Abstract
Segmentation of anatomical structures from chest x ray has an increasing importance in the past four decades and researchers have proposed various techniques and evaluated them using different datasets. In order to evaluate and compare a proposed technique, it is necessary to have knowledge about public datasets available. In this survey, properties and characteristics of different public chest x ray datasets available for segmentation of anatomical structures are studied. Different approaches for segmentation of anatomical structures (lung, heart, clavicles) are summarized. Segmentation techniques for each anatomical structure for a given dataset are compared and analyzed. The paper outlines the issues where further research can be focused.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
With the discovery of x ray [15] in 1895, there is a revolution in the field of diagnostics. With the invention of the modern digital computer in late 1940s, attempts were made to make computers perform tasks which need human intelligence for the completion. In 1960s researchers published articles about radiology report analysis using computer [8]. In 1970s, focus was upon the detection of abnormalities in chest x-ray using a computer.
The traditional chest analysis is the most prevalent radiological procedure, making up a minimum of a third of all exams in a typical radiology division. Moreover, Pulmonary diseases like pneumonia, tuberculosis [20, 21], emphysema and lung cancer can be screened based on the chest radiograph [26]. But, computerized interpretation of a chest radiograph is extremely challenging due to presence of superimposed anatomical structures. The complexity of computerized analysis of chest x-ray along with their prevalence in radiology department is the main reason for the researchers to concentrate on the development of computer algorithms to assist radiologists in reading chest images.
Researchers have developed a variety of algorithms for computer aided analysis of medical images (X-ray, computed tomography, for instance) [17]. Segmentation of organs (like lung, heart, clavicles) has been regarded as one of most important problems in computer aided diagnostics applications [18, 19]. Higher the accuracy in segmentation of the anatomical structures, higher is the accuracy in classification and detection of diseases like cardiomegaly, pneumonia and other lung related diseases.
One of the major problems faced by the researchers was the lack of public chest x-ray datasets which can act as benchmark for the comparison of performance of different techniques proposed. Performance of an algorithm was evaluated on customized x-ray data sets for about three decades from 1970s to late 1990s. In 2000, a public dataset [25] from JSRT was made available to researchers. A few more public datasets were made available which can act as benchmark for the evaluation of proposed algorithms.
Although, in recent years, a few more public datasets [7, 9, 12, 27, 29] of chest x-ray are dedicated, the information about the recent datasets is not available in any of the existing surveys according to our knowledge. Authors in [14] have focused on different segmentation techniques on chest x-ray datasets but the recent techniques are not included. Therefore, the focus of this survey is on the public datasets suited for segmentation of anatomical structures from chest x-rays. The use of publicly available datasets for evaluation of a given approach has two main advantages. First advantage is that the time and resources can be saved as new chest x-ray data set need not be obtained and researchers can spend their efforts on development of their algorithms and implementations. Second advantage is the use of common datasets enables comparison of performance of different approaches proposed for a given task [4].
The scope of the survey is public chest x-ray datasets for segmentation of anatomical structures. All the techniques that are evaluated using a specific dataset are compared in terms of corresponding performance metrics. Section 2 gives description about three public datasets available for segmentation of anatomical structures. Section 3 gives details about commonly used performance metrics for segmentation of anatomical structures. Section 4 compares different techniques based on the common data set used for evaluation. Section 5 concludes the paper by outlining some of the observations which are helpful for future work.
2 Public Datasets of Chest X-Ray for Segmentation of Anatomical Structures
The following are the public datasets available for segmentation of anatomical structures (lung, heart and clavicles).
-
JSRT/SCR for lung segmentation, heart segmentation and clavicle segmentation [27]
-
MC dataset for lung field segmentation [12]
-
CRASS dataset for lung field segmentation [9]
Some datasets like Montgomery County (MC) can be use for multiple purposes. It can be used for lung field segmentation and tuberculosis screening.
2.1 SCR Dataset
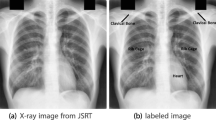
JSRT in cooperation with Japanese Radiological Society has developed a Chest X ray image database of 247 chest radiographs with and without nodule. The images are collected from thirteen distinct institutions in Japan and 1 in the USA in 1988 and made it as a public dataset [25]. Out of 247 images, 154 CXR images have lung nodules, while 93 are actually normal with no nodules. JSRT is the only public dataset available for lung nodule detection (Figs. 1 and 2).
ISI, University Medical Centre Utrecht, The Netherlands has established SCR dataset [27] in order to promote comparision of techniques proposed for segmentation of lung regions, the heart and the clavicles [27]. For each image from JSRT dataset, the borders of both lungs, the heart, and both the clavicles were stored in files with .pfs extension. Individual anatomic structures are stored with .gif extension [27]. SCR dataset is the most common dataset used in studies related to segmentation of anatomic structures (lungs, heart, clavicles) in a CXR as shown in Table 2. Sample masks are shown in the Fig. 3.
2.2 CRASS Dataset
CRASS dataset was collected from African region where tuberculosis is prevalent. It contain a set of 548 PA chest radiographs acquired from adults of age greater than 15 years. Out of 548 images, 333 are abnormal and 225 are normal. Among 333 abnormal images, 220 are abnormal at upper lung area near the clavicle. Among 548 images, 299 are marked as training set and the remaining 249 images are considered as test set. The main purpose of CRASS dataset is to form a benchmark for clavicle segmentation.
Researchers have proposed different techniques for clavicle segmentation and evaluated on CRASS dataset as shown in the Table 5. Human observers performed better than all other techniques [9]. Better techniques for clavicle segmentation need to be developed.
2.3 Montgomery County Dataset
U.S. National Library of Medicine (USNLM) and the Department of Health and Human Services, MC, MD, USA has collected Montgomery County (MC) dataset. There are 138 PA CXRs in this dataset which are collected under TB control programme. 80 CXRs are considered to be normal and 58 are abnormal with manifestations of TB [12].
All images are deidentified and are available in DICOM format. The spatial resolution of the CXR images is either 4020 by 4892 or 4892 by 4020 pixels. All image file names follow the same pattern: MCUC followed by four digit unique identifier. For each CXR, corresponding clinical readings are stored in a file with .txt extension. Clinical reading comprises of age, gender and lung abnormality. For example, a clinical reading of a CXR in the MC appears in the following form: Patient’s Sex: M Patient’s Age: 031Y Cavitary nodular infiltrate in RUL; active TB.
Manual segmentation on images of MC dataset was performed under the supervision of a radiologist and binary lung masks were generated. Mask images for left and right lungs are stored separately with .png extension and are included in seperate folders in the dataset [12]. Montgomery dataset was primarily made available for tuberculosis screening but it is useful for segmentation of lung fields. Table 6 gives different techniques and their performance when MC dataset is used. Lower order region growing technique [5] achieved higher accuracy \( 96.6 \pm 1.8\) when compared to other techniques. Segmentation techniques should be evaluated on multiple datasets (SCR and MC) to achieve better insight about their performance.
3 Performance Metrics for Segmentation of Anatomical Structures
There are different ways to measure the performance of Segmentation technique but the final decision whether the segmentation is sufficiently accurate or not is determined by the requirements of the target application. In general, the problem of segmentation is considered as a relation between lung and background. Most of the research papers consider classical accuracy, sensitivity, and specificity as performance metrics (Table 1).
\(N_{TP}\) denotes the true positive portion and it is equivalent to the portion of image identified correctly as lung region, \( N_{TN}\) denotes the true negative portion of the image which is equivalent to the portion of image correctly identified as background region, \(N_{FP}\)denotes the false positive portion and it is equivalent to the part of the image incorrectly classified as lung region, and \(N_{FN}\) is the false negative fraction which is same as the part of the image incorrectly classified as background region.
The Jaccard similarity coefficient is the overlap measure. It is the measured as the coincidence between the ground truth (GT) and the estimated segmentation mask (S) over all pixels in the image.
where TP (true positives) is the count of pixels which are classified correctly, FP (false positives) is the number of pixels which are identified as part of the object but they belong to background in reality, and FN (false negatives) are the pixels which are identified as background but are in actually part of the object.
Dice coefficient is the metric to measure intersection between the GT and S as given below.
Average contour distance (ACD) is the average distance between the segmentation boundary S and the ground truth boundary GT [3].
4 Comparitive Study of Segmentation Techniques for Each Dataset
4.1 Comparision of Performance of Lung Field Segmentation Techniques on JSRT SCR Dataset
SCR dataset was used to evaluate the performance of different lung segmentation techniques as shown in Table 2. Highest accuracy is \(96.3\pm 1.2\) when lower order adaptive region growing technique [5] is used. Human observer accuracy is calculated as \(94.6\pm 1.8\) and more than half of the segmentation techniques generated an accuracy more than human observer. Accuracy could be improved further and execution time could be decreased.
4.2 Comparision of Performance of Heart Segmentation Techniques on JSRT SCR Dataset
Segmentation of heart from a given chest x-ray is a challenging task as it is difficult to extract the heart region exactly. In spite of the complexity, various techniques were proposed and evaluated on JSRT SCR dataset. Most of them have low accuracy when compared to human observer as shown in Table 3. Highest accuracy \( 89.9\pm 4.4\) was achieved by using Fully Convolutional Networks [28].
4.3 Comparision of Performance of Clavicle Segmentation Techniques on JSRT SCR Dataset
Clavicle segmentation is the most challenging task as it is very difficult to seperate the clavicles from a given chest x-ray. Even though automated techniques were proposed, none of them performed better than human observer as shown in Table 4. Maximum accuracy achieved was \(89.6\pm 3.7\) by the human observer.
4.4 Comparision of Performance of Clavicle Segmentation Techniques on CRASS Dataset
Clavicle segmentation is quite challenging but researchers have addressed the problem by adopting pixel classification based methods, HDAP, Fully Convolution Networks and Active Shape Model. None of the techniques have resulted in better accuracy than human observer as shown in Table 5.
4.5 Comparision of Performance of Lung Field Segmentation Techniques on Montgomery County Dataset
Only a few segmentation techniques are evaluated using Montgomery County Dataset [3, 5, 6]. Lower order region growing approach has reported high accuracy of \( 96.6 \pm 1.8 \) as shown in Table 6. SCAN Technique has recorded an accuracy of \(91.4\pm 0.61\) with MC data set against \(94.7\pm 0.4\) using JSRT SCR dataset.
5 Conclusion and Future Scope
Lung field segmentation has attracted attention from most of the researchers and some of the techniques have attained an accuracy more than the accuracy of human observer. Segmentation of other anatomical structures heart and clavicles was not focused much during the last four decades. The accuracies reported in the automatic segmentation of heart and clavicles were not encouraging due to the reason that medical applications demand an accuracy more than the accuracy of human observer.
Another observation results from the fact that most of the researchers have used JSRT SCR dataset alone for the evaluation of the performance of the technique proposed. It is advisable to evaluate the performance of the proposed technique using all the available datasets to have a better insight.
Eventhough CRASS and JSRT datasets are available for clavicle segmentation, segmentation of clavicle remains as a challenging task. Better techniques should be proposed to increase the accuracy of clavicle segmentation.
As massive datasets of chest x-rays are available, deep learning techniques could play a major role in automatic multiple disease detection.
Paediatric chest x-ray datasets are needed to analyze and process the chest diseases related to children. Hence more paediatric pubic datasets are needed for evaluation of segmentation and disease detection techniques.
References
Bi, L., Kim, J., Kumar, A., Fulham, M., Feng, D.: Stacked fully convolutional networks with multi-channel learning: application to medical image segmentation. Vis. Comput. 33(6–8), 1061–1071 (2017)
Candemir, S., Jaeger, S., Palaniappan, K., Antani, S., Thoma, G.: Graph-cut based automatic lung boundary detection in chest radiographs. In: IEEE Healthcare Technology Conference: Translational Engineering in Health and Medicine, pp. 31–34 (2012)
Candemir, S., et al.: Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 33(2), 577–590 (2014)
Chaquet, J.M., Carmona, E.J., Fernández-Caballero, A.: A survey of video datasets for human action and activity recognition. Comput. Vis. Image Underst. 117(6), 633–659 (2013)
Chondro, P., Yao, C.Y., Ruan, S.J., Chien, L.C.: Low order adaptive region growing for lung segmentation on plain chest radiographs. Neurocomputing 275, 1002–1011 (2018)
Dai, W., et al.: Scan: structure correcting adversarial network for organ segmentation in chest x-rays. arXiv preprint arXiv:1703.08770 (2017)
Demner-Fushman, D., et al.: Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 23(2), 304–310 (2015)
Giger, M.L., Chan, H.P., Boone, J.: Anniversary paper: history and status of cad and quantitative image analysis: the role of medical physics and AAPM. Med. Phys. 35(12), 5799–5820 (2008)
Hogeweg, L., Sánchez, C.I., de Jong, P.A., Maduskar, P., van Ginneken, B.: Clavicle segmentation in chest radiographs. Med. Image Anal. 16(8), 1490–1502 (2012)
Ibragimov, B., Likar, B., Pernuš, F., Vrtovec, T.: Accurate landmark-based segmentation by incorporating landmark misdetections. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 1072–1075. IEEE (2016)
Ibragimov, B., Likar, B., Pernus, F., et al.: A game-theoretic framework for landmark-based image segmentation. IEEE Trans. Med. Imaging 31(9), 1761–1776 (2012)
Jaeger, S., Candemir, S., Antani, S., Wáng, Y.X.J., Lu, P.X., Thoma, G.: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6), 475 (2014)
Li, X., Luo, S., Hu, Q., Li, J., Wang, D., Chiong, F.: Automatic lung field segmentation in x-ray radiographs using statistical shape and appearance models. J. Med. Imaging Health Inform. 6(2), 338–348 (2016)
Mittal, A., Hooda, R., Sofat, S.: Lung field segmentation in chest radiographs: a historical review, current status, and expectations from deep learning. IET Image Process. 11(11), 937–952 (2017)
Mould, R.F.: A Century of X-rays and Radioactivity in Medicine: with Emphasis on Photographic Records of the Early Years. CRC Press, Boca Raton (1993)
Novikov, A.A., Lenis, D., Major, D., et al.: Fully convolutional architectures for multi-class segmentation in chest radiographs. IEEE Trans. Med. Imaging 37(8), 1865–1876 (2018)
Ruikar, D.D., Hegadi, R.S., Santosh, K.C.: A systematic review on orthopedic simulators for psycho-motor skill and surgical procedure training. J. Med. Syst. 42(9), 168 (2018)
Ruikar, D.D., Santosh, K.C., Hegadi, R.S.: Automated fractured bone segmentation and labeling from CT images. J. Med. Syst. 43(3), 60 (2019). https://doi.org/10.1007/s10916-019-1176-x
Ruikar, D.D., Santosh, K.C., Hegadi, R.S.: Segmentation and analysis of CT images for bone fracture detection and labeling (chap. 7). In: Medical imaging: Artificial Intelligence, Image Recognition, and Machine Learning Techniques. CRC Press, Boca Raton (2019). ISBN 9780367139612
Santosh, K., Antani, S.: Automated chest x-ray screening: can lung region symmetry help detect pulmonary abnormalities? IEEE Trans. Med. Imaging 37(5), 1168–1177 (2018)
Santosh, K., Vajda, S., Antani, S., Thoma, G.R.: Edge map analysis in chest x-rays for automatic pulmonary abnormality screening. Int. J. Comput. Assist. Radiol. Surg. 11(9), 1637–1646 (2016)
Seghers, D., Loeckx, D., Maes, F., Vandermeulen, D., Suetens, P.: Minimal shape and intensity cost path segmentation. IEEE Trans. Med. Imaging 26(8), 1115–1129 (2007)
Shao, Y., Gao, Y., Guo, Y., Shi, Y., Yang, X., Shen, D.: Hierarchical lung field segmentation with joint shape and appearance sparse learning. IEEE Trans. Med. Imaging 33(9), 1761–1780 (2014)
Shi, Y., et al.: Segmenting lung fields in serial chest radiographs using both population-based and patient-specific shape statistics. IEEE Trans. Med. Imaging 27(4), 481–494 (2008)
Shiraishi, J., et al.: Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 174(1), 71–74 (2000)
Van Ginneken, B., Romeny, B.T.H., Viergever, M.A.: Computer-aided diagnosis in chest radiography: a survey. IEEE Trans. Med. Imaging 20(12), 1228–1241 (2001)
Van Ginneken, B., Stegmann, M.B., Loog, M.: Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Med. Image Anal. 10(1), 19–40 (2006)
Wang, C.: Segmentation of multiple structures in chest radiographs using multi-task fully convolutional networks. In: Sharma, P., Bianchi, F.M. (eds.) SCIA 2017. LNCS, vol. 10270, pp. 282–289. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59129-2_24
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R.M.: ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3462–3471. IEEE (2017)
Yang, W., et al.: Lung field segmentation in chest radiographs from boundary maps by a structured edge detector. IEEE J. Biomed. Health Inform. 22(3), 842–851 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Jangam, E., Rao, A.C.S. (2019). Public Datasets and Techniques for Segmentation of Anatomical Structures from Chest X-Rays: Comparitive Study, Current Trends and Future Directions. In: Santosh, K., Hegadi, R. (eds) Recent Trends in Image Processing and Pattern Recognition. RTIP2R 2018. Communications in Computer and Information Science, vol 1036. Springer, Singapore. https://doi.org/10.1007/978-981-13-9184-2_29
Download citation
DOI: https://doi.org/10.1007/978-981-13-9184-2_29
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-9183-5
Online ISBN: 978-981-13-9184-2
eBook Packages: Computer ScienceComputer Science (R0)