Abstract
AIM2 (absent in melanoma 2) is a cytoplasmic sensor of double-stranded DNA from pathogens or damaged cellular organelles. It recruits ASC (apoptosis-associated specklike protein containing a CARD) and caspase-1 to form the AIM2 inflammasome, activate caspase-1, and elicit inflammatory responses via cytokine maturation and pyroptotic cell death. Structural studies from X-ray crystallography, NMR, and cryo-EM have revealed many details in AIM2 inflammasome activation, assembly, and regulation. Many principles learned from AIM2 inflammasome also apply to other inflammasomes. In this chapter, we discuss the interactions between dsDNA and AIM2-like receptors, between AIM2 and adaptor protein ASC, and between ASC and caspase-1 with the focus on helical filament assembly formed by PYD and CARD domains.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
Cells face external and internal assaults every day. The first step for cells to initiate protective responses is to detect pathogen- or danger derived signals, the so-called PAMPs (pathogen-associated molecular patterns) or DAMPs (danger-associated molecular patterns). An arsenal of pattern recognition receptors senses the presence of PAMPs or DAMPs both on cell surface and in the cytoplasm. Nucleic acids are prominent PAMPs as they are ubiquitous in living organisms, their presence, especially the presence of double-stranded DNA in cytoplasm, signals pathogen invasion or cellular damage.
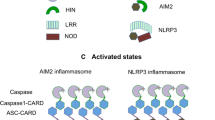
AIM2 (absent in melanoma 2) is the prototype and best characterized member of the AIM2-like receptors (ALRs). Upon dsDNA recognition, AIM2 recruits an adaptor protein ASC (apoptosis-associated specklike protein containing a CARD), which in turn activates caspase-1 to form the multicomponent AIM2 inflammasome [1]. AIM2 inflammasome activates caspase-1 to process cytokines into mature form. ALRs are characterized by the N-terminal PYD domain and one or two C-terminal HIN domains (hematopoietic, interferon-inducible, and nuclear localization) (Fig. 7.1). Mouse p202 is the only known ALR that does not contain an N-terminal PYD domain. PYD domain belongs to the death domain superfamily known for homotypic interactions via three types of interfaces [2]. The main function of HIN domain is to mediate interaction with DNA, but it also participates in homo- and hetero-oligomerization [3].
As the first structurally elucidated inflammasome [4], AIM2 inflammasome serves as a prototype for our understanding of inflammasome assembly and regulation in general.
7.2 HIN Domain and Its Interaction with DNA
A HIN domain consists of two OB (oligonucleotide/oligosaccharide binding) folds which are known to recognize nucleic acids, especially single-stranded nucleic acids [5]. Despite low sequence similarity, OB folds adopt similar three-dimensional structures: a central twisted β barrel composed of five antiparallel β strands capped by an α helix at one end. The loops between β strands are highly variable in terms of length and composition, often contributing to ligand specificity. The first HIN domain structure comes from human AIM2. In hAIM2HIN, the two OB folds are connected by a rigid α helix (Fig. 7.2a). HIN domain framework is highly conserved. All known HIN domain structures, free or in complex with DNA, superimpose well with each other with pairwise Cα RMSD ranging from 0.3 to 2.5 Å [3, 6, 7, 8, 9] (Fig. 7.2b). The most divergent regions are L12 and L45 loops that connect β strands in both OB folds. OB1 and OB2 function as one unit as well. In all known HIN: dsDNA structures, the DNA-binding surface is formed by both OB folds [3, 6, 7, 9] (Fig. 7.2c). The HIN domain contacts both strands in the dsDNA, explaining why dsDNA is preferred over ssDNA [10, 11]. The nature of the interaction is mostly electrostatic, but polar and hydrophobic interactions contribute as well (Fig. 7.2d, e). Positively charged patches formed by arginines and lysines in both OB folds make contact to the backbone phosphates and riboses on DNA molecules. No significant interaction with bases is observed. This interaction with backbone phosphates and riboses is consistent with the sequence-independent binding [6] and in stark contrast to ssDNA-binding OB folds where bases make intimate contacts with OB folds [12, 13]. The overall architecture of HIN domains is highly conserved, but their ways to engage DNA are astonishingly different. AIM2HIN domain binds to dsDNA using both OB folds and the connecting α helix [6] (Fig. 7.2c). IFI16HIN2 domain binds to dsDNA in a similar manner, with contributions from both OB folds and the α helix in between, but the overall surface is slightly tilted [6] (Fig. 7.2c). A similar situation has been observed for a different crystal form of hAIM2HIN: dsDNA complex and mouse AIM2HIN: dsDNA complex [6, 9], suggesting the DNA-binding surface is rather plastic, probably to better accommodate a variety of DNA species. In contrast, the p202 HIN1 (p202HIN1) use a completely opposite surface to bind dsDNA [9, 3] (Fig. 7.2c). In p202HIN1, the DNA-binding interface is formed by loops between β strands in both OB folds, especially the loop between β1 and β2 (L12) and β4 and β5 (L45). OB1 contacts the minor groove while L12 and L45 in OB2 clamp the major groove in the almost ideal B-form dsDNA. This binding mode is reminiscent of ssDNA recognition by OB folds in RPA or BRCA2 [12, 13]. The structural diversity of HIN domains goes further. The HIN2 domain of p202 (p202HIN2) completely sheds DNA-binding capacity, instead, it acquires the capability to tetramerize, forming an oligomeric core for p202HIN1 to append to, increasing DNA-binding affinity in full-length p202 through avidity [3]. The extensive and intimate tetramerization interface is formed by L12 and L45 loops, the equivalents of DNA-binding regions in p202HIN1 (Fig. 7.2f).
Structures of HIN domains. a Cartoon representation of human AIM2 HIN domain showing the tandem OB-fold structure; α helices are colored in red, β strands in yellow, and loops in green. OB1, OB2, and the connecting α-helical linker are labeled. b Superposition of HIN structures of human AIM2 (orange), human IFI16 HIN2 (yellow), mouse AIM2 (green), mouse p202 HIN1 (blue), and mouse p202 HIN2 (cyan). c Different dsDNA-binding surfaces of HIN domains. The superposed HIN domain structure is colored in gray. dsDNA in complex with human AIM2HIN, human IFI16HIN2, mouse AIM2HIN, and mouse p202HIN1 are colored in orange, yellow, green, and blue, respectively. d and e Surface charge distribution of human AIM2HIN (d) and mouse p202HIN1 (e) DNA-binding surfaces. The DNAs are shown as ribbons and colored in gray. f Tetramer structure of mouse p202 HIN shown as a transparent surface. One subunit is also shown as a cartoon representation and colored in cyan. Superposition of p202 HIN1 and HIN2 places dsDNA at HIN2 dimerization interface. HIN domains in (a), (b), (c), and (f) are in the same orientation
7.3 AIM2 PYD Structures and Filament Formation
All ALRs contain one PYD domain at their N-termini except p202 (Fig. 7.1). PYD belongs to the DD (death domain) superfamily that also includes DD, DED (death effector domain), and CARD (caspase recruitment domain) families [2]. Death domains are widely found in proteins mediating inflammation and cell death. Despite low sequence similarity, death domain superfamily is characterized by a six-helix bundle conformation. Both the helices and connecting loops contribute to homotypic interactions. The interfaces can be roughly classified as type I, II, and III surfaces [14, 2]. As another chapter in this book is going to examine death domain interactions in detail, this chapter will focus on general assembly features. The high tendency to self-association makes PYD domains difficulty to study. So far, the monomeric structures obtained for AIM2 PYD domain (AIM2PYD) are through fusion with MBP (maltose-binding protein) [15], surface engineering [16], or in low pH [17]. The long loop connecting α2 and α3 is characteristic of PYD family proteins [18]. α2-α3 loop, together with its neighboring helical regions, is also highly flexible (Fig. 7.3a), indicating structural plasticity that may be important for homotypic interactions. Indeed, a single mutation on α2 abolishes AIM2PYD self-association [16]. The C-terminus of α6 also displays structural divergence. NMR and molecular dynamics simulations suggest AIM2PYD adopts multiple conformations in solution, and the distribution of conformations may shift upon AIM2 oligomerization [19].
AIM2PYD structures. a Superposition of human AIM2PYD fused with MBP (blue), human AIM2PYD with F27G mutation (pink), and mouse AIM2PYD (yellow). For clarity, the MBP portion is not shown. N- and C-termini and the six α helices are labeled. Red arrow: α2-α3 loop. b Side and top views of hAIM2PYD filament structure. In red frame is one subunit in the filament shown in the same orientation as in (a). The vertical arrow at left denotes helical axis
Recent developments in cryo-electron microscopy enabled high-resolution structural studies of self-associated AIM2PYD. AIM2PYD spontaneously assembles into helical filaments in solution [4, 20]. Reconstruction of GFP-AIM2PYD filament revealed a right-handed one-start helical tube with AIM2PYD forming the core and GFP moieties packing outside. Reconstruction of the inner AIM2PYD core to 5.0 Å yielded a hollow tube with the inner diameter of ~20 Å and an outer diameter of ~90 Å (Fig. 7.3b). The rotation and axial rise between adjacent subunits are 138.9° and 6.0 Å, respectively. In comparison to AIM2PYD crystal structures, AIM2PYD in filament shows little conformational change except for the α2-α3 loop [21].
It is not clear how dsDNA: AIM2HIN interaction leads to AIM2PYD helical filament formation. One hypothesis is that AIM2 exists in an autoinhibitory state with intramolecular interactions between PYD and HIN domains. Binding of dsDNA to HIN domain releases PYD domain from autoinhibition to interact with ASC [6]. A second, non-exclusive hypothesis notes that AIM2 is capable of self-association when protein concentration reaches a certain threshold, even in the absence of dsDNA [20]. It is plausible that regardless of free AIM2 conformation, its concentration in DNA free environment is below the self-association threshold. Presence of dsDNA from microbial infection or cell damage reduces the threshold to induce efficient AIM2 self-association and AIM2 inflammasome assembly. A recent publication is in agreement with such a unifying hypothesis [22].
7.4 ASC PYD Structures in Isolation and Filaments
ASC is the common adaptor protein of several inflammasomes including AIM2 and NLRP3 (Fig. 7.1). Formation of micron-sized ASC “speck” is a hallmark of inflammasome assembly in cells [23]. With its bipartite domain organization, ASC bridges upstream sensors to downstream effector caspases via homotypic PYD-PYD and CARD-CARD interactions. Like AIM2PYD and many other death domain superfamily members, ASCPYD and ASCCARD are also prone to self-association. Human ASCPYD (hASCPYD) and full-length ASC (hASC) structures have been determined by NMR at low pH that presumably prefers and stabilizes monomers [24, 25]. A recent study determined the crystal structure of zebrafish ASCPYD (zASCPYD) with an N-terminal MBP fusion tag [26]. Both hASCPYD and zASCPYD conform to the canonical six-helix bundle structure of DD family (Fig. 7.4a). Under more physiological conditions, ASCPYD spontaneously forms long filaments, a process that is greatly accelerated by oligomerized AIM2 or NLRP3 [4]. Cryo-EM reconstruction of human ASCPYD filament at 3.8 Å yielded a distinct right-handed three start helical assembly with C3 point group symmetry. Like hAIM2PYD filament, hASCPYD filament is hollow with outer and inner diameters of ~90 and ~20 Å, respectively (Fig. 7.4b). ASCPYD filaments bear a rotation of ~52.9° and an axial rise of 13.9 Å per subunit along each of the three-start strands. The 4–4.5 Å cryo-EM reconstruction of mouse ASCPYD displayed very similar helical assembly. Mouse ASCPYD filament is right-handed, of C3 symmetry, and with 53° rotation and 14.2 Å axial rise per subunit [27]. Monomeric ASCPYD structure is largely maintained in filaments, suggesting the PYD domain undergoes minimal conformational change when incorporated into the filament. Human and mouse ASCPYD structures in filaments superpose well with each other with a 1.1 Å backbone RMSD. Interestingly, although hASCPYD and zASCPYD monomer structures are well conserved, the interfaces used for filament assembly are overlapping but notably different [26, 4], exemplifying functional adaptability as well as conservation.
ASCPYD structures. a Superposition of human ASCPYD NMR structure (teal) and zebrafish AIM2PYD fused with MBP (pink). For clarity, the MBP portion is not shown. N- and C-termini and the six α helices are labeled. b Side and top views of hASCPYD filament structure. In red frame is one subunit in the filament shown in the same orientation as in (a). C-termini of three subunits in the hASCPYD filaments are labeled to show that they point outwards. The vertical arrow at left denotes helical axis
The close resemblance between AIM2PYD and ASCPYD filaments may be the structural basis of AIM2-induced ASC speck formation as one would assume AIM2 filaments may seamlessly morph into ASC filaments. Experimental data support such a nucleation model. When AIM2PYD and ASCPYD are co-expressed, AIM2PYD is found only at the ends of ASCPYD filaments [4]. However, subunit packing is ~30% denser in ASCPYD filaments than in GFP-AIM2PYD filaments, probably due to steric hindrance introduced by GFP moiety. Nevertheless, GFP-AIM2PYD is still capable of nucleating the filament formation of ASCPYD, albeit to a lesser degree than tag free AIM2PYD, suggesting the intrinsic plasticity and structural tolerance in helical filament assembly [21].
7.5 ASC CARD Structures in Isolation and Filaments
The ASC PYD and CARD domains (ASCPYD and ASCCARD) are linked together by a ~20 amino acid linker. In ASC solution structure the linker adopts an extended conformation with no apparent interactions between ASCPYD and ASCCARD, suggesting each domain may freely self-associate without interference from the other (Fig. 7.5a). In both human and mouse ASCPYD filaments, the C-termini of ASCPYD subunits points outward, leaving ample space to accommodate ASCCARD and potentially other downstream molecules such as caspase-1 (Fig. 7.4b). The six-helix bundle fold is conserved in both human ASCCARD (hASCCARD) and zebrafish ASCCARD (zASCCARD) structures. The two CARD structures superpose with a 3.07 Å RMSD value despite low sequence identity (Fig. 7.5b). The hASCCARD filament refined at 3.2 Å is slightly slimmer than hASCPYD filament. The inner and outer diameters are ~10 and ~80 Å, respectively (Fig. 7.5c). Helical assembly and subunit packing, on the other hand, are very distinct. hASCCARD filaments adopt a left-handed one-start helical symmetry, with −100.6° rotation and 5.0 Å axial rise for each subunit [28]. ASCCARD in isolation and in filament superpose well with each other with an RMSD of 2.3 Å.
ASCCARD and Casp1CARD structures. a Human full-length ASC structure with PYD domain colored in teal and CARD domain colored in red. The flexible linker is in gray. N- and C-termini and the six α helices of ASCCARD are labeled. b Superposition of human ASCCARD NMR structure (red), zebrafish ASCCARD fused with MBP (pink), and human ASCCARD in complex with VHHASC (yellow). For clarity, the MBP and VHH are not shown. c and d Side and top views of hASCCARD (c) and hCasp1CARD (d) filament structures. In red frame is one subunit in the filament shown in the same orientation as in (b). The vertical arrows at left denote helical axis
In theory, full-length ASC may form filaments through its PYD or CARD domains. In cells, it seems the core of ASC speck is made up of ASCPYD while ASCCARD crosslinking ASCPYD filaments into a speck [29]. This finding is corroborated by studies on zASC that shows zASCPYD forms the core of filaments in vitro [26].
7.6 Caspase-1 CARD Helical Assembly
Caspase-1 is recruited to AIM2 inflammasome via CARD: CARD homotypic interaction with ASC. It may be recruited to other CARD containing inflammasomes such as NLRC4 inflammasome. Reconstituted AIM2PYD: ASC: GFP-Casp1CARD ternary complex displays a star shape with ASC residing in the center and Casp1CARD detected all along the arms [4]. Like ASCPYD filament formation is nucleated by AIM2PYD, Casp1CARD forms helical assembly with nucleation provided by ASCCARD or NLRC4CARD [30]. Cryo-EM reconstitution of Casp1CARD filament at 4.8 Å showed a hollow tube with an outer diameter of ~80 Å and an inner diameter of <20 Å [30]. It assumes left-handed, one-start symmetry with −100.2° rotation and 5.1 Å axial rise per subunit (Fig. 7.5d), strikingly similar to that of ASCCARD and NLRC4CARD filaments [28, 31]. When a single layer of Casp1CARD is examined as an entity, its “top” and “bottom” charge distribution patterns are complementary to each other, again very similar to those of ASCCARD and NLRC4CARD filaments, indicating a recruitment mechanism of caspase-1 to elongating ASCCARD and NLRC4CARD filaments.
7.7 Regulators of AIM2 Inflammasome
7.7.1 HIN Regulators
p202 is a mouse ALR protein with tandem HIN domains but no PYD (Fig. 7.1). While p202HIN1 binds to dsDNA using the opposite surface when compared to AIM2HIN (Fig. 7.2c), p202HIN2 mediates tetramerization but does not bind dsDNA at all [3] (Fig. 7.2f). In addition to competing with AIM2 for dsDNA, p202 specifically interacts with AIM2 and disrupts its clustering on dsDNA [3]. p202HIN2 binds to AIM2HIN in a 4:2 stoichiometry, hinting a dimer interface is required for AIM2HIN interaction. Regulation of AIM2 inflammasome activation by p202 was once believed to be unique in mouse, as human genome does not contain HIN domain-only proteins, but a recent study identified a HIN domain-only IFI16 transcript form in human cells, suggesting such regulation mechanism may be more prevalent than initially thought [32].
7.7.2 DD-only Regulators
The human genome encodes several PYD-only proteins (POPs) and CARD-only proteins (COPs) that interfere with inflammasome assembly and activation [33]. Structural information exists for human POP1 (also known as ASC2) [18] and ICEBERG, a CARD-only protein [34]. POP1 and ICEBERG both assume the canonical six-helix bundle structure, POP1 even features the characteristic loop between α2 and α3 of PYD family (Fig. 7.6a, b). POP3 interacts with AIM2PYD and IFI16PYD while POP1 and POP2 interact with ASCPYD [35]. On the COP side, INCA and ICEBERG have both been shown to disrupt inflammasome activation by engaging Casp1CARD, but the mechanisms are distinct. While ICEBERG is filamentous, INCA is monomeric in solution. INCA does not associate with ASCCARD but preferentially caps Casp1CARD filaments, thus terminating caspase-1 polymerization and activation [30]. It is postulated that two surfaces critical for Casp1CARD filament formation are defective in INCA, therefore INCA may interact with Casp1CARD filament but is incapable of elongation. ICEBERG does not interact with ASCCARD either, but it can be incorporated into Casp1CARD filaments [30], potentially reducing caspase-1 catalytic domain dimerization. High-resolution interactions between POPs or COPs with their counterparts in inflammasome are yet to be structurally captured. Viruses also encode POPs and COPs but their structural and mechanistic information is even less known.
7.7.3 Single-Chain Antibody Regulator
Aside from naturally occurring cellular and viral POPs and COPs, antibodies may be engineered to interfere with inflammasome assembly and activation. Indeed, a single-chain antibody generated from alpaca (VHHASC) has been shown to selectively interact with ASCCARD and blocks its polymerization [36]. VHHASC binds to a composite surface formed by α6 and α1 of ASCCARD and causes steric hindrance to preclude filament elongation (Fig. 7.6c).
7.8 Conclusions
Polymerization is prevalent in AIM2 inflammasome assembly, whether it requires DNA as a platform or stems solely from an intrinsic property of PYD or CARD domains. In AIM2 inflammasome assembly polymerization takes the form of helical assemblies. Upstream proteins often “nucleate” unidirectional polymerization of downstream proteins via homotypic interactions between PYD or CARD domains. Structurally, downstream helical assemblies often follow the symmetry, subunit packing, and charge distribution patterns of their upstream nucleating filaments, but certain degree of deviation is allowed. The nucleated helical assembly mechanism is found in RIG-I-MAVS-mediated intracellular double-stranded RNA sensing as well as in other inflammasomes [30, 21, 4, 37], but the helical assembly symmetry may not always pass from upstream to downstream proteins [28, 31]. Helical filament formation provides an effective way to mobilize numerous proteins in a short period of time for rapid and robust activation of signaling, for signal amplification, and for reduction of stochastic activation.
References
Broz P, Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16(7):407–420
Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H (2007) The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol 25:561–586
Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, Walz T, Stacey KJ, Wu H (2013) Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep 4(2):327–339
Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH (2014) Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156(6):1193–1206
Theobald DL, Mitton-Fry RM, Wuttke DS (2003) Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32:115–133
Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS (2012) Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36(4):561–571
Li H, Wang J, Wang J, Cao LS, Wang ZX, Wu JW (2014) Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr F Struct Biol Commun 70(Pt 1):21–29
Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, Xiao T, Tempel W, Zuo X, Wang YX, Chirgadze NY, Arrowsmith CH (2011) Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure 19(3):418–429
Ru H, Ni X, Zhao L, Crowley C, Ding W, Hung LW, Shaw N, Cheng G, Liu ZJ (2013) Structural basis for termination of AIM2-mediated signaling by p202. Cell Res
Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G (2009) An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10(3):266–272
Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ (2009) HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323(5917):1057–1060
Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385(6612):176–181
Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297(5588):1837–1848
Ferrao R, Wu H (2012) Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol 22(2):241–247
Jin T, Perry A, Smith P, Jiang J, Xiao TS (2013) Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem 288(19):13225–13235
Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H (2014) Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol 426(7):1420–1427
Hou X, Niu X (2015) The NMR solution structure of AIM2 PYD domain from Mus musculus reveals a distinct alpha2-alpha3 helix conformation from its human homologues. Biochem Biophys Res Commun 461(2):396–400
Natarajan A, Ghose R, Hill JM (2006) Structure and dynamics of ASC2, a pyrin domain-only protein that regulates inflammatory signaling. J Biol Chem 281(42):31863–31875
Wang H, Yang L, Niu X (2016) Conformation switching of AIM2 PYD domain revealed by NMR relaxation and MD simulation. Biochem Biophys Res Commun 473(2):636–641
Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J (2015) Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun 6:7827
Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, Wu H (2015) Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov 1
Matyszewski M, Morrone SR, Sohn J (2018) Digital signaling network drives the assembly of the AIM2-ASC inflammasome. Proc Natl Acad Sci USA 115(9):E1963–E1972
Man SM, Kanneganti TD (2015) Regulation of inflammasome activation. Immunol Rev 265(1):6–21
de Alba E (2009) Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J Biol Chem 284(47):32932–32941
Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G (2003) The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol 332(5):1155–1163
Li Y, Huang Y, Cao X, Yin X, Jin X, Liu S, Jiang J, Jiang W, Xiao TS, Zhou R, Cai G, Hu B, Jin T (2018) Functional and structural characterization of zebrafish ASC. FEBS J 285(14):2691–2707
Sborgi L, Ravotti F, Dandey VP, Dick MS, Mazur A, Reckel S, Chami M, Scherer S, Huber M, Bockmann A, Egelman EH, Stahlberg H, Broz P, Meier BH, Hiller S (2015) Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc Natl Acad Sci USA 112(43):13237–13242
Li Y, Fu TM, Lu A, Witt K, Ruan J, Shen C, Wu H (2018) Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc Natl Acad Sci USA 115(43):10845–10852
Dick MS, Sborgi L, Ruhl S, Hiller S, Broz P (2016) ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat Commun 7:11929
Lu A, Li Y, Schmidt FI, Yin Q, Chen S, Fu TM, Tong AB, Ploegh HL, Mao Y, Wu H (2016) Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol 23(5):416–425
Matyszewski M, Zheng W, Lueck J, Antiochos B, Egelman EH, Sohn J (2018) Cryo-EM structure of the NLRC4(CARD) filament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly. J Biol Chem 293(52):20240–20248
Wang PH, Ye ZW, Deng JJ, Siu KL, Gao WW, Chaudhary V, Cheng Y, Fung SY, Yuen KS, Ho TH, Chan CP, Zhang Y, Kok KH, Yang W, Chan CP, Jin DY (2018) Inhibition of AIM2 inflammasome activation by a novel transcript isoform of IFI16. EMBO Rep 19(10)
Dorfleutner A, Chu L, Stehlik C (2015) Inhibiting the inflammasome: one domain at a time. Immunol Rev 265(1):205–216
Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM (2000) ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell 103(1):99–111
Indramohan M, Stehlik C, Dorfleutner A (2018) COPs and POPs patrol inflammasome activation. J Mol Biol 430(2):153–173
Schmidt FI, Lu A, Chen JW, Ruan J, Tang C, Wu H, Ploegh HL (2016) A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J Exp Med 213(5):771–790
Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S (2014) Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell 55(4):511–523
Acknowledgements
This work was supported by the US National Institutes of Health grant R00AI108793 and start-up funds from Florida State University.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Wang, B., Tian, Y., Yin, Q. (2019). AIM2 Inflammasome Assembly and Signaling. In: Jin, T., Yin, Q. (eds) Structural Immunology. Advances in Experimental Medicine and Biology, vol 1172. Springer, Singapore. https://doi.org/10.1007/978-981-13-9367-9_7
Download citation
DOI: https://doi.org/10.1007/978-981-13-9367-9_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-9366-2
Online ISBN: 978-981-13-9367-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)