Abstract
Mitochondria (mt) are double-membraned, dynamic organelles that play an essential role in a large number of cellular processes, and impairments in mt function have emerged as a causative factor for a growing number of human disorders. Given that most biological functions are driven by physical associations between proteins, the first step towards understanding mt dysfunction is to map its protein-protein interaction (PPI) network in a comprehensive and systematic fashion. While mass-spectrometry (MS) based approaches possess the high sensitivity ideal for such an endeavor, it also requires stringent biochemical purification of bait proteins to avoid detecting spurious, non-specific PPIs. Here, we outline a tagging-based affinity purification coupled with mass spectrometry (AP-MS) workflow for discovering new mt protein associations and providing novel insights into their role in mt biology and human physiology/pathology. Because AP-MS relies on the creation of proteins fused with affinity tags, we employ a versatile-affinity (VA) tag, consisting of 3× FLAG, 6 × His, and Strep III epitopes. For efficient delivery of affinity-tagged open reading frames (ORF) into mammalian cells, the VA-tag is cloned onto a specific ORF using Gateway recombinant cloning, and the resulting expression vector is stably introduced in target cells using lentiviral transduction. In this chapter, we show a functional workflow for mapping the mt interactome that includes tagging, stable transduction, selection and expansion of mammalian cell lines, mt extraction, identification of interacting protein partners by AP-MS, and lastly, computational assessment of protein complexes/PPI networks.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Affinity purification
- Protein complex
- Versatile-affinity tagging
- Lentivirus
- Mass spectrometry
- Mitochondria
- Protein-protein interaction
6.1 Introduction
Mitochondria (mt) are intimately involved in many biological processes including energy metabolism, apoptosis, and cell signalling [1]. Therefore, it is unsurprising that many important human pathologies, especially those of neurodegenerative [2, 3], metabolic [4], cardiovascular [5] and malignant [6] origins, often have mt dysfunction as a common hallmark. Given its disease relevance, mt research has become a “hot spot” in different fields, driving the discovery of novel physiological and pathological mechanisms as well as the development of new techniques and paradigms.
In recent years, proteomic approaches have moved to the forefront of the exploration of molecular mechanisms involved in mt function and disease [1, 7,8,9]. Unlike the classical reductionist approach focusing exclusively on one particular protein or process, proteomics is the large-scale study of complete groups of proteins sharing a common characteristic, such as subcellular, nuclear, or membrane localization, in a living organism, to gather systems level insights in protein expression, physical and consequently functional relationships, and potential role in pathogenesis in an unbiased manner [1]. More importantly, given that many biological processes are mediated through the physical association between proteins, the systematic mapping of protein-protein interaction (PPI) networks is becoming an increasingly powerful means of studying how molecular events drive biological systems. In the context of mt health and disease, mapping PPIs can be especially valuable in the development of new diagnostic biomarkers and elucidating drug targets. As well, the differences in the PPI network between healthy and diseased states enable researchers to design models and make predictions about the underlying biological processes contributing to pathogenesis.
The construction of large-scale human PPI networks were traditionally conducted by systematically examining pair-wise interactions identified through several experimental means, including yeast-two hybrid [10, 11] and biochemical fractionation coupled with mass spectrometry (MS) [12, 13]. However, affinity-purification using the tag-based approach or immunoprecipitation (IP) using a protein-specific antibody coupled with MS [7, 8, 14, 15] has become the method of choice for purification of a single protein and interactors at a time. These methods are well-suited to identify and characterize transiently or stably interacting proteins.
The recent successful purification of mt proteins using a lentivirus and cell culture-based AP-MS framework appeared to be a promising approach to isolate stable protein complexes [7, 8]. This flexible and robust AP-MS procedure allows the tagged proteins to be expressed at a level similar to the native physiological state and then purified via a two-step enrichment procedure. Although its high sensitivity makes MS-based approach crucial for large-scale PPI research [13, 15, 16], great care must be exercised to minimize non-specific (spurious) PPIs and contaminants [17]. Thus, the AP procedure provides a comprehensive yet selective strategy of protein purification to minimize false negative and positives, allowing for accurate and sensitive MS identification of distinct complexes and PPIs.
As an alternative to AP-MS, immunoprecipitation coupled with MS (IP-MS) has been commonly used to purify mammalian protein complexes [18]; however, the scale of such studies is often limited by the availability of validated antibodies against specific bait targets. Therefore, for large-scale, systematic identification of PPI by AP-MS, a more suitable strategy that can be integrated into a high-throughput workflow would be necessary. To this end, since the main requirement for AP-MS is the creation of proteins fused with affinity tags, rather than relying on protein-specific antibodies, transgenic cells could be generated with an affinity tag introduced on the C-terminus (to avoid interference with mt protein import signal peptides, typically located at the N-terminal) of the open reading frame (ORF) of interest. In this manner, the tagged protein can still be expressed at a near physiological level.
The purification method described here uses the versatile-affinity (VA) tag [19], a 12-kDa triple affinity tag consisting of 3× FLAG, 6 × His, and Strep III epitopes. Here, the 3× FLAG is spaced from the rest of the sequence by a dual tobacco etch virus (TEV) protease cleavage site, thus allowing for its proteolytic cleavage, and the use of two other tags, permitting a highly selective two-stage protein purification and enrichment. A widely used epitope for purification, 3× FLAG is not only small in size, which reduces the chance of functional perturbation of the tagged proteins, but also highly amenable to sensitive immunodetection with commercially available high-affinity anti-FLAG antibodies. In addition, the inclusion of 6 × His and Strep III adds considerable versatility to the VA-tag. The widely used 6 × His gives researchers the option of purifying proteins under denaturing conditions, while Strep III is highly selective, tightly binds to Strep-tactin and is efficiently displaced (competed) with biotin for elution.
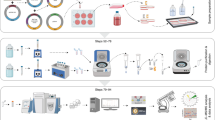
The overall workflow proceeds as outlined in Fig. 6.1. Briefly, a Gateway cloning procedure is first used to introduce the affinity tag coding sequence on the C-terminus of the mt protein of interest (also called “baits”) on a lentiviral expression backbone. The resulting expression constructs are packaged into lentiviral particles and used to transduce target mammalian cells at multiplicity of infection (MOI) of ≈0.3, thereby generating cell lines stably expressing the VA-tagged mt protein at near-physiological expression level. Transgenic cells are cross-linked, lysed, and from the lysates derived from these expanded cell lines the tagged bait proteins can be isolated by tandem affinity purification and then processed for analysis by MS. The identification of co-purifying proteins (also called “preys”) is achieved by searching the resulting mass/charge spectra against peptide sequence databases, which then enables computational filtration, scoring and clustering algorithms to be used to generate a mt interaction network.
Outline of the AP-MS workflow. Schematic of Lentiviral-based versatile-affinity (VA) tagging of MP using the MAPLE lentiviral transfer vector (a), followed by purification of VA-tagged proteins and their interactors from mt fractions isolated from transgenic HEK293 cells (b). Proteins co-purified with the target bait protein identified using MS and analyzed by clustering algorithm to predict protein complexes (c). MAPLE mammalian affinity purification and lentiviral expression, MP mitochondrial protein, AP affinity purification, VA versatile-affinity, PPIs protein-protein interactions
In the rest of this chapter, we describe detailed step-by-step procedures for the entire workflow outlined above. It is noteworthy that while the overall context for methodologies recorded here deals primarily with mt, the basic approach can be adapted for analysis of the proteomes of other organelles and subcellular compartments or whole cells.
6.2 Materials
6.2.1 Vector Construction and Generation
-
pLD-puro-Cc-VA (Addgene# 24588) maintained in ccdA-containing strains such as one shot ccdB survival or DB3.1 E. coli.
-
Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific, cat# 11791-020)
-
Stbl3 (Thermo Fisher Scientific, cat# C737303) or NEB Stable (NEB, cat# C3040H)
-
Expression clones from Harvard Plasmid ID repository
-
Roche High Pure RNA Isolation kit (Sigma-Aldrich, cat# 12033674001)
-
iScript cDNA Synthesis kit (BioRad, cat# 1708890).
6.2.2 Lentiviral Transduction
6.2.2.1 Lentiviral Production
-
HEK 293T cells (ATCC, cat# CRL-3216)
-
Lentiviral coat protein-encoding plasmid, e.g. pMD2.G (Addgene# 12259)
-
Lentiviral packaging plasmid: the third-generation packaging plasmids pMDLg/pRRE (Plasmid # 12251) and pRSV-Rev (Plasmid# 12253)
-
Lipofectamine LTX with Plus reagent (Thermo Fisher Scientific, cat# 15338100)
-
Opti-MEM I (Thermo Fisher Scientific, cat# 31985070)
-
Low-protein binding 0.4 μm membrane syringe filter (e.g. polyether sulfone, Sarstedt, cat# 83.1826)
-
0.2 μm membrane syringe filter (Sarstedt, cat# 83.1826.001)
6.2.2.2 Lentiviral Infection
-
Six-well plate (Sarstedt, cat# 83.3920.005)
-
Hexadimethrine bromide (Sigma-Aldrich, cat# H9268)
6.2.2.3 Antibiotic Selection
-
Blasticidin S (Fisher Scientific, cat# BP2647-25)
-
Puromycin (Fisher Scientific, cat# BP2956-100)
-
Presto Blue (Thermo Fisher Scientific, A13261)
6.2.3 Confirmation of Tagging by Immunoblotting
-
Trans-Blot ® Cell system (Bio-Rad, cat# 170-3853)
-
Nitrocellulose membrane (Bio-Rad, cat# 162-0115)
-
Transfer stock buffer (10X): 120 g Tris -HCl and 576 g glycine in distilled water
-
1X transfer buffer: 400 mL of 10X transfer stock buffer with 800 mL methanol and 2800 mL of sterile distilled water. Store at room temperature.
-
TBS stock buffer (5X): 48.44 g Tris and 584.4 g NaCl in distilled water
-
Wash buffer: 1X TBS buffer containing 0.05% Tween-20
-
Blocking buffer: 5% (w/v) non-fat dry milk in 1X TBS buffer containing 0.1% Tween-20
-
Primary antibody: M2 antibody (Sigma-Aldrich, cat# F3165)
-
Primary antibody buffer: TBS buffer supplemented with 1% gelatin, 0.05% Tween-20, and 0.02% sodium azide
-
Secondary antibody: Anti-mouse IgG conjugated to horseradish peroxidase (HRP; SantaCruz, cat# sc-516102)
-
Secondary antibody buffer: 5% (w/v) non-fat dry milk in TBS buffer containing 0.1% Tween-20
-
Chemiluminescence reagent (PIERCE, cat# 1856136), prepared (0.125 mL of chemiluminescence reagent per cm2 of membrane) by mixing equal volumes of the enhanced luminol reagent and the oxidizing reagent
-
Kodak X-OMAT autoradiography film (Clonex Corp., cat# CLEC810)
-
Stripping buffer: 62.5 mM Tris-HCl (pH 6.7), 2% (w/v) SDS, and 100 mM β- mercaptoethanol. Store at room temperature.
6.2.4 Freezing, Reviving and Differentiating Mammalian Cells
6.2.4.1 Cryogenic Preservation of Successfully Tagged Mammalian Cells
-
DMSO (Sigma-Aldrich, cat# C6164)
-
Trypsin-EDTA solution (Thermo Fisher Scientific, cat# 15400054)
-
CryoPure Storage System (Sarstedt, cat# 72.377)
-
Mr. Frosty™ Freezing Container (Thermo Fisher Scientific, cat# 5100-0001)
6.2.4.2 Reviving and Large-Scale Culturing of Mammalian Cells
-
One hundred millimeter (Sarstedt, cat# 83.3902) and 150 mm (Sarstedt, cat# 83.3903) polystyrene culture dishes.
-
DMEM complete: DMEM high glucose (Thermo Fisher Scientific, cat# 11995073), supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, cat# 12483020), 100 units/mL of penicillin and 100 μg/mL of streptomycin (Thermo Fisher Scientific, cat# 15140122).
6.2.4.3 Differentiation of SH-SY5Y Cells
-
Human neuroblastoma SH-SY5Y (ATCC, cat# CRL-2266)
-
All-trans retinoic acid (at-RA, Sigma-Aldrich; cat# R2625)
-
Brain-derived neurotrophic factor (BDNF, Thermo Fisher Scientific, cat# 14-8366-80)
6.2.5 Cell Harvesting, Mt Fractionation and Protein Purification
6.2.5.1 Cell Preparation
-
Phosphate-Buffered Saline (PBS, pH 7.4) (Thermo Fisher Scientific, cat# 10010023)
6.2.5.2 Cross-Linking with DSP
-
Dithiobis succinimidyl propionate (DSP, Thermo Fisher Scientific, cat# PG82081)
-
Dimethyl sulfoxide (DMSO, Sigma-Aldrich, cat# D8418)
-
One molar Tris-HCl (pH 7.5) stock solution: 121.1 g of Tris base in 800 mL of distilled water, adjusted to pH 7.5 with concentrated HCl and brought to a final volume of 1 L.
-
Quenching solution: 100 mM Tris-HCl (pH 7.5) supplemented with 2 mM EDTA.
6.2.5.3 Mt Purification
-
Homogenization buffer: 10 mM NaCl, 1.5 mM MgCl2 and 10 mM Tris-HCl.
-
Mitochondria stabilization buffer: 210 mM mannitol, 70 mM sucrose, 5 mM Tris-HCl (pH 7.5), and 1 mM EDTA.
6.2.5.4 Tandem Affinity Purification Using VA-Tag
-
Mitochondria lysis buffer: 30 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% digitonin (Sigma-Aldrich, cat# D140), and supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, cat# 78440).
-
Bio-Spin chromatography columns (Bio-Rad, cat# 7326008)
-
Anti-Flag M2 agarose beads (Sigma-Aldrich, cat# M8823)
-
TBS buffer: 50 mM Tris (pH 7.5) and 150 mM NaCl
-
TEV buffer: 10 mM Tris-HCl (pH 7.9), 125 mM NaCl, and 0.1% digitonin (Sigma-Aldrich, cat# D140)
-
TEV protease (Sigma-Aldrich, cat# T4455), Dilute to working concentration of 2 mg/mL
-
3× FLAG peptide (Sigma-Aldrich, cat# F4799)
-
Strep-Tactin-Sepharose bead (IBA, cat# 2-1201-010)
-
Ammonium bicarbonate (Sigma-Aldrich, cat # 09830)
-
D-biotin (Sigma-Aldrich, B4501)
-
Tris (2-carboxyethyl) phosphine (TCEP-HCl, Sigma-Aldrich, cat# C4706)
6.2.6 Sample Preparation for MS Analysis
-
Digestion buffer: 50 mM NH4HCO3 and 1 mM CaCl2
-
Alkylating solution: 10 mM iodoacetamide (Sigma-Aldrich, cat# I1149)
-
Trypsin gold (Promega, cat# V5280)
-
C18 Zip-Tip (Millipore, cat# Z720046)
-
HPLC-grade H2O in 0.1% formic acid (Fisher Scientific, cat# LS118)
-
Elution solution (1% acetic acid, 65% acetonitrile, in HPLC H2O in 0.1% formic acid)
-
Washing solution (1% acetic acid, 2% acetonitrile in HPLC H2O in 0.1% formic acid)
6.2.7 MS for Identifying Interacting Proteins
-
EASY nano liquid chromatography (nLC) 1000 (Proxeon, Mississauga, ON, Canada)
-
Velos Pro Ion Trap and Orbitrap Elite Mass Spectrometer (Thermo Fisher Scientific)
-
P-2000 Laser-based Micropipette Puller System (Sutter Instruments)
-
Luna 3 μm C18 100A from Phenomenex (cat# 04A-4251)
-
Fused-silica capillary tubing (ID 75.4 mm) from Polymicro technologies (cat# 1068150019)
-
Ninety-six-well plate from Thermo Fisher Scientific (AB-0800)
-
Methanol from Thermo Fisher Scientific (A456-4)
-
Formic acid from Thermo Fisher Scientific (117-50)
-
Water from Thermo Fisher Scientific (W6-4)
-
Sample buffer: 1% formic acid in water
-
MS Buffer A: 0.1% formic acid in water from Thermo Fisher Scientific (LS118-1)
-
MS Buffer B: 0.1% formic acid in acetonitrile from Thermo Fisher Scientific (LS120-1)
6.3 Methods
6.3.1 Vector Construction and Generation
In this section, we will specifically address cloning of Gateway-compatible entry clone into pLD-puro-Cc-VA (Addgene# 24588). The protocol can be modified and applied to a variety of other destination vectors. Note: (1) pLD-puro-Cc-VA is maintained in either One shot ccdB survival or DB3.1 Escherichia coli since it contains gene ccdB; (2) since pLD-puro-Cc-VA is a lentiviral vector, host bacteria is grown at 30 °C for no longer than 16 h and in case of liquid media at rotating speed of no more than 180 RPM; (3) The recipe below can be downscaled to save ingredients.
Procedure:
-
1.
In a 1.5 mL Eppendorf tube, 25–75 ng entry plasmid is combined with 75 ng pLD-puro-Cc-VA, and made the volume up to 4 μL with TE buffer (pH 8.0).
-
2.
LR Clonase II is vortexed briefly and 2 μL is added to the mixture above and the reaction is incubated at 25 °C for 1 h.
-
3.
Proteinase K (0.5 μL) is added and mixed prior to incubation at 37 °C for 10 min.
Transformation:
-
1.
Of the above mixture, 1–5 μL is pipetted into 50 μL of competent bacteria suitable for propagation of lentiviral plasmids (have low potential to mutate or recombine long terminal repeats (LTR)) such as Stbl3 or NEB Stable. The tube is mixed by gentle flicking a few times.
-
2.
Tube is incubated on ice for 30 min followed by heat shock at 42 °C for 20–30 s.
-
3.
After heat shock, tube is returned to ice briefly and 950 μL SOC media is added.
-
4.
The tube is shaken at 37 °C for 1 h at 225 RPM.
-
5.
With the above mixture, 50–100 μL is plated on ampicillin-containing Luria-Broth agar plate and incubated at 30 °C for 16 h.
-
6.
Individual colonies are picked and grown in liquid media with ampicillin at 30 °C for 16 h with shaking at 180 RPM.
-
7.
Plasmid is extracted and sanger-sequenced in the forward direction using CMV-F and reverse using the primer 5′- CTTTCCCCTGCACTGTACCC-3′, to validate cloning.
6.3.1.1 Vector Generation
-
1.
Where available, we obtained lentiviral expression vectors from Harvard Plasmid ID repository. If not available, Gateway-compatible vectors cloned into pLD-puro-Cc-VA vector using Gateway cloning.
-
2.
If neither lentiviral nor Gateway-compatible clones of the coding sequence (CDS) of choice were available, the target cDNA can be cloned as follows.
-
3.
Cloning of target CDS from cDNA library:
-
(A)
cDNA library generation: Total RNA is extracted from human cells using a compatible kit such as Roche High Pure RNA Isolation kit as per manufacturer’s instructions. Note: Care must be taken to avoid contaminating RNA sample with RNAse, which can lead to RNA degradation and poor cDNA library quality, resulting in failure of cDNA synthesis.
-
(B)
Next, cDNA is synthesized from RNA using iScript cDNA Synthesis kit. Using Primer3Plus tool, gene-specific primers are designed to amplify from the mRNA sequence. Note: The following rules should be taken into account while designing primers. Use default parameters whenever possible. Following Primer3Plus instructions, the CDS sequence is flanked to ensure that the designed primers amplify the full sequence. In general settings, under “Mispriming/Repeat Library” menu, choose Human. In the advanced settings, it is preferable to choose primers with a 1 or 2 CG clamp nucleotides at the 3′ end of the primer under “Advanced Settings” menu. If default settings failed to identify selective primers, adjust the “Product Size” setting under “Advanced Settings” menu so that the Min size matches the desired CDS length, and the Opt and Max is set reasonably above that. For example, if the CDS sequence is 1000 nucleotides, then Min size should be set to 1000, Opt to 1100, and Max to 1200. The Opt and Max sizes can be further adjusted accordingly until appropriate primers are identified.
-
(C)
If the above primers flank the specific CDS, then a second should be manually designed to amplify the exact desired CDS with the STOP codon at its 3′ end removed to allow in-frame fusion with the VA-tag in the pLD-puro-Cc-VA vector. This second PCR can be performed using the former set of primers designed using Primer3Plus. Note: Primers designed manually as above should be prepended on its 5′ end with either restriction sites of extra 6 nucleotides (on the 5′ end of the primer) that match restriction sites on the MAPLE vector. Alternately, the manually-designed primers can be used when cloning into a donor (pDONR) vector, rendering it into a suitable entry (pENTR) vector that is Gateway-compatible with MAPLE. For details, review Gateway® Technology Guide from Thermo Fisher Scientific.
-
(A)
6.3.2 Lentiviral Transduction
6.3.2.1 Lentiviral Production
-
1.
A fresh vial of healthy frozen 293T cells is thawed, plated, and regularly passaged in media composed of DMEM complete media.
-
2.
After three passages, 293T cells are plated in standard 100 mm diameter dishes with 30–40% confluence at the time of transfection.
-
3.
Media is replaced with antibiotic-free DMEM or Opti-MEM I 1 h before transfection.
-
4.
Before transfection, individual plasmids are decontaminated by heating in a thermal cycler at 85 °C for 30 min followed by slow cooling at the rate of 1 °C/min. Note: Alternatively, plasmid extraction can be performed under aseptic condition and eluted from the plasmid mini-prep column in sterile tubes using filter-sterilized nuclease-free water or elution buffer.
-
5.
After 8–12 h post plating, cells are transfected with a mixture of the following plasmids: 7.5 μg lentiviral expression (transfer) plasmid, 1.9 μg lentiviral coat protein encoding plasmid (e.g. pMD2.G), and third-generation lentiviral packaging plasmids (3.75 μg pMDLg/pRRE; 1.9 μg pRSV-Rev). Using the transfection reagent Lipofectamine LTX with Plus reagent, DNA is mixed according to the following recipe: (Tube A: DNA variable, 15 μl Plus reagent, up to 750 μl Opti-MEM I; Tube B: 52.5 μl Lipofectamine LTX, up to 750 μl Opti-MEM I). Tube A is added drop-wise to Tube B with frequent shaking and DNA-transfection reagent mixture is incubated at room temperature for 15 min. Media is replaced again with antibiotic-free DMEM or Opti-MEM I and DNA-transfection reagent mixture is added drop-wise to the cells and mixed gently. After 6–12 h, media is changed to complete media containing antibiotic and 30% FBS and returned to the incubator. Media is harvested after 24 h and again 48 h post-transfection. Media from both harvests are pooled and filtered through a low-protein binding 0.4 μm membrane syringe filter. Hexadimethrine bromide is freshly dissolved at 10 mg/mL, filter-sterilized through 0.2 μm syringe membrane filter and added to the lentiviral supernatant obtained in the previous step to a final concentration of 10 μg/mL.
6.3.2.2 Lentiviral Infection
Adherent Cells:
-
1.
Target cells are plated in six-well plate at 10–20% confluence and allowed to fully adhere prior to exposure to lentiviral supernatant.
-
2.
Determination of multiplicity of infection (MOI):
-
(A)
Target cells are plated at ~10 to 20% confluence
-
(B)
Serially diluted lentiviral supernatant in complete media is added to target cells
-
(C)
Cells are centrifuged at 140 xg for 0.5–1 h at 30 °C.
-
(D)
After 24–48 h of infection, selection antibiotic (in case of pLD-puro-Cc-VA, puromycin 2 μg/ml) is added and a kill curve is constructed by plotting percent viability at different lentiviral dilutions versus control.
-
(E)
Lentiviral supernatant dilution that results in 30–50% cell viability provides highest probability that any given cell is infected with a single lentiviral particle and thus ensures near-physiological level of expression.
-
(A)
-
3.
Media replaced with lentiviral supernatant (0.3–0.5 MOI) is centrifuged at 140 xg, for 0.5–1 h at 30 °C, and returned to the incubator.
-
4.
After 24 h, lentiviral supernatant is replaced with fresh media, and antibiotic selection can be carried out 24–48 h after infection.
-
5.
If toxicity is evident, lentiviral supernatant can be mixed with complete medium at 1:1 ratio. Exposure duration can be also minimized anywhere from 6 to 16 h. These measures will reduce the number of stably transduced cells, and will limit toxicity in difficult-to-work with cell lines.
Suspension Cells:
-
1.
Approximately 6 × 106 cells are centrifuged at 140 xg for 5 min at room temperature.
-
2.
Media is aspirated and replaced with lentiviral supernatant at 0.3–0.5 MOI as defined above.
-
3.
Cells are centrifuged at 140 xg for 0.5–1 h at 30 °C, and resuspended cells are then returned to the incubator for 24 h.
-
4.
After 24 h, cells are again centrifuged at 140 xg for 5 min at room temperature and lentiviral supernatant is replaced with fresh media, followed by antibiotic selection 24–48 h after infection.
6.3.2.3 Antibiotic Selection
Because mammalian cell lines differ in their antibiotics sensitivity, prior to antibiotic selection, a preliminary experiment should be performed to determine the lowest antibiotic concentration and duration of exposure required to efficiently kill 100% of uninfected cells and thus selection of successfully lentivirus-transduced cells. Here, we provide general guideline for establishing a kill curve using puromycin or blasticidin. The same principle can be extended to other mammalian selection antibiotics, such as zeocin, hygromycin, and neomycin with adaptation to ensure longer incubation as the latter antibiotics have slower effect on non-transduced cells.
Establish Antibiotic Kill Curve:
-
Day 0
-
1.
Seed cells at 50% confluence per well, in 24-well plate with blasticidin or puromycin selection, respectively.
-
2.
Incubate cells overnight.
-
1.
-
Day 1
-
1.
Make three sets of eight concentrations of antibiotic in growth media. For puromycin: 0, 0.06, 0.2, 0.6, 2, 6 and 20 μg/mL. For blasticidin: 0, 0.15, 0.5, 1.5, 5, 15 and 50 μg/mL. Each dilution should have at least 110 μL as the final volume.
-
2.
Remove cells from incubator, aspirate media, and replace with selection media supplemented at varying concentrations of antibiotics.
-
1.
-
Day 3–15
-
1.
Cell viability is determined using Presto Blue (Thermo Fisher Scientific, A13261). Optimum effectiveness is typically reached within 1–2 days for puromycin and 5–7 days for blasticidin.
-
2.
Change media every day with the appropriate selection media. If cells become too confluent, they should be split and allowed to adhere and recover before resuming the antibiotic selection.
-
3.
The minimum concentration resulting in complete cell death after selection period (depending on the antibiotic) should be used for antibiotic selection of transduced cells.
-
4.
Validate transgene expression using immunoblotting.
-
1.
6.3.3 Confirmation of Successful Tagging with Immunoblotting
6.3.3.1 SDS-Polyacrylamide Gel Electrophoresis
-
1.
The following protocol uses the Mini Protean 3 Cell (Bio-Rad cat# 165-3301) gel system. Before use, clean the glass plates, spacers, and combs to ensure they are free of dried gel fragments, grease, and dust.
-
2.
For each gel, assemble one small and one large glass plate so that they are separated by spacers and an alignment card. Make sure to have the small glass plate on top of the spacers so that both sides and the bottom of the plates and the spacers are even.
-
3.
Slide glass plates into the holder without tightening the screws. Make sure the glass plates are pushed all the way to the bottom before tightening the screws. If the assembly is correct, then the whole setup should snap into place when placed above the gasket.
-
4.
Take out the alignment card, slide in the comb and mark a line at 2–3 cm from the bottom of the comb. The comb should be of the same thickness as that of the spacers.
-
5.
Gently remove the comb. Pour 12% polyacrylamide resolving gel up to the marked line. Pipette a thin layer of ethanol on top of the gel and allow to polymerize for about 30 min.
-
6.
Pour 4% polyacrylamide stacking layer to fill up the remaining volume between glass plates. Insert the comb and leave the gel to polymerize for another 30 min.
-
7.
Remove the comb once the stacking gel has set, then use a 3 mL syringe fitted with a 22-gauge needle to rinse the wells with running buffer.
-
8.
Place the gel in the electrophoresis unit and add running buffer.
-
9.
Load each well with 10–20 μl samples in 1X SDS sample buffer. Reserve one lane for precision plus protein dual color standards (Bio-Rad cat#1610374). Complete the assembly of the electrophoresis unit and attach the power cords to the power supply.
-
10.
Turn the power supply on and run the gel at 120 V. Stop running when the blue dye front reaches the bottom.
-
11.
Once the electrophoresis is completed, remove the gel plate assembly and place on a paper towel. Carefully pry the glass plates apart using a spatula.
-
12.
Immediately transfer the gel to the nitrocellulose membrane and follow the transfer protocol described below.
6.3.3.2 Immunoblotting Using Chemiluminescence
Note: These instructions assume the use of a Trans-Blot ® Cell system. Wear gloves at all times when handling the membrane.
-
1.
Fill the cooling unit with water and store at −20 °C until ready to use.
-
2.
Cut the nitrocellulose membrane to be slightly larger than the gel.
-
3.
Equilibrate the gel by soaking it in transfer buffer for 15 min.
-
4.
Briefly soak the membrane, four pieces of Quick Draw TM blotting paper (Sigma Aldrich cat#P7796), and two scotch-brite® fiber pads in transfer buffer.
-
5.
Mark one side of the membrane for future reference.
-
6.
Preparing the gel sandwich:
-
(a)
On a clean, even surface, place the cassette with the gray side down
-
(b)
Place one pre-wetted fiber pad on the gray side of the cassette.
-
(c)
Then put two pieces of wet blotting paper on top of the fiber pad.
-
(d)
Place the equilibrated gel on top of the filter paper.
-
(e)
Carefully place the pre-wetted membrane on top of the gel.
-
(f)
Place the other two pieces of blotting paper over the membrane, creating a ‘sandwich’.
-
(g)
Remove any air bubbles trapped between layers by rolling a pipet over the sandwich.
-
(a)
-
7.
Complete the assembly of the sandwich with the second fiber pad.
-
8.
Close and lock the cassette firmly with minimal disturbance to the gel sandwich.
-
9.
Insert the sandwich into the transfer apparatus. Make sure the membrane is positioned between the gel and anode as most polypeptides eluted from the gels are anions.
-
10.
Add transfer buffer into the transfer apparatus, pour slowly to avoid forming bubbles. Once filled, put the lid on and turn on the power supply set at either 30 V overnight or 100 V for 1 h. To maintain even buffer temperature and ion distribution, add one stir bar.
-
11.
After the transfer, unclamp the apparatus and remove the membrane, and allow it to air dry at room temperature. Mark the side of the membrane that is facing the gel as well as the positions of the pre-stained markers.
6.3.3.3 Confirm Tagging with Anti-FLAG M2 Antibody
-
1.
After transferring the proteins to the membrane, block the membrane by incubating in blocking buffer for 1 h at room temperature on a C2 platform rocking shaker (New Brunswick Scientific, Edison, NJ, USA).
-
2.
Wash the membrane twice for 5 min with wash buffer.
-
3.
Dilute the anti-FLAG M2 primary antibody at 1:5000 in primary antibody buffer, and incubate with the membrane for 1 h on the rocking shaker.
-
4.
Wash the membrane with wash buffer for 10 min three times.
-
5.
Dilute HRP-labelled secondary antibody at 1:20,000 in secondary antibody buffer, and incubate with the membrane for 45 min at room temperature on the rocking shaker.
-
6.
Wash the membrane with wash buffer three times with 10 min for each wash.
6.3.3.4 Imaging Blots
-
1.
Transfer the membrane to a shallow tray and incubate in the Western lightning™ chemiluminescence reagent plus for 5 min by gently shaking in the dark.
-
2.
Drain off the excess chemiluminescence reagent.
-
3.
Cover the membrane with Saran wrap.
-
4.
Expose the membrane to Kodak X-OMAT blue autoradiography film for 30 s. If necessary, expose the membrane for up to another 30 min.
6.3.3.5 Stripping and Re-probing the Blots
-
1.
After imaging, wash the membrane for 15 min in TBS buffer.
-
2.
Incubate the membrane for 30 min at 50 °C in stripping buffer.
-
3.
Wash the membrane for 20 min in TBS buffer.
-
4.
Incubate the membrane for 1 min in the Western lightning ™ chemiluminescence reagent plus. Expose the membrane to Kodak X-OMAT blue autoradiography film for 30 min to 1 h to ensure that the original signal is removed.
-
5.
Wash membrane again for 20 min in TBS buffer. The membrane is now ready for reuse.
-
6.
If successful tagging of the target bait protein is confirmed by immunoblotting, frozen stocks of mammalian cells can then be made and preserved in liquid nitrogen following the steps in the next section.
6.3.4 Freezing, Reviving, and Differentiating Mammalian Cells
Note: Successful use of cell line models is pivotal for studying the structure and function of the mt. While mt fractions can be obtained from other sources such as animal or human tissues, cell line models continue to be the mainstay for studying mt as they not only provide an abundant source of mt samples, but also allow for easy transfection and tagging of ORFs. Here, we will cover how to freeze and grow mammalian cells. In addition, given the link between mt and neurodegenerative diseases, we will cover the steps needed to differentiate neuronal-like cells from the model human SH-SY5Y neuroblastoma cell line.
6.3.4.1 Cryogenic Preservation of Successfully-Tagged Mammalian Cells
-
1.
Take culture dishes from the incubator, aspirate media, and wash twice with 10 mL PBS.
-
2.
Add Versene solution to gently detach cells (3 mL Versene for 150 mm cell culture dish). Put the culture dishes back into the incubator for 10–15 min.
-
3.
Meanwhile, make freezing media by supplementing complete media with 10% DMSO.
-
4.
Take out the dishes, gently tap on the sides to dislodge the cells.
-
5.
Transfer the cell suspension into a 15 mL falcon tube.
-
6.
Rinse the dish with another 6 mL of PBS, add this to the 15 mL falcon tube as well.
-
7.
Centrifuge the cell suspension at 200 xg for 2–5 min, aspirate the supernatant while being careful not to disturb the pellet.
-
8.
Resuspend the cells in freezing media by gently flicking the tube and slowly pipetting up and down.
-
9.
Gently aliquot the cell suspension in cryogenic tubes.
-
10.
Gradual freezing at the rate of 1 °C per minute can be achieved by using a Mr. Frosty freezing container in a − 80 °C freezer overnight.
-
11.
Transfer to liquid nitrogen when feasible.
6.3.4.2 Reviving and Large-Scale Culturing of Mammalian Cells
-
1.
Transfer 10 mL of complete growth medium in a 100 mm diameter tissue culture dish, place it in a 37 °C incubator with 5% CO2, and allow to equilibrate for 15 min.
-
2.
Retrieve one vial containing the frozen cells from liquid nitrogen storage.
-
3.
Immediately thaw the cells by placing it in the 37 °C water bath until only small silver of ice remains in the vial.
-
4.
Dry the vial thoroughly, wipe its outside with 70% ethanol, and then place it into a biological safety cabinet.
-
5.
Retrieve the pre-warmed 100 mm diameter tissue culture dish from the incubator, transfer the thawed cells to the dish by gentle pipetting.
-
6.
Rock the plate gently a few times to distribute the cells evenly, and put back the plate into the incubator for overnight incubation.
-
7.
Change media the next day, and afterwards change every 3 days to maintain the culture.
-
8.
Passage into 150 mm tissue culture dishes when cells have reached ~80% confluence.
6.3.4.3 Differentiation of SH-SY5Y Cells
Note: The SH-SY5Y cell line can be differentiated into neuron-like cells by the addition of all-trans-retinoic acid (at-RA), and brain-derived neurotrophic factor (BDNF) essentially as previously described with minor modifications [20]. Undifferentiated cells can be tagged with lentiviral transduction first and then differentiated.
-
1.
For differentiation, plate cells at a density of 4 × 104 cells/cm2 in DMEM complete media containing 5% FBS.
-
2.
The next day, add differentiation medium containing 5% FBS and 10 μM at-RA to the cells.
-
3.
Change the differentiation medium every day for 5 days.
-
4.
On the 6th day, switch to DMEM supplemented with penicillin (50 u/mL), streptomycin (50 μg/mL), L-glutamine (2 mM), and BDNF (20 ng/mL) but without FBS.
-
5.
Change media every 2–3 days for an additional 7 days.
-
6.
Differentiation can be monitored microscopically for neuron-like morphology.
-
7.
To obtain sufficient mt yield, about 8 × 150 mm dishes of differentiated cells are needed.
6.3.5 Cell Harvesting, Mt Fractionation, and Protein Purification
Note : Prior to harvesting, cells expressing VA-tagged target proteins can be cross-linked using the cell membrane-permeable reversible cross-linking reagent, dithiobis succinimidyl propionate (DSP). Our cross-linking protocol is ideal for attached cells growing in 150 mm diameter dishes.
6.3.5.1 Cell Preparation
-
1.
Remove media from the tissue culture dish.
-
2.
Wash each plate with 10 mL ice-cold PBS twice to remove traces of media.
6.3.5.2 Cross-Linking with DSP
-
1.
Prepare 0.25 M stock of DSP in DMSO.
-
2.
Dilute 0.25 M DSP stock 1:500 in PBS, to a working concentration of 0.5 mM, immediately before use.
-
3.
Add 4 mL of working solution to each plate.
-
4.
Incubate plates at room temperature for 30 min.
-
5.
Quench the cross-linking reaction by adding 400 μL quenching solution, mix the solutions well by pipetting or rocking each plate, and then incubate at room temperature for 10 min, and periodically tap the plate for cells to dissociate.
-
6.
Gently pipette the liquid from the slanted plates to wash cells off the dish, and centrifuge the cell suspension at 1300 xg for 5 min. After discarding the supernatant, either flash-freeze the cell pellet with liquid nitrogen for storage or continue onto mt extraction.
6.3.5.3 Mt Extraction
-
1.
Mt isolation is based on methods previously described [21], with minor modifications.
-
2.
Depending on the cell line, cross-linked cells should be harvested from 2 to 5 × 150 mm tissue culture dishes. After harvesting, decant supernatant and resuspend cells in 1.1 mL of ice-cold hypotonic buffer. Allow cells to swell by incubating for 10 min on ice.
-
3.
Homogenize cells by passing through a syringe fitted with a 22-gauge needle for 30 strokes. Note: alternatively, a pre-chilled Dounce glass tissue homogenizer can be used.
-
4.
Add the homogenate into a conical centrifuge tube containing 800 μL of ice-cold mt stabilization buffer and mix gently. Pellet unbroken cells, nuclei, and large debris by centrifuging at 1300 x g for 5 min at 4 °C.
-
5.
Transfer the supernatant to another conical tube, and centrifuge at 17,000 xg for 15 min at 4 °C. The cytosolic (supernatant) and mt (pellet) fractions can be now used for affinity purification or frozen for future use.
6.3.5.4 Tandem Affinity Purification Using VA-Tag
-
1.
Resuspend the mt pellet in mt lysis buffer, and incubate on ice for 30 min. Centrifuge the sample mixture at 23,000 xg for 20 min at 4 °C. Transfer 100 μL of packed anti-Flag M2 agarose beads (Sigma-Aldrich) or 200 μL of bead slurry into a fresh Eppendorf tube, and wash the beads twice with TBS buffer.
-
2.
Add the harvested supernatant to the tube containing pre-washed anti-FLAG M2 agarose beads, and incubate at 4 °C for 4 h with gentle rotation. Transfer the supernatant along with the beads to a Bio-spin disposable chromatography column.
-
3.
Wash the column four times with 1 mL ice-cold TBS buffer, and wait for 10 min between washes.
-
4.
Incubate the beads with 100 μL of TEV cleavage buffer, 5 μL TEV protease (2 mg/mL), and 2 μL 3×FLAG peptide (5 mg/mL), and rotate overnight at 4 °C.
-
5.
The next day, in a fresh column, add 20 μL Strep-Tactin-Sepharose bead slurry and wash three times with 1 mL TEV cleavage buffer. Remove the top and bottom caps of the TEV cleavage column and drain the eluates into the column containing pre-washed Strep-Tactin-Sepharose beads. Incubate the column at 4 °C for 4 h, followed by washing the column four times with 1 mL TEV buffer, and 10 min wait between washes.
-
6.
Elute with 50 mM ammonium bicarbonate and/or 2 mM D-biotin. Dry the eluted samples with a Speedvac prior to MS analysis, which is described below.
6.3.6 Sample Preparation for MS Analyses
6.3.6.1 Digestion
-
1.
Dilute the dried samples with 50 μL digestion buffer. Add 0.9 μL of 2 mM Tris (2-carboxyethyl) phosphine and incubate at room temperature for 45 min to reduce protein disulfide bonds.
-
2.
Add 1 μL of alkylating solution and incubate the samples in the dark for 40 min. Digest the mixture with trypsin gold (Promega, cat # V5280). Note: typically, we use 12 μg per digestion.
-
3.
Incubate the samples overnight at room temperature with gentle agitation by a rocking shaker. The next day, stop the digestion by adding 1 μL of acetic acid.
6.3.6.2 Desalting
-
1.
Prepare a 96 well plate with 350 μL of wash solution and elution solution. Equilibrate the zip-tip by aspirating 20 μL of elution solution and discarding to waste three times. Wash by aspirating and dispensing three times the washing solution. After washing, draw 20 μL of digested sample into the tip, and then expel the liquid back into the sample, repeat this five to ten times. This will allow peptides to bind to the zip-tip.
-
2.
Wash the zip-tip by drawing 20 μL of wash solution into the tip, then expelling into a waste beaker. Repeat this step three times. This step should remove all unwanted salts and detergents.
-
3.
Add 5–10 μL of the fresh elution solution into a new, clean tube, and draw this solution through the zip-tip before expelling back into the same tube. Repeat this step two to three times in order to disassociate as much of the bound peptide as possible. Note: Be careful not to draw air into the zip tip, which may make it difficult to expel all the solution.
-
4.
Repeat the equilibration, washing, binding and elution steps for two more times. Dry the samples by evaporation. This can be achieved either by air-drying for 2–3 h in a fume hood or under vacuum (e.g. Speedvac).
-
5.
Resuspend the peptides in 20 μL of 0.1% formic acid, and then subject to MS. Note: At this point either proceed with MS or store peptides at −80 °C (or on dry ice) for future use.
6.3.7 MS for Identifying Interacting Proteins
6.3.7.1 Preparation of Nanocolumn
-
1.
Cut a 30–40 cm length of fused-silica capillary tubing. Place the length of tubing in the P-2000 laser puller (Sutter Instrument) so that the clear section is in the mirrored chamber of the puller. The laser is then focused on the center of the tubing and the fused silica can be melted. Select the program on the laser puller that will result in a 2–5 mm tip.
6.3.7.2 Packing the Column
-
1.
Place a small amount (5 mg) of C18 RP packing material (3 mm) in a microcentrifuge tube. Add 1 mL of methanol. Agitate the tube to create a slurry of the packing material. Place the open microcentrifuge tube into the pressure injection cell, Model PC1000, (Next Advance) and secure the lid by tightening the screws.
-
2.
Insert the flat end of the pulled capillary column through the ferrule until it reaches the bottom of the microcentrifuge tube. Tighten the ferrule until the capillary does not move when gently tugged.
-
3.
Adjust the pressure on the helium tank to 800–900 psi. Slowly pressurize the injection cell by opening the valve on the high-pressure line. If the injection cell is pressurized too rapidly, the microcentrifuge tube can rupture. Stop filling the capillary when the packing material reaches 10 cm of the capillary length.
6.3.7.3 LC-MS/MS Analysis
-
1.
Place the packed column in line with the LC-MS instrument. A Proxeon nanoLC pump is used to deliver a stable tip flow rate of 300 nL/min during the peptide separations.
Resuspend the desalted peptide samples in 25 μL of 1% formic acid.
-
2.
Inject 2 μL of each sample onto a Proxeon EASY nLC 1000 nano high-performance liquid chromatography connected to an Orbitrap Elite mass spectrometer.
-
3.
Directly inject samples onto a nano column (C18 column, 10 cm × 75 μm ID, 3 μm, 100 Å) employing a water/acetonitrile/0.1% formic acid gradient as following. Over the course of 100 min, peptides can be separated using: 1% acetonitrile and increasing to 3% acetonitrile in the first 2 min; and a linear gradient is set up from 3% to 24% acetonitrile for 74 min, and then from 24% to 100% acetonitrile for 14 min, followed by 10 min wash at 100% acetonitrile.
-
4.
Eluted peptides can then be directly sprayed into mass spectrometer using positive electrospray ionization (ESI) at an ion source temperature of 250 °C and an ion spray voltage of 2.1 kV.
-
5.
Acquire full-scan MS spectra (m/z 350–2000) in the Orbitrap at 60,000 (m/z 400) resolution.
-
6.
Using the Thermo Fisher Scientific XCalibur software, set the automatic gain control settings at 1e6 for full FTMS (Fourier Transform Mass Spectrometry) scans and 5e4 for MS/MS scans. Sequentially isolate the 15 most intense peptide ions with charge states ≥2 and fragment in the linear ion trap by low-energy collision-induced dissociation (CID).
-
7.
Perform CID when ions intensity exceed 1500 counts, using a normalized collision energy set at 35%, activation Q at 0.250 and an activation time of 10 ms.
6.3.7.4 Protein and Peptide Identification
-
1.
The identified spectra can be obtained from the MS run as raw files. For database searching, convert the raw files into the standard mzXML format. Subject each identified spectrum to peptide spectrum matching against candidate peptides from human protein sequences using the SEQUEST ver. 2.7 rev. 9 search engine.
-
2.
Run the putative matches identified by SEQUEST through the STATQUEST [22] filtering algorithm to assign statistical confidence.
6.3.8 Bioinformatics Pipeline
To detect high-confidence PPIs, we followed two major steps: (1) pre-processing, as well as (2) scoring, benchmarking, and clustering.
6.3.8.1 Pre-processing
-
1.
Filter the identified prey (or interacting) proteins from STATQUEST search engine at 90% probability cut-off.
-
2.
Retain interacting proteins from AP-MS experiments in which targeted bait protein was captured.
-
3.
Remove the most commonly reoccurring prey proteins (e.g., ribosomes, elongation or splicing factors) found in 80% or more in the AP-MS experiments. As well, prey proteins present in both the negative control and AP-MS experiments should be eliminated.
6.3.8.2 Scoring, Benchmarking, and Clustering
-
1.
Apply CompPASS [15, 23] or hypergeometric [13] algorithms to compute statistical scores for each interacting protein pairs.
-
2.
The scored PPIs are then benchmarked against the reference set that is compiled either from the manually curated interactions from literatures or curated human protein complexes containing PPIs from CORUM (Comprehensive Resource of Mammalian Protein Complexes) database. To do so, we consider PPIs within complexes as true positive (TP), while PPIs between complexes as true negative (TN) dataset. Using the TP and TN data, the scored PPIs were then plotted using a Receiver Operate Characteristic (ROC). The area-under-the curve from the ROC plot is then computed and compared to evaluate the scoring performance, where 0.5 indicating completely random and 1 indicating perfect scoring.
-
3.
After eliminating associations below a stringent threshold cut-off from the ROC analysis, the quality of scored PPIs can be assessed by cross-referencing to external sources such the experimentally derived interactions from literature sources or to those deposited in BioGRID database. As well, the newly-derived PPIs from the network can be compared to the computationally predicted interactions from STRING, GeneMANIA, and HumanNet. Interactions from the network can also be validated against the existing large-scale PPI studies by computing the functional coherence and similarity based on Gene Ontology annotations, and benchmarked against literature-curated PPIs (e.g. CORUM) for the performance measures using fivefold cross validation.
-
4.
The selected high-confidence PPIs were then used to generate macromolecular protein complexes using clustering algorithms, such as coreMethod [24] or MCL (Markov Clustering) [25] for downstream analysis.
6.4 Conclusion
In recent years, network biology has risen to the forefront in the exploration of the molecular basis of human health and disease [1, 7,8,9, 12, 13, 15]. Since macromolecular protein complexes are the direct mediators of most cellular functions [26], the systematic elucidation of PPIs and subunit composition of these complexes provides a powerful means for studying the pathophysiology of mt in the context of human health and disease [12, 27]. However, the construction of such a large-scale PPI networks requires reliable and efficient methods for elucidating proteome-wide mt interactions in a human cellular context, so in this chapter, we outlined a tagging-based AP-MS approach that can provide a viable, systematic workflow for discovering new mt links and providing future directions for understanding the role of mt in human physiology and pathology. To confirm the successful integration and expression of VA-tagged mt proteins, immunoblotting was performed in human embryonic kidney (HEK293) cells, using anti-FLAG antibody against the FLAG epitope, as shown in Fig. 6.2.
A key component of the AP-MS workflow is the cross-linking step. Functionally relevant protein interactions in a biological system can be transient, and cross-linking chemicals provide means for capturing the transient interactions by covalently binding the interacting proteins together, facilitating the subsequent capture and identification of the interacting proteins [28]. Given this transient nature of most PPIs, the number of co-purified proteins, or “preys”, of the tagged bait can depend on whether cross-linking was performed. In other words, chemical cross-linking is a critical step towards the detection of transient or weak interactions. As seen in Fig. 6.3, the mt protein, dynamin 1-like protein (DNM1L) was only recovered after cross-linking. This is consistent with past evidence suggesting that cross-linking can help to capture transient interactions in large-scale purifications [29, 30].
Detection of VA-tagged DNM1L in HEK293 cells with or without cross-linking. Silver stained SDS-PAGE gel showing affinity purified VA-tagged mt DNM1L in the presence (+) and absence (−) of chemical cross-linking step. Asterisk indicate the gel band corresponding to the recovery of DNM1L with cross-linking
The AP-MS approach outlined in this chapter has been employed in a 2017 study from our group to purify a set of 27 nuclear-encoded human mt proteins associated with various neurodegenerative disorders. These mt bait proteins were selected broadly in terms of mt processes and were localized to the mt matrix, inner or outer membrane, and intermembrane space. In the assessment of the FLAG mt protein network generated by VA-tagging coupled with MS, 1964 high-confidence associations encompassing 772 prey proteins were identified using precision Orbitrap MS [8]. The overall reliability of the FLAG mt protein network was supported by the observation that nearly 75% of these interactions were confirmed in human HEK293 and SH-SY5Y cell lines by primary antibody immunoprecipitation and MS, as well as the significant enrichment (p < 2.2 × 10−16) for literature-curated human mt PPIs in the FLAG MP network [8]. Notably, among the binding partners of the mt protein baits, an enrichment for 14-3-3 and canonical RNA-binding domains were observed. These domains are associated with mt biogenesis, apoptosis, and stress response, which provides support for the mt relevance of the network generated using the AP-MS workflow [8].
In summary, in this chapter we outlined a tagging-based AP-MS workflow that can be used to effectively identify mt PPIs and elucidate the subunit compositions of mt protein complexes. This system is compatible with Gateway cloning, and importantly, enables efficient and stable delivery of affinity-tagged ORFs into most mammalian cell types. Successful implementation of this work-flow will help to expand the human mt interaction landscape, thereby generating valuable insight into the molecular interactions underlining human mt health and disease.
References
Jiang Y, Wang X (2012) Comparative mitochondrial proteomics: perspective in human diseases. J Hematol Oncol 5:11. https://doi.org/10.1186/1756-8722-5-11
Cassarino DS, Bennett JP (1999) An evaluation of the role of mitochondria in neurodegenerative diseases: mitochondrial mutations and oxidative pathology, protective nuclear responses, and cell death in neurodegeneration. Brain Res Rev 29:1–25. https://doi.org/10.1016/S0165-0173(98)00046-0
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nat Lond 443:787–795. https://doi.org/10.1038/nature05292
Ren J, Pulakat L, Whaley-Connell A, Sowers JR (2010) Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J Mol Med 88:993–1001. https://doi.org/10.1007/s00109-010-0663-9
Ballinger SW (2005) Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med 38:1278–1295. https://doi.org/10.1016/j.freeradbiomed.2005.02.014
Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39:359–407. https://doi.org/10.1146/annurev.genet.39.110304.095751
Floyd BJ, Wilkerson EM, Veling MT et al (2016) Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell 63:621–632. https://doi.org/10.1016/j.molcel.2016.06.033
Malty RH, Aoki H, Kumar A et al (2017) A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-κB signaling. Cell Syst 5:564–577.e12. https://doi.org/10.1016/j.cels.2017.10.010
Basso M, Giraudo S, Corpillo D et al (2004) Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics 4:3943–3952. https://doi.org/10.1002/pmic.200400848
Sokolina K, Kittanakom S, Snider J et al (2017) Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol Syst Biol 13:918
Yao Z, Darowski K, St-Denis N et al (2017) A global analysis of the receptor tyrosine kinase-protein phosphatase interactome. Mol Cell 65:347–360. https://doi.org/10.1016/j.molcel.2016.12.004
Havugimana PC, Hart GT, Nepusz T et al (2012) A census of human soluble protein complexes. Cell 150:1068–1081. https://doi.org/10.1016/j.cell.2012.08.011
Wan C, Borgeson B, Phanse S et al (2015) Panorama of ancient metazoan macromolecular complexes. Nature 525:339–344. https://doi.org/10.1038/nature14877
Hein MY, Hubner NC, Poser I et al (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163:712–723. https://doi.org/10.1016/j.cell.2015.09.053
Huttlin EL, Ting L, Bruckner RJ et al (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162:425–440. https://doi.org/10.1016/j.cell.2015.06.043
Krogan NJ, Cagney G, Yu H et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643. https://doi.org/10.1038/nature04670
Morgenstern M, Stiller SB, Lübbert P et al (2017) Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19:2836–2852. https://doi.org/10.1016/j.celrep.2017.06.014
ten Have S, Boulon S, Ahmad Y, Lamond AI (2011) Mass spectrometry-based immuno-precipitation proteomics – the user’s guide. Proteomics 11:1153–1159. https://doi.org/10.1002/pmic.201000548
Mak AB, Ni Z, Hewel JA et al (2010) A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol Cell Proteomics MCP 9:811–823. https://doi.org/10.1074/mcp.M000002-MCP201
Encinas M, Iglesias M, Liu Y et al (2000) Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 75:991–1003
Clayton DA, Shadel GS (2014) Purification of mitochondria by sucrose step density gradient centrifugation. Cold Spring Harb Protoc 2014:pdb.prot080028. https://doi.org/10.1101/pdb.prot080028
Kislinger T, Cox B, Kannan A et al (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125:173–186. https://doi.org/10.1016/j.cell.2006.01.044
Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. https://doi.org/10.1016/j.cell.2009.04.042
Leung HCM, Xiang Q, Yiu SM, Chin FYL (2009) Predicting protein complexes from PPI data: a core-attachment approach. J Comput Biol J Comput Mol Cell Biol 16:133–144. https://doi.org/10.1089/cmb.2008.01TT
van Dongen S, Abreu-Goodger C (2012) Using MCL to extract clusters from networks. Methods Mol Biol Clifton NJ 804:281–295. https://doi.org/10.1007/978-1-61779-361-5_15
Westermarck J, Ivaska J, Corthals GL (2013) Identification of protein interactions involved in cellular signaling. Mol Cell Proteomics MCP 12:1752–1763. https://doi.org/10.1074/mcp.R113.027771
Sahni N, Yi S, Taipale M et al (2015) Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161:647–660. https://doi.org/10.1016/j.cell.2015.04.013
Yang B, Tang S, Ma C et al (2017) Spontaneous and specific chemical cross-linking in live cells to capture and identify protein interactions. Nat Commun 8:2240. https://doi.org/10.1038/s41467-017-02409-z
Vlasblom J, Jin K, Kassir S, Babu M (2014) Exploring mitochondrial system properties of neurodegenerative diseases through interactome mapping. J Proteome 100:8–24. https://doi.org/10.1016/j.jprot.2013.11.008
Hyung S-J, Ruotolo BT (2012) Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics 12:1547–1564. https://doi.org/10.1002/pmic.201100520
Acknowledgments
We thank Babu’s group for their helpful comments. This work was funded by National Institutes of Health (R01GM106019) and Canadian Institutes of Health Research (MOP-125952; RSN-124512, 132191; FDN-154318) to MB. MB is a CIHR New Investigator (MSH-130178).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Wu, Z., Malty, R., Moutaoufik, M.T., Zhang, Q., Jessulat, M., Babu, M. (2019). A Tag-Based Affinity Purification Mass Spectrometry Workflow for Systematic Isolation of the Human Mitochondrial Protein Complexes. In: Urbani, A., Babu, M. (eds) Mitochondria in Health and in Sickness. Advances in Experimental Medicine and Biology, vol 1158. Springer, Singapore. https://doi.org/10.1007/978-981-13-8367-0_6
Download citation
DOI: https://doi.org/10.1007/978-981-13-8367-0_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8366-3
Online ISBN: 978-981-13-8367-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)