Abstract
Non-coding RNA molecules have become increasingly more important in the realm of understanding the transcriptome. The functional role of the non-coding transcriptome has only relatively recently become an area of expansive research, largely through the methods of next-generation sequencing. Circular RNAs (circRNAs), a subtype of mostly non-coding RNAs, have recently been shown to have considerable abundance and cell-type- and development-specific expression patterns, in particular in human tissues. CircRNAs also show a distinct expression pattern in complex human diseases such as neurodegenerative diseases and cancer. This chapter focuses on the role of circRNAs as a biomarker in cancer using hepatocellular carcinoma (HCC) as an example. CircRNAs, as potential biomarkers of cancer, could not only play a significant role in diagnosis and monitoring of malignant growth but also might serve as primary targets for therapeutic intervention.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
Advancements in high-throughput RNA sequencing (RNA-seq) technologies and related bioinformatic tools have revolutionised our understanding of transcriptome. This has helped define molecular features of malignancies without the limitations of preceding transcriptomic tools such as microarrays, which are limited by their inability to detect transcripts de novo, or Sanger sequencing of cDNA/expressed sequence tag libraries, which is limited by its high costs and low throughput. In order to understand the principals behind this revolution in transcriptomics characterisation, it is important to understand the different types of RNA produced by human cells and the methodology of linear RNA-seq.
5.2 Types of RNA
There are different types of RNA that can be produced by human cells, which have distinct functions. Approximately 3% of RNA produced encodes a protein template (messenger RNA), while the remaining 97% is non-coding RNA (ncRNA). The most abundant ncRNAs are transfer RNA (tRNA) and ribosomal RNA (rRNA), which are both vital to the process of mRNA translation. This chapter focuses on a type of ncRNA called circular RNAs (circRNAs), its role within the cell and how it can be involved in human disease (Palazzo and Lee 2015).
5.2.1 Circular RNAs
CircRNAs are single-stranded, generally non-coding RNA molecules formed by backsplicing: the process in which the 5′ and 3′ ends of linear precursor mRNA (pre-mRNA) are covalently linked to create a circular structure (Salzman et al. 2012). The site of circRNA linkage, forming a phosphodiester bond, is termed the backsplice junction. In addition to their shape, circRNAs are distinguished from linear RNAs by the absence of a poly(A) tail and a 5′ cap (Szabo and Salzman 2016). Due to their circular structure, exons within circRNAs appear out of order from their genomic template and linear RNA counterparts (Nigro et al. 1991). CircRNAs can exist as exonic circRNAs—circRNAs that contain single or multiple exons; intronic circRNAs/RNA lariats—circRNAs that contain intronic regions only; and exon–intron circRNAs—circRNAs that contain both exons and introns (Jeck and Sharpless 2014; Memczak et al. 2013).
The size of circRNAs can range from smaller than 100 nucleotides to larger than 4 kb, although in humans they are commonly composed of a few hundred nucleotides (Lasda and Parker 2014). CircRNAs have been identified for thousands of human genes, accounting for 20–30% of the human transcriptome (You et al. 2015). Increasing circRNA diversity is the ability for multiple circRNA isoforms to arise from the same gene, a phenomenon known as alternative circularisation (Zhang et al. 2014). Although alternative circularisation can produce tens of distinct circular products from a single gene, most genes only produce one or two distinct circRNAs (Szabo and Salzman 2016). CircRNAs are considerably more stable than linear RNAs. For mammalian cells, the median half-life of circRNAs (18.8–23.7 h) is at least 2.5 times longer than their linear counterparts (4.0–7.4 h). Nevertheless, the half-life of circRNAs can vary considerably and can be greater than 48 h (Enuka et al. 2016; Jeck et al. 2013).
Sequence analysis coupled with splice inhibitor assays has shown that both linear mRNAs and circRNAs are derived from pre-mRNA and its interactions with the canonical spliceosome (Starke et al. 2015; Guo et al. 2014). The co-expression of circRNAs and linear mRNAs could be competitive, leading to a negative correlation in expressions between the two RNA subtypes (Ashwal-Fluss et al. 2014); however, several studies also observed independent expressions of circular and linear RNA (Gao et al. 2015; Salzman et al. 2013; Veno et al. 2015). Although for most genes expressing circRNAs the abundance of circles is roughly 5–10% of linear counterparts (Salzman et al. 2013), for many other genes, however, circular transcripts are the primary RNA species transcribed from the host locus (Salzman et al. 2012; Veno et al. 2015; Rybak-Wolf et al. 2015). Moreover, some genes express circular transcripts preferentially in certain tissues. For example, CDR1as expression is exclusively circular in the brain (Memczak et al. 2013).
A good body of evidence has uncovered specific expression of many circRNAs across different cell types and developmental stages (Memczak et al. 2013; Salzman et al. 2013; Veno et al. 2015; Rybak-Wolf et al. 2015). For example, circRNA hsa_circRNA_2149 is specific to human leukocyte cell line CD19+ and is absent in CD34+ leukocytes, neutrophils or the embryonic kidney cell line HEK293 (Memczak et al. 2013). These observations are not exclusive to humans, with a study in mice reporting the exclusive presence of circRNA derived from gene Rims2 in the granular layer of the cerebellum with an expression ~20-fold higher than its linear RNA counterpart (Rybak-Wolf et al. 2015).
5.3 Circular RNAs in Human Disease
Many circRNAs exhibit differential expressions in a diverse range of human disease such as neurodegenerative diseases and cancers (Chen et al. 2018, 2016; Li et al. 2015a; Zheng et al. 2016). For example, (1) in brains with multiple system atrophy (MSA), five circRNAs: IQCK, MAP 4K3, EFCAB11, DTNA and MCTP1 have specific overexpression in the frontal cortex (Chen et al. 2016); (2) exosomal circRNAs in peripheral blood of colon cancer patients have unique expression patterns compared to those found in controls (Li et al. 2015a); (3) transcriptome analysis of endometrial cancer data reported 120 differentially expressed circRNA between tumour and healthy tissue samples (Chen et al. 2018). These examples of differential expression of circRNAs in diseases suggest regulatory roles of circRNAs and additionally make them potential biomarkers for non-invasive diagnosis of diseases, especially those of the brain.
5.4 Linear RNA-seq
In order to understand the recent advances in the circRNA research field, it is important to comprehend the RNA-seq technique in the context of linear RNA. A typical high-throughput (also known as next-generation sequencing) linear RNA-seq workflow can be divided into three core stages: (1) library preparation, (2) sequencing and (3) data analysis.
5.4.1 Library Preparation
Library preparation is conducted pre-sequencing and involves selecting the desired RNA species, fragmenting it into 200–500 nucleotides, and attaching adaptors to its cDNA product. Selecting the desired RNA species often includes the removal of rRNA, which forms a high percentage of total RNA, which would otherwise consuming sequencing reads and reduce sequencing quality (Costa et al. 2010). rRNA removal can be achieved with poly(A) selection or rRNA depletion. RNA species can also be selected via gel electrophoresis that distinguishes based on RNA size and shape. Fragmentation of a given sequence is a necessary step during library preparation. Phred scores determine the estimated error of probability of bases called during downstream processing by assigning a quality score value to each base called (Cock et al. 2010). The higher the Phred score, the higher the base call accuracy (Endrullat et al. 2016). Fragmentation can occur using several methods such as nebulisation, sonification and enzymatic DNA digestion (Knierim et al. 2011). For small RNAs such as miRNAs and piRNAs, fragmentation is not necessary. A limitation of RNA-seq is that standard cDNA conversion does not maintain strand orientation. In order to maintain strand of origin information for each transcript, alternative library preparation is required but this is both time consuming and costly (Levin et al. 2010).
5.4.2 Sequencing
Over the years, a variety of high-throughput sequencing platforms have been developed. Despite sharing similar basic principles, Illumina platforms have dominated the sequencing industry (https://sapac.illumina.com/?langsel=/au/). Samples loaded into Illumina platforms first undergo cluster generation before sequencing can begin—each cDNA molecule is amplified in a separate well located on a slide, known as a flow cell. Clusters are generated via bridge amplification, which involves forming a cDNA bridge structure, synthesising the complementary strand with polymerase and denaturing the bridge to leave twice the amount of cDNA. In order to facilitate bridge formation, cDNA adaptors are hybridised to complementary oligos lining the surface of the flow cell. Once these clusters are generated, sequencing-by-synthesis (SBS) can begin. During SBS, RNA strands from each cluster have their complementary strand synthesised by polymerase. However, the nucleotides incorporated into the complementary strand are fluorescently tagged and contain a reversible terminator group. The reversible terminator group prevents more than one nucleotide from incorporating at a time to allow for the detection of each fluorescent signal. After the addition of a nucleotide, the fluorescent tag and terminator group are cleaved, allowing the next nucleotide to be incorporated. A single read is generated when polymerase finishes incorporating all nucleotides into a complementary strand.
5.4.3 Data Analysis
Data analysis involves several steps, broadly grouped into assembly and expression analyses. Assembly involves aligning (mapping) sequencing reads to a reference genome and transcript assembly, while expression analysis can involve differential expression assessment and pathway analysis. Alignment tools primarily determine the genomic origin of each read based on sequence similarity. Selected alignment parameters should be flexible enough to account for sequencing and reference genome errors, while stringent enough to align without losing specificity. Reputable alignment tools include TopHat, GSNAP and STAR. By utilising different algorithms and pre-existing knowledge about isoform structure, transcript assembly can be achieved by tools such as Cufflinks, SLIDE and StringTie. Counting the number of reads corresponding to each transcript also allows assembly tools to quantify expression levels. There are typically three units by which expression is calculated: (1) RPKM (reads per kilobase million), (2) FPKM (fragments per kilobase million) and (3) TPM (transcripts per kilobase million). Further analysis can include a determination of differential expression between samples or comparison of transcript expression profiles within a sample, for which raw read counts must be normalised, then statistically determined to be significant. Normalisation corrects for bases which can arise from variation in gene length (Oshlack and Wakefield 2009), library fragment size and sequencing read depth (Mortazavi et al. 2008). Bias arising from read depth can be observed from sequencing identical samples at different depths; samples sequenced at a greater depth will generate more reads, and wrongly appear to have increased expression. After differential expression analysis, pathways analysis is a further assessment, which can be conducted to determine potential biological pathways affected by the differentially expressed genes.
5.5 CircRNA Detection Tools, Challenges and Solutions
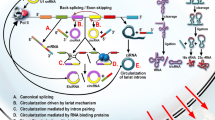
CircRNAs can be identified by various biochemical tools and bioinformatic pipelines following RNA-seq. These analytical pipelines require two bioinformatic tools: a standard alignment tool and a circRNA detection tool. Unless RNase R treatment is applied (see below), reads sequenced and aligned in circRNA detection are derived from fragmented linear RNAs and circRNAs and then converted to cDNA. However, only canonically spliced RNA reads are aligned by standard alignment tools, while circRNA reads remain aside due to their out-of-order exon arrangement. To identify and characterise circRNAs, these separate reads are aligned by circRNA detection tools (Fig. 5.1).
Schematic illustration of the generation of a linear mRNA (left) via canonical splicing and a circular RNA (right) via backsplicing in which the splice donor of exon 3 splices to the acceptor of exon 2, forming a circlular RNA molecule without 5′–3′polarity and a poly(A) tail, and forms back-spliced junction (BSJ)
Detection tools can be broadly classified as candidate-based (also known as pseudo-reference-based) or segmented-read-based (also known as fragmented-based) (Chen et al. 2015). For candidate-based tools such as NCLScan and PTESFinder, reference genome annotation is required as exon boundaries must be known. Annotation allows circRNA reads to be compared to different combinations of out-of-order exons derived from a reference genome. A downside of candidate-based tools is that they cannot identify circRNAs transcribed from unannotated genomic origins (KNIFE is an exception). Segmented-read-based tools do not require gene annotation to identify circRNAs.
For detection tools such as find_circ, PTESFinder and UROBORUS, whole circRNA reads are not aligned to the reference genomes. Instead, only short nucleotide sequences from each read are mapped (known as anchors). To reduce the number of false-positive circRNAs, detection tools also possess read filtering strategies which can be modified by the user. Filtering can include selecting for flanking GU/AG motifs, setting a minimum number of supporting reads, removing reads mapping to the mitochondrial genome and setting a minimum circRNA size. Detection tools can be also classified on read compatibility. Some detection tools are restricted to paired-end (PE) data (reads are generated from sequencing cDNA molecules in both directions) or single-end (SE) data (reads are generated from sequencing cDNA molecules in one direction), but most are compatible with both. For SE data, if only one read is generated from a circRNA, then that read must cross the backsplice junction for circRNA identification. As a result, the read corresponds to two distinct genomic origins. However, if two or more SE reads are generated from the same circRNA, then those reads can come from any sequence within the circRNA. Similarly, for PE data, each read can come from any sequence within the circRNA.
A major challenge of circRNA detection is that there is no gold-standard detection tool. Different detection tools implement different detection strategies, each with their own bias, strengths and weaknesses. For example, KNIFE can identify circRNAs at both annotated and un-annotated exon boundaries, but it is restricted in detecting circRNAs in regions of genomic variation. For each experiment, the selected detection tool should be based on research aims and available computational capacity. Computational capacity includes random access memory, physical disk space and processor speed. Research aims should encompass the trade-offs between each detection tool’s accuracy and sensitivity. As detection accuracy increases, sensitivity tends to decrease. Even for highly accurate detection tools such as MapSplice, a significant proportion (11%) of circRNAs are likely false positives. Accuracy is further decreased for circRNAs detected by a single detection tool. These circRNAs are known as exotic circRNAs and are detected with a false-positive rate as low as 20% (by MapSplice) and as high as 79% (by find_circ). Nonetheless, updated detection tools such as CIRI2 have improved overall circRNA detection (Hansen 2018). If further accuracy is required, a multi-bioinformatic approach can be implemented—circRNAs common between numerous detection tools are likely to be bona fide (Hansen et al. 2016).
The relatively low abundance of circRNAs in total RNA reduces the quality of sequencing data. To overcome this challenge, library preparation must include circRNA enrichment through ribosomal RNA (rRNA) depletion. Poly(A) selection is not suitable for circRNAs due to the absence of a poly(A) tail. Still, a few circRNAs will be present in poly(A) selection because the process is not completely efficient and because some circRNAs contain A-rich sequences. Further enrichment can be achieved with RNase R, although its use is debatable (Jeck et al. 2013). As a 3′ to 5′ exonuclease, RNase R degrades linear RNA molecules, but it may also degrade particular circRNAs (Jeck et al. 2013). Suspected circRNAs can be validated with probes for Northern blotting and fluorescent in situ hybridisation, as well as inverse or outward-facing primers for reverse transcriptase quantitative PCR (qPCR) (Capel et al. 1993; Barrett and Salzman 2016).
5.6 Mechanisms of CircRNA Action
The most established circRNA function to date is as a microRNA (miRNA) sponge. Many circRNAs, including zinc finger (ZNF) gene family of circRNAs and mouse circular Sry, are enriched with target sites for a number of miRNA families (Guo et al. 2014; Hansen et al. 2013). MiRNAs are ~21-nucleotide-long non-coding RNA transcripts that function in RNA silencing and post-translational regulation by guiding the effector protein Argonaute (AGO) to coding mRNAs, consequently repressing the latter’s protein production (Ambros 2004; Bartel 2009). As a well-studied case, a human circRNA transcribed antisense to the CDR1 gene harbours >60 conserved binding sites for miR-7 (Memczak et al. 2013; Guo et al. 2014). miR-7 targets many important players implicated in various pathways and diseases such as several oncogenic factors in cancer-associated signalling pathways, including EGFR, IRS-1 and IRS-2 (Kefas et al. 2008), and α-synuclein in Parkinson’s disease (Junn et al. 2009). It has been proposed that as a miRNA sponge, the CDR1 antisense transcript (CDR1as) binds to miR-7, sequestering it away from its targets, resulting in an upregulation of miR-7-targeted mRNA. Indeed, PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) experiments for human AGO showed that CDR1as is densely bound by the miRNA effector protein AGO in the cytoplasm. Further, both miR-7 loss-of-function study and ectopic delivery of CDR1as caused significant reduction in brain sizes in zebrafish and other animals (Memczak et al. 2013). These results strongly support the notion that CDR1as inhibits miR-7 via sponging. Another example from a recent study found that the circRNA HIPK3 contains 18 potential binding sites that were observed to sponge to nine miRNAs. More specifically, circular HIPK3 binds to miR-124 and inhibits its activity; silencing of circular HIPK3 significantly inhibits cell growth in humans (Zheng et al. 2016).
Some other studies suggested that circRNAs can also directly bind to other RNA-binding proteins (RBP), and consequently sequester and transport target RBPs (Hentze and Preiss 2013; Wilusz and Sharp 2013). However, it is still unclear if miRNA sponges or protein decoys are common functions of circRNA. The fact that only few circRNAs share properties similar to well-defined miRNA sponges and that circRNAs exist in species that lack RNA interference (RNAi) pathways strongly suggests other mechanisms of circRNAs action that have yet to be discovered (Guo et al. 2014; van Rossum et al. 2016).
5.7 Current Understanding of CircRNAs as Biomarkers
Emerging evidence has implicated a role for circRNAs in disease states, including neurodegenerative diseases and cancers. The high stability and enrichment in exosomes mean that circRNAs have the potential to serve as biomarkers for disease detection, diagnosis and progress monitoring (Li et al. 2015a). In addition to circRNAs in MSA, colon and endometrial cancers discussed earlier, several other groups have investigated circRNA expression for use as biomarkers in various cancer types. For example, in colorectal and ovarian cancer, the circular-to-linear ratio of RNA transcripts is lower in tumour tissues and there is an inverse relationship between this ratio and tumour cell growth rate (Bachmayr-Heyda et al. 2015). Other studies have reported differentially expressed individual circRNAs in laryngeal squamous cell cancer and gastric cancer tissues (Li et al. 2015b; Xuan et al. 2016). Taken together, there is mounting evidence that circRNA represents a potential model for development of novel biomarkers.
To gain an in-depth and holistic understanding of cancer, metastatic phenotypes must be observed beyond what is morphologically available. Characterising RNA expression levels can allow interpretation of the consequences of the genetic and epigenetic changes driving metastasis, thus enabling the links between different phenotypes and molecular characteristics to be identified. Compared to the healthy cells in an individual, those with tumourigenic transformation display a clear difference in transcriptome profiles. Here, we will be using hepatocellular carcinoma (HCC) as an example. HCC is an aggressive primary liver cancer (PLC) which responds poorly to treatment. The difficulties in treating HCC are reflected by a high mortality-to-incidence ratio and low survival rate, particularly for patients beyond early-stage diagnosis (White et al. 2016). As a largely preventable disease, recent years have seen considerable progress in eliminating HCC risk factors, including obesity, diet, diabetes, alcohol and tobacco use, in addition to hepatitis B and C (White et al. 2016), whereas the development of diagnostic tools allowing early identification of the cancer has not been a focus of attention. It has been recently suggested that a number of different non-coding RNA species including circular RNAs (circRNAs), microRNAs (miRNAs) (Thurnherr et al. 2016), P-element Induced WImpy testis (PIWI)–interacting RNAs (piRNAs) (Rizzo et al. 2016) and small interfering RNAs (siRNAs) (Farra et al. 2015) are differentially expressed in HCC and are involved in the molecular pathology. Given the high stability of circRNAs, this differential expression in tumourigenic cells may be detectable in tissues such as saliva, blood and exosomes as a result of shedding of the tumour cells (Meng et al. 2017). Unlike linear RNAs, which are degraded by exonucleases and virtually absent in peripheral whole blood, circRNAs are enriched to levels comparable to that in the brain, >15-fold higher expression than non-neuronal organs like the liver (Memczak et al. 2015). This makes circRNA biomarkers of HCC easier to detect in whole blood, where peripheral cellular expression levels are minimal (Memczak et al. 2015).
This theory has come to fruition in recent years, with potential circRNA biomarkers of HCC having been discovered based on their abnormal expression levels in tumours. Yu et al. demonstrated that cSMARCA5 (hsa_circ_0001445), a circRNA transcribed from the SMARCA5 gene, is significantly downregulated in HCC resulting in a worsened prognosis, whereas mRNA and protein levels of SMARCA5 are upregulated (Yu et al. 2018). Han et al. demonstrated a similar pattern of findings based on the downregulation of circMTO1 in HCC tissue resulting in a worsened prognosis (hsa_circRNA_0007874/hsa_circRNA_104135) and suggested the possibility of circMTO1 upregulation resulting in the inflation of mRNA and protein expression levels of tumour suppressor gene, p21 (Han et al. 2017). For use as biomarkers, however, circRNA upregulation is considered to be more useful than downregulation, since expression is easier to quantify with detection tools that lack sensitivity and it can be more difficult to identify, interpret and distinguish the clinical significance of low-expressed circRNAs. Upregulation of circRNAs has been observed in individuals with HCC and appears more frequently than downregulation. Huang et al. demonstrated the upregulation of circRNA_10038 (hsa_circRNA_100338) and circRNA_104075 (hsa_circRNA_104075), both of which were positively correlated with metastatic properties of HCC cell lines. Further study by Huang et al. looked at circRNA_10038 and the association with a specific miRNA, miR-141-3p, and found cancer-inhibiting antagonistic properties associated with miR-141-3p directed at circRNA_10038 that were positively correlated with the regulation of liver cancer cell metastasis (Huang et al. 2017).
CircRNAs are not by-products of alternative splicing. Hansen et al. demonstrated that circRNAs can act as miRNA sponges; circRNAs bind miRNA, preventing their repression of mRNA and resulting in further control of gene expression, as determined by their results with circRNA ciRS-7 acting as a sponge for miRNA miR-138 (Hansen et al. 2013).
Beyond interacting with miRNA, circRNAs have also been shown to interact with proteins and spliceosomal machinery, further influencing expression levels and cell proliferation (Ashwal-Fluss et al. 2014; Yu et al. 2018; Han et al. 2017; Du et al. 2016; Dudekula et al. 2016; Yu et al. 2016). The role of circRNAs as miRNA sponges, and the accumulating evidence to suggest circRNAs are efficient biomarkers in the disease profile for HCC is increasingly propagating the notion behind circRNA involvement in HCC disease pathology, and ultimately as significant molecular components of the disease profile.
Additionally, as more research is undertaken, it is likely that additional functions of circRNAs will be discovered. Where the relationship between circRNAs and a disease is due to a direct role in driving the pathogenic process, the circRNAs are a distinct feature of disease, meaning their use as a biomarker does not rely on a correlation between factors. In addition to their use as an early diagnostic tool, such a relationship allows the potential use of circRNAs as biomarkers for further clinical parameters, such as predicting prognosis and assessing the effectiveness of therapeutic agents.
5.8 Future Requirements
CircRNAs show tremendous potential as clinical biomarkers of disease diagnosis, prognosis and treatment assessment. Encouraging circRNA biomarker usage is the development of new circRNA detection tools to identify previously undiscovered circRNA types. One promising detection tool is accurate circRNA_finder suite (acfs), the first circRNA detection tool to identify fusion circRNAs (You and Conrad 2016). To further progress the use of circRNA biomarkers, the establishment of reputable pipelines with high sensitivity and precision is essential. For such pipelines, performance standards should be comparable to current pipelines available in linear RNA-seq analysis. In the meantime, multi-bioinformatic approaches should be adopted. Also, spurring the use of circRNA biomarkers is the introduction of third-generation sequencing, which enables long-read sequencing and direct RNA-seq. Implementing third-generation sequencing removes artefacts and bias which would arise from cDNA conversion and simplifies computational analysis by generating full-length reads. What’s more is that direct RNA-seq technologies provides real time, massive parallel sequencing, with portability and affordability. Taken together with data produced to date, it is highly likely that the discovery of more disease-driving circRNAs will occur in the future, which opens the door to their routine clinical use as early detection biomarkers, prognostic indicators and therapeutic efficacy indicators.
References
Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355. https://doi.org/10.1038/nature02871
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D (2015) Correlation of circular RNA abundance with proliferation—exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 5:8057. https://doi.org/10.1038/srep08057
Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143(11):1838–1847. https://doi.org/10.1242/dev.128074
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73(5):1019–1030
Chen I, Chen CY, Chuang TJ (2015) Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 6(5):563–579. https://doi.org/10.1002/wrna.1294
Chen BJ, Mills JD, Takenaka K, Bliim N, Halliday GM, Janitz M (2016) Characterization of circular RNAs landscape in multiple system atrophy brain. J Neurochem 139(3):485–496. https://doi.org/10.1111/jnc.13752
Chen BJ, Byrne FL, Takenaka K, Modesitt SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL, Janitz M (2018) Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget 9(5):5786–5796. https://doi.org/10.18632/oncotarget.23534
Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM (2010) The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38(6):1767–1771. https://doi.org/10.1093/nar/gkp1137
Costa V, Angelini C, De Feis I, Ciccodicola A (2010) Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol 2010:853916. https://doi.org/10.1155/2010/853916
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44(6):2846–2858. https://doi.org/10.1093/nar/gkw027
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42. https://doi.org/10.1080/15476286.2015.1128065
Endrullat C, Glokler J, Franke P, Frohme M (2016) Standardization and quality management in next-generation sequencing. Appl Transl Genom 10:2–9. https://doi.org/10.1016/j.atg.2016.06.001
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y (2016) Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 44(3):1370–1383. https://doi.org/10.1093/nar/gkv1367
Farra R, Grassi M, Grassi G, Dapas B (2015) Therapeutic potential of small interfering RNAs/micro interfering RNA in hepatocellular carcinoma. World J Gastroenterol 21(30):8994–9001. https://doi.org/10.3748/wjg.v21.i30.8994
Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16:4. https://doi.org/10.1186/s13059-014-0571-3
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7):409. https://doi.org/10.1186/s13059-014-0409-z
Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66(4):1151–1164. https://doi.org/10.1002/hep.29270
Hansen TB (2018) Improved circRNA Identification by combining prediction algorithms. Front Cell Dev Biol 6:20. https://doi.org/10.3389/fcell.2018.00020
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388. https://doi.org/10.1038/nature11993
Hansen TB, Veno MT, Damgaard CK, Kjems J (2016) Comparison of circular RNA prediction tools. Nucleic Acids Res 44(6):e58. https://doi.org/10.1093/nar/gkv1458
Hentze MW, Preiss T (2013) Circular RNAs: splicing’s enigma variations. EMBO J 32(7):923–925. https://doi.org/10.1038/emboj.2013.53
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY (2017) Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep 7(1):5428. https://doi.org/10.1038/s41598-017-05432-8
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461. https://doi.org/10.1038/nbt.2890
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157. https://doi.org/10.1261/rna.035667.112
Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM (2009) Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 106(31):13052–13057. https://doi.org/10.1073/pnas.0906277106
Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10):3566–3572. https://doi.org/10.1158/0008-5472.CAN-07-6639
Knierim E, Lucke B, Schwarz JM, Schuelke M, Seelow D (2011) Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS One 6(11):e28240. https://doi.org/10.1371/journal.pone.0028240
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20(12):1829–1842. https://doi.org/10.1261/rna.047126.114
Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A (2010) Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7(9):709–715. https://doi.org/10.1038/nmeth.1491
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015a) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981–984. https://doi.org/10.1038/cr.2015.82
Li PF, Chen SC, Chen HL, Mo XY, Li TW, Shao YF, Xiao BX, Guo JM (2015b) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta 444:132–136. https://doi.org/10.1016/j.cca.2015.02.018
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338. https://doi.org/10.1038/nature11928
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. Plos One 10(10):e0141214. https://doi.org/10.1371/journal.pone.0141214
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16(1):94. https://doi.org/10.1186/s12943-017-0663-2
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B (1991) Scrambled exons. Cell 64(3):607–613. https://doi.org/10.1016/0092-8674(91)90244-s
Oshlack A, Wakefield MJ (2009) Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4(1):14. https://doi.org/10.1186/1745-6150-4-14
Palazzo AF, Lee ES (2015) Non-coding RNA: what is functional and what is junk? Front Genet 6:2. https://doi.org/10.3389/fgene.2015.00002
Rizzo F, Rinaldi A, Marchese G, Coviello E, Sellitto A, Cordella A, Giurato G, Nassa G, Ravo M, Tarallo R, Milanesi L, Destro A, Torzilli G, Roncalli M, Di Tommaso L, Weisz A (2016) Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget 7(34):54650–54661. https://doi.org/10.18632/oncotarget.10567
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–885. https://doi.org/10.1016/j.molcel.2015.03.027
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7(2):e30733. https://doi.org/10.1371/journal.pone.0030733
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777. https://doi.org/10.1371/journal.pgen.1003777
Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10(1):103–111. https://doi.org/10.1016/j.celrep.2014.12.002
Szabo L, Salzman J (2016) Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 17(11):679–692. https://doi.org/10.1038/nrg.2016.114
Thurnherr T, Mah WC, Lei Z, Jin Y, Rozen SG, Lee CG (2016) Differentially expressed miRNAs in hepatocellular carcinoma target genes in the genetic information processing and metabolism pathways. Sci Rep 6:20065. https://doi.org/10.1038/srep20065
van Rossum D, Verheijen BM, Pasterkamp RJ (2016) Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci 9:74. https://doi.org/10.3389/fnmol.2016.00074
Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J (2015) Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16:245. https://doi.org/10.1186/s13059-015-0801-3
White DL, Kanwal F, Jiao L, El-Serag HB (2016) Epidemiology of hepatocellular carcinoma. In: Carr BI (ed) Hepatocellular carcinoma: diagnosis and treatment. Springer, Cham, pp 3–24. https://doi.org/10.1007/978-3-319-34214-6_1
Wilusz JE, Sharp PA (2013) Molecular biology. A circuitous route to noncoding RNA. Science 340(6131):440–441. https://doi.org/10.1126/science.1238522
Xuan LJ, Qu LM, Zhou H, Wang P, Yu HY, Wu TY, Wang X, Li QY, Tian LL, Liu M, Sun YN (2016) Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 8(2):932–939
You X, Conrad TO (2016) Acfs: accurate circRNA identification and quantification from RNA-Seq data. Sci Rep 6:38820. https://doi.org/10.1038/srep38820
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18(4):603–610. https://doi.org/10.1038/nn.3975
Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L (2016) The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 11(7):e0158347. https://doi.org/10.1371/journal.pone.0158347
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP (2018) Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 68(6):1214–1227. https://doi.org/10.1016/j.jhep.2018.01.012
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147. https://doi.org/10.1016/j.cell.2014.09.001
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. https://doi.org/10.1038/ncomms11215
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Grouse, L., Curry-Hyde, A., Chen, B.J., Janitz, M. (2019). Circular RNAs in Human Health and Disease. In: Hesson, L., Pritchard, A. (eds) Clinical Epigenetics. Springer, Singapore. https://doi.org/10.1007/978-981-13-8958-0_5
Download citation
DOI: https://doi.org/10.1007/978-981-13-8958-0_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8957-3
Online ISBN: 978-981-13-8958-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)