structures of ssRNA.
Cytosolic PRRs include RIG-I like receptors (RLR) and nucleotide-binding oligomerization domain-containing (NOD)—like receptors (NLR). RLRs are cytosolic helicases which detect viral RNA, ds RNA, short 5′ppp RNA and RNase L cleaved RNA. Retinoic acid-inducible gene I product (RIG-I), melanoma differentiation-associated antigen 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicases belonging to the SF2 superfamily. Other RNA helicases like SNRNP200 which belongs to the Ski-2 superfamily, or DDX60 belonging to the Ski-2-like helicase family are also involved in viral RNA sensing. NLRs like NLRP3 and NOD2 are cytosolic RNA-sensing PRRs. In addition to these, RNA-binding proteins like RNase L, protein kinase R (PKR), and interferon-inducible transmembrane protein (IFIT) are also present which play pertinent roles to enhance the antiviral immunity during a viral infection.
Recognition of PAMPs or DAMPs by PRRs activate various signaling cascades which ultimately triggers the transcription of type I/III IFN, production of various pro-inflammatory cytokines, and various other genes that can ensure an intracellular antiviral state which will aid in containing the viral infection. Furthermore, these interferons can lead to the expression of multiple interferon-stimulated genes (ISGs) which induce antiviral activities controlling the life cycle of the virus, restricting its replication and transmission to the surrounding cells. This chapter will elaborate on these RNA-sensing PRRs, their structures, agonists, mechanism of activation, and response mounted against viral infection.
2 Innate Immunity: Soldiers Against Viruses
Innate immunity provides the first line of defence against the invading pathogens. It exerts its role by distinguishing between self and non-self, subsequently initiating a host response against the invading pathogen. Innate immunity is triggered by the identification of PAMPs (pathogen-associated molecular patterns) via the PRRs (pattern recognition receptors). PRRs are germline-encoded proteins expressed by a variety of cells which can carry out surveillance to detect any pathogenic invasions. PRRs have been classified into various families based on protein domain homology; Toll-like receptors (TLRs), RIG-I like receptors (RLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), AIM2-like receptors (ALRs), and cytosol DNA sensing PRR cyclic GMP-AMP synthase (cGAS). PRRs not only survey the PAMPS but also the DAMPS (danger-associated molecular patterns), which are typically the cellular products produced as a response to cell stress (Anafu et al. 2013; Chen et al. 2017; Chow et al. 2018; Said et al. 2018).
Recognition of PAMPs or DAMPs by PRRs leads to transcription of genes involved in pro-inflammatory responses. The responses mounted against a viral infection, upregulates type I and III interferons, pro-inflammatory chemokines, and various other genes that can ensure an intracellular antiviral state in order to contain the infection. Furthermore, these interferons can lead to the expression of multiple interferon-stimulated genes (ISGs) which induce antiviral activities controlling the life cycle of the virus, restricting its replication and transmission to the surrounding cells.
Viruses are known for their utilization of the host machinery to aid their replication within the infected cells. The viral nucleic acid, their intermittent forms contain PAMPs which enable the PRRs to distinguish them from self and mount a response against them. Highly robust targets for the PRRs are the cell entry points and phases of viral replication. Both enveloped and non-enveloped viruses utilize endosomal compartments to undergo proteolytic cleavage or pH-dependent conformational changes to ensure membrane fusion (Said et al. 2018). Thus, monitoring the endosomal compartments in order to restrict the virus from penetrating into the cytoplasm is a key antiviral strategy. Endosomal compartments are guarded by Toll-like receptors (TLRs) which are the earliest discovered PRRs. TLR3, 7, and 8 specifically cater to viral RNA sensing. Cytosolic PRRs include RIG-I, MDA5, LGP2, NLRP3, and NOD2. RNA sensing is also carried out by other RNA helicases like SNRNP200 and DDX60 which will be discussed in details. In addition to these, RNA-binding proteins like RNase L, PKR, and IFITs are also present which play pertinent roles to enhance the antiviral immunity during a viral infection.
3 RNA-Sensing Endosomal PRRs
3.1 Toll-Like Receptors (TLRs)
TLRs are a very important and earliest detected type of PRRs. They comprise of 11 genes on the human genome. Majority of the TLRs are associated with the plasma membrane. However, the TLRs associated with RNA sensing namely, TLR3, 7, and 8 are present in the endosomal compartment. TLRs are type I transmembrane proteins which consists of N-terminal ectodomain or extracellular leucine-rich domain (ECD), middle transmembrane domain (TM), and C-terminal cytoplasmic Toll/IL-1 receptor (TIR) domain. Leucine-rich regions (LRRs) in the ECD are attached into a horseshoe-shaped solenoid structure. α-helices of each LRR forms into the convex structure of the solenoid and the β sheets assemble into the concave surfaces of the solenoid structure. It is a unique property of the TLRs to bind their agonists to the lateral convex surface instead of concave surface.
The formation of M-shaped dimer or multimer is needed for all TLR activation, so that the C-terminal regions of the two TLR ECDs are brought into proximity. It in turn causes the multimerization of cytoplasmic TIR domains, which will recruit downstream adaptors TRIF or MyD88 through homotypic interaction, further forming signaling complex called signalosome and activating downstream transcription factors like NF-κB which induces pro-inflammatory cytokines and interferon regulatory factor (IRF) which in turn induces type I interferon.
3.1.1 TLR3: Expression, Structure, and Signaling Pathway
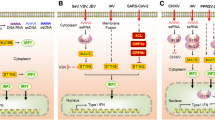
Immune cells like monocytes, macrophages, conventional dendritic cells, NK cells, T and B lymphocytes, mast cells, eosinophils, and basophiles demonstrate the expression of TLR3 in their endosomal compartments (Chen et al. 2017; Said et al. 2018; Gantier and Williams 2011; Hewson et al. 2005; Thomas et al. 2007). They are also expressed in nonimmune cells like keratinocytes, fibroblasts, hepatocytes, astrocytes, and microglia. TLR3 senses dsRNA; however, stable stem structures of ssRNA are also recognized by TLR3. Its structure consists of N-terminal ectodomain or extracellular 23 leucine-rich domain (ECD), middle transmembrane domain (TM), and C-terminal cytoplasmic Toll/IL-1 receptor (TIR) domain. The 23 LRR ECD is responsible for the viral dsRNA binding. Dimerization of the ECD initiates the signaling cascade wherein TIR domain-containing adaptor protein-inducing IFN-β (TRIF) is then recruited and undergoes slight conformational changes to form a signaling complex together with TNF receptor-associated factor 6 (TRAF6), TRAF3, TBK1, IKKε, and IKK. This leads to the activation of IRF3/IRF7 and NF-κB, which results in the production of type 1 IFNs and inflammatory cytokines, respectively.
Further, there are positive regulators to this pathway like S100A9 which acts during the early stages of TLR3 activation by easing the maturation of TLR3-containing early endosomes into late endosomes, etc. Similarly, there are negative regulators such as Rho proteins that decrease the production of pro-inflammatory cytokines upon TLR3 triggering. The pathway is also regulated via post-translational modifications.TLR3 has been implicated in pathogenesis of HCV, infection, HSV-1, HBV, etc. This makes TLR3 an interesting candidate to prod into antiviral therapies.
3.1.2 TLR7 and 8: Expression, Structure, and Signaling Pathway
TLR7/8/9 subfamily members are localized at endosomes of the cells. They are present on the endosomes of monocytes, macrophages, plasmocystoid dendritic cells, NK cells, and T and B cells. TLR8 is also present on mast cells and Tregs. Both have very similar ligand recognition and intracellular signaling. TLR7 and 8 are activated by small molecular agonists and GU or U rich single-stranded RNA (ssRNA) (Chen et al. 2017; Gantier and Williams 2011; Crozat and Beutler 2004; Jensen and Thomsen 2012; Jurk et al. 2002; Patel et al. 2014). In viral genomes, untranslated terminal regions (UTR) are GU or U rich. They are highly conserved sequences as they are involved in viral protein translation and RNA replication. Further, they can also recognize phagocytosed vRNA, by the adenosine-to-inosine (A-to-I) editing, which is an important arm of the antiviral response. But 2′-O-methylation within an RNA sequence leads to the triggering of TLR8 but not TLR7.
The ECDs of both TLR7 and 8 have 26 LRR motifs in their extracellular domain, which contain multiple insertions such as the Z-loop or undefined region situated between LRRs 14 and 15. Cathepsins and arginine endopeptidase cleave the TLR7 and 8 in the endosomes, at this z-loop. Once cleaved, the fragments are still associated to each other by the intermolecular interactions as they both are crucial for the receptor activation. Recent crystal structures have demonstrated the presence of two binding sites: one for a small chemical stimuli or degradation product of ssRNA and the other which recognizes the viral ssRNA. At steady state they both exist as dimers. However, on association with the agonists, the conformation changes causing multimerization of the cytoplasmic TIR domains. A downstream adaptor MyD88 is recruited through homotypic interaction; a signaling complex called myddosome is formed involving IRAK4, IRAK1, TRAF6, and TRAF3 and downstream transcription factors NF-κB and IRF7 are activated to induce pro-inflammatory cytokines and IFNs, respectively (Fig. 15.1). Furthermore, TLR7 recognizes Streptococcus Group B (SGB) RNA and TLR8 recognize the RNAs from Escherichia coli, Mycobacteria bovis, Helicobacter pylori, and Borrelia burgdorferi.
4 RNA-Sensing Cytosolic PRRs
The first line of cytosolic RNA sensors include the PRR families RLRs and NLRs. RLRs consists of cytosolic helicases: RIG-I, MDA5, and LGP2; other DEXD/H-box helicases like DDX3, DDX60, and SNRP20 (Chow et al. 2018; Dang et al. 2018).
4.1 RIG-I Like Receptors (RLRs)
Cytosolic helicases are ubiquitously expressed in low levels in most cell types. They belong to a family of aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicases (SF2 superfamily). RLRs are also known as double-stranded RNA (dsRNA)-dependent ATPases. Their function includes sensing the viral RNA in the cytosol, binding to the non-self RNA stably, and imposing the innate immune response against the target RNA. Three members widely studied are the retinoic acid-inducible gene I product (RIG-I), melanoma differentiation-associated antigen 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2).
Overall structure of the RLRs are as follows:
-
1.
Zinc-binding C-terminal domain (CTD), otherwise known as the repressor domain (RD)
-
2.
Attached to the CTD is a central RNA helicase core consisting of two RecA-like helicase domains promotes dsRNA recognition.
-
3.
Further, RIG-I and MDA5 contain at the N-terminus tandem caspase activation and recruitment domains (CARDs) that enables signaling capabilities. LGP2 uniquely lacks this domain.
4.1.1 RIG-I
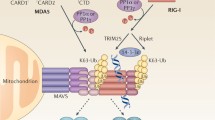
When disengaged, the RIG-I CARDs and RD are bound to the helicase region and this leads to autoinhibition (Chow et al. 2018; Dang et al. 2018; Pichlmair et al. 2006). During viral replication the CTD/RD detects and associates with the viral RNA along with the helicase, exposing the CARDS. These CARDs are no longer autorepressed; the RLRs oligomerize and further associate with the mitochondrial antiviral signaling adaptor called MAVS (also referred to as IPS-1/VISA/cardif). MAVS are located at the cytosolic face of the outer mitochondrial membrane, and this mitochondrial association is necessary for initiating further signaling events. On association the MAVS undergo a prion-like aggregation. This leads to the recruitment of E3 ubiquitin ligases and the downstream effector proteins TNF receptor-associated factor 2 (TRAF2), TRAF3, and TRAF6 which assembles into an active “signalosome.” The resulting cascade leads to the phosphorylation and nuclear translocation of key innate immune transcription factors IRF3 and IRF7. Further, it leads to the activation of NF-κB to drive the expression of type I and III interferons, innate immune genes, pro-inflammatory cytokines, and chemokines that will aid in containing the virus infection.
The RD domain is not only critical for RNA sensing but also confers it with the displayed selectivity. RIG-I can recognize 5′ppp dsRNA, 5′pp dsRNA, pU/UC genomic RNA, AU-rich 3 UTR, RNase L cleavage products and circular viral RNA (Chow et al. 2018; Said et al. 2018; Dang et al. 2018; Liu and Gale Jr 2011; Yoneyama and Fujita 2007; Zou et al. 2009). The distinguishing between self and non-self RNA is based on the 5′-7-methylguanosine cap, which is subject to 2′-O-methylation.
4.1.2 MDA5
RIG-I and MDA5 being homologues showcase the same overall domain structures. The RD domain however recognizes long dsRNA which under normal circumstances is absent in an uninfected cell. The presence of this PAMP signifies vial invasion which activates MDA5 and stimulates a cytokine response similar to the RIG-I. MDA5 binds to the long dsRNA replicative intermediates generated by picornavirus, consisting of its positive-sense genome annealed to the negative-sense antigenome, AU-rich motifs and RNase L cleavage products (Said et al. 2018; Dang et al. 2018; Bruns et al. 2014; Rodriguez et al. 2014).
On binding with the target RNA it undergoes a conformational change and the 2 CARD domains are exposed. The 2 CARDS assemble into a stable tetramer due to its binding with the dsRNA and they form a filament structure. They therefore are independent on the polyubiquitin chain binding; the tetramer itself nucleates the MAVS, which in turn activates TRAF3/TBK1/IKKe/IRF3 and TRAF6/IKK/NF-jB, which drive IFN and pro-inflammatory cytokine expression, respectively.
4.1.3 LGP2
Although LGP2 is homologues to RIG-I and MDA5, it lacks the CARD domain which restricts it from binding to the MAVS. However, LGP2 is shown to bind to the termini of blunt-ended dsRNA of different lengths with high affinity, forming complexes with 2:1 stoichiometry. LGP2 has been both implicated as a positive and negative effector of RIG-I and MDA5 response (Said et al. 2018; Jensen and Thomsen 2012; Bruns et al. 2014; Rodriguez et al. 2014). Studies have shown that CTDs of LGP2 and RIG-I are analogous because of which LGP2 CTD interacts with RIG-I to abolish its ability to initiate antiviral signaling. RIG-I also undergoes attenuation of viral sensing. LGP2 has also been believed to utilize its ATP dependent/RNA helicase activity to increase the interaction of nucleic acid PAMPS for improved antiviral RLR signaling. LGP2 has also been implicated for its activity against DICER protein to maintain the cytosolic PAMPs in intact condition for detection by the cytosolic RNA sensors. Therefore, LGP2 overall is a modulator of the innate immune response to a viral infection and not a sensor of PAMPs. LGP2 does not initiate antiviral gene expression.
4.1.4 Other DEXD/H-Box Helicases
Certain helicases which are a member of the DEXD/H-box helicases family have been anticipated to function in the RLR-based RNA sensing. DDX3 is one such helicases which associates itself with MAVS, and when it encounters transfected dsRNA, it induces interferon production. DDX3 has also been suggested to aid as a scaffold for the assembly of an RLR signaling complex, promoting innate immunity signaling (Chow et al. 2018; Said et al. 2018; Jensen and Thomsen 2012). DDX3 is being studied further to understand the function and mechanism of action in RNA sensing.
Another DEXD/H-box Helicase DDX60 (Ski-2-like helicase) is implicated in the cytosolic antiviral response. DDX60 maintains the quality of host RNA by acting as an exosome complex to degrade other foreign RNAs. Exosome-mediated degradation also enables the degraded vRNA to be recognized by RIG-I/MDA5. DDX60 also functions as an ISG to suppress viral infection in an infected cell. vRNA and RIG-I/MDA5 interactions are brought together by DDX60 in order to enhance the type 1 interferon production. Thus, DDX60 functions on two levels augmenting the antiviral response by detecting the vRNA and subject them to the RIG-I/MDA5 based RLR pathway; further, they aid in revealing the molecular signature (PAMP) of the vRNA via the exosome-mediated degradation, making DDX60 a immune-stimulatory molecule.
SNRNP200 is another member of the Ski-2 RNA helicase family. It has been recently discovered for promoting vRNA sensing and IRAF3 activation by interacting with TBK-1, found in the RIG-I/MAVS signaling pathway. SNRP200 is a member of the spliceosome complex for removal of introns. During viral infection, the amino terminal of SNRP200 binds to the vRNA, perinuclear relocation occurs and it aids as an adaptor o trigger IRF3 signaling.
4.2 NLRs
Nucleotide-binding oligomerization domain-containing (NOD)-like receptors (NLR) are divided into five subfamilies (based on the N terminal effector domains): NLRA (acid activation domain), NLRB (baculovirus inhibitor of apoptosis repeats), NLRC (caspase activation and recruitment domain CARD), NLRP (pyrin domain PYD), and NLRX (unknown domain). NLR is another family of cytosolic RNA-sensing PRRs. All NLRs upon activation lead to inflammasome formation except NOD1 and NOD2. General structure of NLRs are as follows:
-
1.
N-terminal effector domain
-
2.
Followed by nucleotide-binding and oligomerization domain (NOD)
-
3.
C-terminal leucine-rich repeats (LRRs).
NLRC2 (NOD2) recognizes ssRNA viruses. Recognition leads to association with MAVS. This interaction is LRR-dependent and nucleotide binding domains. This triggers the MAVS-dependent pathway which leads to type I IFN and pro-inflammatory cytokine release.
NLRC5 is another NLR which has a distinguished longer LRR domain which thereby forms a helical structure instead of the horseshoe shape. Upon interaction with the viral DNA it induces IFN-mediated JAK-STAT (Janus kinase-signal transducer and activator of transcription) pathway. NLRC5 is an amplifier of the antiviral responses.
NLRP3 on the other hand has a pyrin domain in its N-terminal which enables it to interact with the PYD in the N terminal of apoptosis-associated speck-like protein containing a CARD (ASC). The CARD domain in the C-terminal of ASC further activates caspase-1, leading to the formation of fully functional IL-1 and IL-18. NLPR3 can be activated by lysosomal disintegration, membrane disruption, or generation of ROS caused due to viral PAMPS and DAMPS; therefore, it serves as an indirect sensor of viral invasion (Fig. 15.2).
4.3 RNA-Binding Proteins
Multiple RNA-binding proteins are also involved directly or indirectly in the process of vRNA sensing.
4.3.1 RNase L/Oligoadenylate Synthase (OAS)
RNase L is an antiviral endoribonuclease. Oligoadenylate synthetase (OAS) catalyzes the conversion of ATP into 2-5-linked oligoadenylates (2-5A) that in turn become the second messengers that bind to and activate RNase L (Chakrabarti et al. 2015; Malathi et al. 2007; Silverman 2007). vRNAs, cellular self-RNA are substrates to RNase L. The cleavage products are utilized for increasing the activation of RIG-I/MDA5. These are being studied in order to use them as targets for antiviral therapies (Sarkar 2014).
4.3.2 Protein Kinase R (PKR)
PKR functions by inhibiting translation of the viral infected cell (Dauber and Wolff 2009; Zhu et al. 2008). Being a serine/threonine kinase it gets activated on binding to a viral dsRNA/short dsRNA with 5′ppp ends and limited secondary structures (Dauber and Wolff 2009; Gil and Esteban 2000), ultimately inhibiting the eukaryotic translation initiation factor 2A (eIF2A). Antiviral response by PKR is also mediated by stabilizing MDA5.
4.3.3 Interferon-Inducible Transmembrane Protein (IFIT)
Multiple viruses induce the interferon response via both innate and adaptive immune responses. This leads to the stimulation of ISGs like IFIT which triggers antiviral cell-intrinsic restriction factors. Though the exact molecular mechanism is yet to be elucidated, they have showcased inhibition of viral translation or the sequestration of viral RNA to inhibit either virus replication or its packaging into new virions (Anafu et al. 2013; Diamond 2014; Vladimer et al. 2014). Furthermore, IFIT acts on the late endosomes blocking the endosomal viral entry. IFIT1 and IFIT5 recognize 5′ppp ssRNA. IFIT2 recognizes U-rich RNAs in vitro, independent of a 5′ppp.
Endosomal RNA sensors | Cytosolic RNA sensors | RNA-binding proteins | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
TLRs | Rlrs-SF2 family cytosolic helicases | NLRs | ||||||||
TLR3 | TLR7 | TLR8 | RIG-I | MDA5 | NLRC2 (NOD2) | NLRP3 | RNase L | PKR | IFIT | |
Cellular localization, distribution | Endosomes of immune cells except, non-immune cells like keratinocytes, fibroblasts, hepatocytes, astrocytes and microglia | Endosomes of monocytes, macrophages, plasmocystoid dendritic cells, NK cells, T and B cells | Endosomes of monocytes, macrophages, dendritic cells, NK cells, T and B cells also in mast cells and Tregs | Cytoplasm of all cell types | Cytoplasm of all cell types | Cytoplasm. Macrophages, monocytes, DCs | Cytoplasm Ubiquitously expressed | Nucleoplasm | Activated PKR is fund in both cytoplasm and nucleus | Mitochondria and cytoplasm |
Agonists | dsRNA, stable stem structures of ssRNA | GU or U rich single-stranded RNA (ssRNA) | GU or U rich single-stranded RNA (ssRNA) | 5′ppp-dsRNA, short dsRNA | Long dsRNA | Bacterial and virus RNA | Pathogen ssRNA/dsRNA and other distinct set of ligands | vRNAs, cellular self-RNA | Viral dsRNA, short dsRNA with 5′ppp ends and limited secondary structures | 5′ppp ssRNA, U-rich RNAs in vitro |
Mechanism of activation | dsRNA induced dimerization | Z-loop proteolytic cleavage, and receptor dimer conformational change | Z-loop proteolytic cleavage, and receptor dimer conformational change | Polyubiquitin chain binding mediated receptor tetramerization | Filament formation mediated receptor tetramerization | Tetramerisation | Potassium efflux-NEK7 involved NLRP3 inflammas-ome complex | Binding to the agonist, apoptosis ad interferon dependent pathways | Dimersiation on encountering the agonists leads to auto-phosphorylation | Interferon dependent pathways |
Cell signaling pathways | TRIF-TRAF3-TBK1/IKKe-IRF3 TRIF-TRAF6-IKKs-NF-κB | MyD88-IRAK4/IRAK1-IRF7. MyD88-IRAF6-IKKs-NF-κB | MyD88-IRAK4/IRAK1-IRF7. MyD88-IRAF6-IKKs-NF-κB | MAVS-TRAF3-TBK1/IKKe-IRF3. MAVS-FADD/TRAF6-IKKs-NF-κB | MAVS-TRAF3-TBK1/IKKe-IRF3. MAVS-FADD/TRAF6-IKKs-NF-κB | RIP2-IKKs-NF-κB | ASC-inflammasome caspase-1-IL-1/IL-18 | |||
Recognized pathogens | MCMV, HSV-1, EMCV, WNV and enteroviruses | SeV, flu virus, coxsackie virus, vaccinia virus, MV, RSV, retrovirus, SGB | SeV, flu virus, coxsackie virus, vaccinia virus, MV, RSV, retrovirus, E. coli, M. bovis, H. pylori, B. burgdorferi | BOV, MV, SeV, NDV, RSV, flu virus, hantavirus, VSV, RV, HCV, JEV, adenovirus, vaccinia virus, HSV, L. monocytogenes, H. pylori, S. flexneri. Rotavirus, dengue virus, WNV, murine hepatitis virus | EMCV, poliovirus and coxasackie virus. Rotavirus, dengue virus, WNV, murine hepatitis virus | RSV, IAV, and HCMV | Flu virus, SeV and bacteria | Influenza A, H5N1, Rotavirus | ||
Crosstalk | Positive regulation by TLR8 | Positive regulation by NOD2 | Positive regulation by TLR3, NOD2 Negative regulation by TLR7 | Negative regulation by TLR3, NOD2 | Negative regulation by TLR3 | Negative regulation by RIG-I | Positive regulation by MDA5 | Positive regulation of RIG-I/MDA5 | Positive regulation by interferon inducing mechanisms | |
References
Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH (2013) Interferon inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J Biol Chem 288(24):17261–17271
Bruns AM, Leser GP, Lamb RA, Horvath CM (2014) The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell 55(5):771–781
Chakrabarti A, Banerjee S, Franchi L, Loo Y-M, Gale M Jr, Núñez G et al (2015) RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17(4):466–477
Chen N, Xia P, Li S, Zhang T, Wang TT, Zhu J (2017) RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 69(5):297–304
Chow KT, Gale M Jr, Loo Y-M (2018) RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 36:667–694
Crozat K, Beutler B (2004) TLR7: a new sensor of viral infection. Proc Natl Acad Sci 101(18):6835–6836
Dang W, Xu L, Yin Y, Chen S, Wang W, Hakim MS et al (2018) IRF-1, RIG-I and MDA5 display potent antiviral activities against norovirus coordinately induced by different types of interferons. Antivir Res 155:48–59
Dauber B, Wolff T (2009) Activation of the antiviral kinase PKR and viral countermeasures. Viruses 1(3):523–544
Diamond MS (2014) IFIT1: a dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 25(5):543–550
Gantier MP, Williams BR (2011) Making sense of viral RNA sensing. Mol Ther 19(9):1578–1581
Gil J, Esteban M (2000) Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5(2):107–114
Hewson CA, Jardine A, Edwards MR, Laza-Stanca V, Johnston SL (2005) Toll-like receptor 3 is induced by and mediates antiviral activity against rhinovirus infection of human bronchial epithelial cells. J Virol 79(19):12273–12279
Jensen S, Thomsen AR (2012) Sensing of RNA viruses–a review on innate immune receptors involved in recognizing RNA virus invasion. J Virol 86(6):2900–2910
Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H et al (2002) Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 3(6):499
Liu HM, Gale M Jr (2011) ZAPS electrifies RIG-I signaling. Nat Immunol 12(1):11
Malathi K, Dong B, Gale M Jr, Silverman RH (2007) Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448(7155):816
Patel MC, Shirey KA, Pletneva LM, Boukhvalova MS, Garzino-Demo A, Vogel SN et al (2014) Novel drugs targeting Toll-like receptors for antiviral therapy. Futur Virol 9(9):811–829
Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F et al (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314(5801):997–1001
Rodriguez KR, Bruns AM, Horvath CM (2014) MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J Virol 88(15):8194–8200
Said EA, Tremblay N, Al-Balushi MS, Al-Jabri AA, Lamarre D (2018) Viruses seen by our cells: the role of viral RNA sensors. J Immunol Res 2018:9480497
Sarkar SN (2014) Could boosting the oligoadenylate synthetase-like pathway bring a new era of antiviral therapy? Futur Virol 9(12):1011–1014
Silverman RH (2007) Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81(23):12720–12729
Thomas A, Laxton C, Rodman J, Myangar N, Horscroft N, Parkinson T (2007) Investigating Toll-like receptor agonists for potential to treat hepatitis C virus infection. Antimicrob Agents Chemother 51(8):2969–2978
Vladimer GI, Górna MW, Superti-Furga G (2014) IFITs: emerging roles as key anti-viral proteins. Front Immunol 5:94
Yoneyama M, Fujita T (2007) Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem 282(21):15315–15318
Zhu R, Zhang Y-B, Zhang Q-Y, Gui J-F (2008) Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol 82(14):6889–6901
Zou J, Chang M, Nie P, Secombes CJ (2009) Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol 9(1):85
Acknowledgments
Authors are grateful to NIV Pune and NMIMS Pune for the support extended.
Conflict of Interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Nair, P., Sapre, S.U. (2020). Significance of RNA Sensors in Activating Immune System in Emerging Viral Diseases. In: Bramhachari, P. (eds) Dynamics of Immune Activation in Viral Diseases. Springer, Singapore. https://doi.org/10.1007/978-981-15-1045-8_15
Download citation
DOI: https://doi.org/10.1007/978-981-15-1045-8_15
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-1044-1
Online ISBN: 978-981-15-1045-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)