Abstract
As a type of novel noncoding RNAs, circular RNAs (circRNAs) have attracted great interest due to its different characteristics from linear RNAs. They are abundantly and stably present in the transcriptome of eukaryotic cells, with development stage specificity and high conservatism. Because circRNAs are not easily degraded by exonuclease RNase R, they can exist more stably in body fluids than linear RNAs. Based on these unique conditions, circRNAs have great potential value as clinical diagnostic and prognostic markers. As the research deepens, more and more evidences suggest that circRNAs may be closely associated with many diseases, especially cancer. Numerous studies have demonstrated the abnormal expression of circRNAs in cancer, and they can regulate the occurrence and progression of cancer by targeting key genes. Abundant circRNAs in tissues and cells can be released into saliva and blood. It is undeniable that circRNAs are a class of promising future biomarkers for cancer diagnosis and prognosis. Here we summarize the researches on circRNAs and cancer over the past few years. We expect this summary to be a stepping stone to further exploration of possible circRNAs as cancer biomarkers.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
With the development of biotechnology and computer technology, more and more “invisible substances” are exposed in the organism, such as circular RNAs (circRNAs). Although do not encode proteins, they are indispensable in regulatory processes. As the latest research hotspot, circRNAs are a group of newly validated noncoding RNA molecules which form a covalent ring structure instead of having a cap structure at the 5′ end and poly (A) tail at the 3′ end. It was found in RNA viruses as early as the 1970s and once thought to be by-products of aberrant RNA splicing due to low expression levels [1, 2]. With the rapid development of bioinformatics and sequencing technologies, large-scale analysis of transcriptome data becomes possible, and the characteristics and functions of circRNAs, a sort of ancient and conservative molecules, are being gradually unveiled. It is well known that circRNAs are richly expressed in various tissues and cells throughout the human body and play vital roles in regulating physiological and pathological processes [3, 4]. CircRNAs are abundant in the transcriptome of eukaryotic cells and conservative in species. They are also stable in expression and exhibit tissue and development stage specificity [5,6,7]. In 2013, professor William R. Jeck’s team detected more than 25,000 circRNAs in human fibroblasts, while in the same year, Memczak et al. identified 1950 human circRNAs and 1903 mice circRNAs through RNA-seq data combined with the human leukocyte database [3]. Unlike linear RNAs, circRNAs are not easily degraded by exonuclease RNase R in body fluids; therefore they have the potential application value as clinical diagnostic and prognostic markers [8].
Cancer is characterized by high mortality, and its incidence is increasing in recent years [9]. It is one of the major diseases threatening human life and social economic development and therefore brings the focus of attention [10, 11]. A survey on the incidence of global cancer in 2012 showed that there are approximately 14.1 million new cancer cases and 8.2 million cancer deaths worldwide each year. Lung cancer and breast cancer are the most common types of cancer [9, 12]. The essence of tumor formation is a kind of genetic disease, of which tissues infiltration and metastasis are the major features [13,14,15]. Its pathogenesis refers to multiple steps and mechanisms. New approaches for early diagnosis and treatment of cancer have been sought for a long time. Biomarkers for cancer have emerged as a class of molecules closely related to the development and progression of tumors [16]. They play key roles in many aspects such as early diagnosis, therapeutic monitoring, and prognostic evaluation [17].
It is universally known that microRNAs (miRNAs) control a large number of biological processes by direct interaction with their target mRNAs. This regulation can be achieved by inhibiting translation or by triggering the degradation of the target mRNA [18, 19]. Multiple researches have shown that miRNAs play an important regulatory role in the development of tumors [20,21,22]. Recently independent experiments have demonstrated that circRNAs can serve as sponges for miRNAs, which means that they can bind with miRNAs and thus suppress the function of miRNAs [3, 23]. These findings trigger the exploration of the potential regulatory effect of circRNAs on cancer. Technologies such as circRNAs chip and qRT-PCR have been widely used in the studies related to circRNAs and cancer. So far, many circRNAs have been found to be closely related to cancer and bring new dawn of the early diagnosis and treatment of cancer [24].
2 The Characteristics of CircRNAs
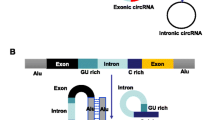
There are numerous types of circRNAs, which are huge in number and widely distributed in the organisms. The circRNAs currently found are mainly divided into three categories according to their origin: exonic circRNAs (ecRNAs), intronic circRNAs (ciRNAs), and exon-intron circular RNAs (EIciRNAs) composed of exons and introns. They are characterized by extremely high stability, strong evolutionary conservation, and unique temporal and spatial expression. Studies have found that the expression levels of circRNAs are ten times higher than their linear isomers [25]. The half-life of circRNAs with the peculiar loop structure is more than 48 h, which is far much longer than the linear mRNAs (10 h in average); this is why the circRNAs can be more stable in tissues, cells, and body fluids [25, 26]. However, the expression of the circRNAs is not dependent on the linear mRNA expression of its parental genes but changes with the life activities such as growth and senescence. Moreover, the type and content of circRNAs in different cells and tissues are also distinct [27, 28]. When the abundance of the circRNAs with conservative nucleotide sequences increases, they can compete with other RNA or miRNAs by competitively binding to the RNA-binding protein (RBP) [3]. These properties give circRNAs a unique advantage as diagnostic and prognostic markers for clinical diseases. CircRNAs are widely involved in the processes of physiological and pathological regulation of human beings. Large amounts of circRNAs in organisms influence basic life activities such as cell proliferation, cycle progression, cell senescence, and apoptosis by regulating gene expression.
The mechanisms of the discovered circRNAs mainly include the following three types:
-
1.
As molecular sponges of miRNAs: this is a relatively common function of circRNAs and the most promising direction in circRNAs researches. CircRNAs themselves contain at least one miRNA binding site and therefore can serve as miRNA sponges to regulate the expression of target genes which are inhibited by miRNAs through ceRNAs [3]. The ceRNAs network in the living body is complex, and any minor changes can affect gene expression and induce tumorigenesis. Therefore, the competitive combination of circRNAs is essential to maintain the balance of the ceRNAs network.
-
2.
Regulate gene transcription and cleavage: the mechanism of this process is diverse. It can not only enhance the expression of parental gene through miRNAs but also exert a positive feedback on its parental genes through interaction with RNA polymerase II. It is also possible to regulate parental gene expression through competitive splicing factors. CircRNAs can regulate parental genes through positive feedback and negative feedback control at different expression levels [5, 29, 30].
-
3.
Interact with RNA-binding proteins: circRNAs can bind stably with RNA-binding proteins such as AGO (argonaute), RNA polymerase II, muscleblind protein (MBL), variable factor QKI (Quaking), and eukaryotic translation initiation factor 4A3 (EIF4A3). They can store and transport RBP to compete with RBP substrate for its binding site and thereby regulate the activity of RBP and interfere with the normal function of a protein in a direct or indirect way [3, 5, 30,31,32].
Since the first discovery of circRNAs in 1976, more and more mature technologies such as high-throughput sequencing and gene chip have been applied to explore their biological functions. On the other hand, the establishment and enrichment of the database also provide extremely convenient conditions for the in-depth study of the circRNAs. The current databases of circRNAs include circRNABase, circBase, Circ2Traits, and CircNet. Through these databases, we can query circRNAs sequences, genome annotations, expression profiles, miRNA-circRNA interaction networks, and related disease information. With these supports we have made tremendous progress in the study of circRNAs.
3 CircRNAs in Cancer
3.1 Lung Cancer
The changes in lifestyle, dietary pattern, and deteriorating environment have led to a rising trend in the prevalence of cancer patients. Worldwide, the incidence and mortality of lung cancer account for the third most severe human tumor. Meanwhile it is one of the most malignant tumors in the world [33]. According to the World Cancer Report 2014 released by WHO, there are 1.82 million new lung cancer cases in 2012 and 1.59 million deaths caused by lung cancer, accounting for 13% and 19.4% of the incidence and deaths of all malignant tumors [34]. The epidemiological data manifest that the morbidity and mortality rates of lung cancer are both the highest among global malignant tumors. More than 220,000 cases were diagnosed with lung cancer in 2015, and the number of patients who died of lung cancer in the same year exceeded 158,000 [35]. Moreover, about 85% of lung cancers are non-small cell lung cancer, and 25–30% of non-small cell lung cancers are squamous cell carcinomas [36]. At present, the common clinical treatment methods, including surgical resection, radiation therapy, and chemotherapy, cannot reduce the incidence of lung cancer. The 5-year survival rate of lung cancer patients is only 15.9% [37]. Therefore, innovative new treatments are urgently needed.
A variety of circRNAs with high expression in lung cancer have been found. Hsa_circ_0000064 is upregulated in lung cancer tissues and lung cancer cell lines A549 and H1229. The abnormal expression of hsa_circ_0000064 is closely related to the clinical features such as tumor lymph node metastasis and TNM staging [38]. The high expression of circRNA-100876 in non-small cell lung cancer (NSCLC) is closely related to the lymph node metastasis and tumor stage of lung cancer. Additionally, the overall survival time of NSCLC patients with high circ_100876 expression is significantly shorter [39]. Circ-HIPK3 can be detected in the NSCLC cell lines H1299, H827, H1975, H2170, H520, and H1650. Experiments have shown that circ-HIPK3 can regulate the expression level of insulin-like growth factor1(IGF1) and promote cell proliferation by binding to miR-379 in NCI-H1299 and NCI-H2170 cells [40]. Hsa_circ_0013958 is significantly upregulated in histiocytes and plasma of lung adenocarcinoma patients and shows statistically significant correlation with lymph node metastasis and tumor staging [41]. By applying high-throughput sequencing on tumor and adjacent tissue from four cases with nonsmoking early lung adenocarcinoma, Zhao et al. detected more than 300 circRNAs differentially expressed in tumor tissues and later verified 5 of them by RT-qPCR. Consistent with the results of the chip, it provided potential targets for early diagnosis and treatment of early-stage lung adenocarcinoma [42]. Cdr1as (ciRS7) contains more than 70 selective binding sites for miRNAs, which can strongly inhibit the activity of miR-7 and hence activate the miR-7-regulated genes. It has a regulatory effect on many diseases [23]. Importantly, similar to liver cancer, Cdr1as can also inhibit tumors in lung cancer by binding to miR-7 [43]. The circ-ITCH is significantly reduced and can act as miR-7 and miR-214 sponge in lung cancer. It can inhibit the activation of Wnt/β-catenin signaling pathway by enhancing the expression of the ITCH gene and thereby suppresses the proliferation of lung cancer cells [44]. A new mechanism of cinnamaldehyde intervention on NSCLC through hsa_circ_0043256/miR-1252/ITCH axis was proposed in another work. The gene expression of ITCH is found positively correlated with hsa_circ_0043256. As a miR-1252 sponge, hsa_circ_0043256 is significantly downregulated, weakening the inhibitory effect of cinnamaldehyde, when cinnamaldehyde is used to block the Wnt/β-catenin signaling pathway [45].
3.2 Gastrointestinal Cancer
Gastrointestinal cancer is one of the most common malignancies which seriously threatens the life and health of human beings [46]. The gastric and colorectal cancers are the most common in this kind of diseases. Studies have shown that the incidence and mortality of gastric cancer rank second in China, which is second only to lung cancer, while colorectal cancer ranks fifth among all malignancies [47]. The 5-year survival rate of advanced gastric cancer is less than 30% [48]. Tumor markers including AFP, CEA, CA19-9, and CA50 are commonly used to assist the clinical diagnosis and prognosis of gastrointestinal tumors. In order to compensate for the lack of tumor detection markers, attempts are made to find circular RNAs with diagnostic and prognostic values.
3.2.1 Gastric Cancer
A study of circRNAs in gastric cancer (GC) tissues and paracancerous tissues identified 467 differentially expressed circRNAs, among which expression of 214 circRNAs were significantly increased and expression of 253 were significantly decreased. Most of the circRNAs could be detected with corresponding miRNA binding sites [49]. Circ-PVT1 is a highly expressed circRNA in GC and a potential independent index for evaluating the prognosis of GC. By analyzing clinical data and tumor tissues of 187 patients with GC, it was found that the survival rate of patients with high circ-PVT1 expression was markedly higher. The promotive effect of circ-PVT1 on GC may act through attenuating the inhibitory effect of miR-125 on cell proliferation by combining with it. The experiment also demonstrated that reducing the expression of circ-PVT1 could inhibit the proliferation of GC cells [50]. In contrast, hsa_circ_0000096 was found obviously downregulated in GC cell lines and tissues compared with normal gastric epithelial cells and paired adjacent non-tumor tissues. It is supposed that hsa_circ_0000096 might interact with miRNAs through endogenous competition and thereby affect GC cell growth and migration by interfering with cell cycle and expression of migration-related protein [51]. Screened by database and verified by qRT-PCR, hsa_circ_002059 was found significantly downregulated in GC tissue compared with the adjacent non-tumor tissue by Li et al. The authors noticed that 10 days after GC tissue resection, the expression of plasma has_circ_002059 was detected to be higher than that before surgery. Low expression level of has_circ_002059 was significantly associated with distant metastases, TNM staging, gender, and age [52]. The combination of hsa_circ_0000096 and hsa_circ_002509 can considerably improve the diagnosis of gastric cancer [51]. Zhang et al. established a prediction system of early recurrence for patients with stage III GC based on four selected circRNAs: hsa_circ_101308, has_circ_104423, hsa_circ_104916, and hsa_circ_100269. The area under the curve (AUC) could reach 0.763 and 0.711 by the test of two centers and could rise to 0.866 and 0.818 by joining TNM staging, which indicated that this circRNA-based predictive model is an effective assessment of the risk of early recurrence after radical gastrectomy for GC. However, this system still needs to be further verified by prospective and multicenter research [53]. Other circRNAs with abnormal expression profiles in GC include has_circ_0001649 (the expression level is significantly downregulated in GC tissue and upregulated in serum after operation) [54], has_circ_0044516 (the expression level is significantly upregulated in GC tissue) [55], has_circ_0014717, and hsa_circ_0000190 (the expression level is significantly downregulated in GC tissues and considered related to distant metastasis, tumor staging, CA19-9) [56, 57].
3.2.2 Colorectal Cancer
Dietmar Pils’s team compared circRNAs expression levels between fibrotic lung and normal lung tissue in patients with idiopathic pulmonary fibrosis and ovarian tumor cells and normal ovarian epithelial cells and confirmed that circRNA abundance was negatively correlated with cell proliferation. The expression of circRNAs in colorectal cancer (CRC) tissues were significantly lower than that in normal tissues, and the expression level were also lower in CRC cell lines. The investigators tested four circRNAs (circ_0817, circ_3204, circ_6229, circ_7374) low-expressed in clinical specimens in colon cancer cell lines together with their corresponding linear RNAs. The expression ratio of the four circRNAs to their parental linear RNAs was lower. Finally, the researchers verified the relationship between the expression of circRNAs/corresponding linear RNAs and cell proliferation, confirming that the cell proliferation rate was negatively correlated with the expression circRNAs [58]. As a circRNA generated from the exon 5–11 of BANP gene, circ-BANP is highly expressed in CRC. Knocking down of circ-BANP can significantly reduce the proliferation of CRC cells. Circ-BANP may play an important regulatory role in CRC cells and may serve as a marker for prognosis and treatment of CRC [59]. Recently, Guo et al. discovered a novel abnormal circRNA, hsa_circ_0000069, by employing unsupervised hierarchical clustering analysis. They determined that hsa_circ_0000069 is highly expressed in CRC through quantitative PCR analysis of 30 paired CRC tissues and adjacent noncancerous tissues and is closely related to the patient’s age, tumor size, lymph node metastasis, and TNM staging. Functional analysis by using specifically designed siRNA in CRC cells confirms that knocking down hsa_circ_0000069 markedly inhibits cell proliferation, migration, and invasion and induces G0/G1 arrest in vitro [60]. Another study has confirmed that hsa_circ_001569 plays a positive regulatory role in cell proliferation and invasion of CRC. The authors found that the expression of has_circ_001569 is elevated in CRC tissues and correlated with tumor volume, TNM staging, and prognosis. It was further confirmed that hsa_circ_001569 can also be used as a “sponge” to adsorb miR-145. This is why the expression of has_circ_001569 and miR-145 are negatively correlated in CRC tissue. Upregulated circ_001569 increases the expression of miR-145 target genes E2F5, BAG4, and FMNL2 and hence enhances the proliferation and invasion of CRC cells, which in turn promotes the progression of colorectal cancer [61]. Circ-CCDC66 is a newly discovered circRNA which is encoded by the CCDC66 gene. Hsiao et al. found that the expression of circ-CCDC66 was increased in polyps and CRC and was negatively correlated with the prognosis. Inhibition of circ-CCDC66 expression in vitro significantly reduces the tumor volume in nude mice, suggesting that circ-CCDC66 can regulate multiple pathological processes including cell proliferation, invasion, migration, and anchorage-independent growth. This function is achieved by acting as the sponge of miR-33b and miR-93. Experiments showed that knocking down circ-CCDC66 can impede tumor proliferation and invasion in vivo. All of these findings indicate that circ-CCDC66 plays an important role in progression and metastasis of CRC [62]. Other circRNAs such as circ-ITCH can also exert the function of miRNA sponge to suppress tumor proliferation. A research based on 45 specimens of CRC found that circ-ITCH expression is abnormally lower than that of paracancerous tissue. The bioinformatics analysis predicted that circ-ITCH and its parent gene ITCH possess the same miR-214, miR-7, and miR-20a binding sites, and the firefly luciferase reporter assay in CRC cell lines HCT116 and SW480 confirmed that circ-ITCH can obstruct the Wnt signaling pathway to inhibit cancer cell proliferation by competitively absorbing miR-7 and miR-20a [63].
3.2.3 Esophageal Cancer
Researches on the regulation mechanism of circRNAs in esophageal cancer suggest that many of them can function as miRNAs sponges. The radiation resistance obtained during radiotherapy is considered to be the most important factor that affects the therapeutic effect and stimulates local tumor recurrence. Based on this, Su et al. explored the differentially expressed circRNAs in radioresistant esophageal cancer cells by using expression profiling and bioinformatics analysis, in order to reveal the possible circRNAs involved in the generation of radiation resistance during treatment of esophageal cancer. They initially identified 57 remarkable upregulated and 17 remarkable downregulated circRNAs in 3752 candidate circRNAs (fold change ≥ 2.0 and P < 0.05), of which 9 were validated by real-time qPCR. Supplemented by gene ontology analysis, the authors confirmed a large number of target genes (including most miRNAs) participating in this biological process. Among them, more than 400 target genes are enriched in the Wnt signaling pathway. Circ_001059 and circRNA_000167 are the two largest nodes of the co-expression network of circRNA/microRNA [64]. Based on the detection of circRNAs expression in 684 cases of esophageal squamous cell carcinoma (ESCC) patients and their matched paracancerous tissues, circ-ITCH was also found to play a role as molecular sponge of miR-7, miR-17, and miR-214 in esophageal cancer. Consistent with the mechanism of action in colorectal cancer, increased ITCH promotes ubiquitin-mediated degradation of Dvl2 and reduces the expression of the oncogene c-myc that inhibits the Wnt signaling pathway and ultimately suppresses the tumor growth [65]. In 51 cases of ESCC patients with different staging, the expression of hsa_circ_0067934 in the tumor tissue is obviously higher than that in paired paracancerous tissues; and the high expression level of hsa_circ_0067934 is associated with the tumor stage (P = 0.025). ] Higher stage of the tumor tissue is companioned with higher expression of hsa_circ_0067934. Interference of hsa_circ_0067934 expression by siRNA in vitro suppresses the proliferation and migration of ESCC cells and blocks cell cycle progression. Cell component analysis and fluorescence in situ hybridization confirm that the circRNAs are mainly located in the cytoplasm [66].
3.2.4 Pancreatic Cancer
Pancreatic cancer (PC) is a common cancer, but there is still lack of reliable biological markers for early diagnosis and evaluation of prognosis. Current studies on the relationship between circRNAs and PC are insufficient. Recently, researchers have explored the expression profiles of circRNAs in four pancreatic ductal adenocarcinoma (PDAC) samples and matched adjacent normal tissues. The result revealed that a large number of circRNAs are abnormally expressed in PDAC, suggesting that they may be involved in the initiation and progression of PDAC. This discovery provides potential biological targets for the diagnosis and treatment of PDAC [67]. Li et al. analyzed the tissue samples from six patients with PDAC by microarray technology and found that compared to normal pancreatic tissues, abnormal expression of circRNAs cluster was gathered in pancreatic cancer tissues. 209 upregulated and 142 downregulated circRNAs were screened out. Subsequently, 7 circRNAs were analyzed with quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) in 20 groups of PC and paracancerous tissues to confirm that the results were consistent with the microarray. GO analysis and pathway analysis suggested that some of the dysregulated circRNAs are involved in molecular biological processes in pancreatic cancer, including influence on endocytosis and meditation of abnormal expression of VEGF pathway. These results reveal the indispensable role of circRNAs in the malignant biological behavior of PC [68].
3.3 Hepatic Cancer
Hepatic cancer generally refers to malignant tumor which originates in hepatocytes or intrahepatic bile duct epithelium [47]. As one of the common malignancies of the digestive system, hepatic cancer is the fifth most common cancer with the third most common mortality in the world [69]. About 250,000 people worldwide are estimated to lose their lives every year due to hepatic cancer, among which China accounts for about 45% [70, 71]. The 5-year survival rate of advanced hepatic cancer is about 10%. Due to the lack of effective early diagnostic tools, only 30–40% of the patients can be diagnosed and take appropriate treatment in early stage [72, 73].
A series of circRNAs associated with hepatic cancer are gradually discovered. A noteworthy downregulation of hsa_circ_0001649 was found associated with inhibition of cell proliferation [74]. In contrast, the upregulated expression of hsa_circ_0005075 and Cdr1as promotes cell adhesion and proliferation, respectively [75, 76]. As the inhibitor and sponge of miR-7, Cdr1as can indirectly interrupt the PI3K/Akt/mTOR signaling pathway by targeting miR-7 [77]. It has been suggested that Cdr1as may be employed as a prognostic biomarker for hepatic carcinoma and a therapeutic target for microvascular infiltration [77]. Huang et al. detected another potential marker hsa_circ_100338 by applying a circRNA microarray and verified miR-141-3p as its direct downstream target through computer calculation and experimental analysis. High expression level of hsa_circ_100338 indicates the metastasis progression and meanwhile makes an influence on the cumulative survival rate [78]. A circRNA-miRNA-mRNA network is constructed with five upregulation circRNAs in hepatic cancer. This network proposes that high levels of circFUT8 (hsa_circ_101368, hsa_circ_0003028), circ-ZFR (hsa_circ_103809, hsa_circ_10072088), and circ-IOP11 (hsa_circ_103847, hsa_circ_0007915) expression are probably associated with the progression of hepatic cancer [79]. Researchers focusing on the genetics and epidemiology of hepatocellular carcinoma (HCC) discovered that the expression of cir-ITCH in HCC tissues is significantly lower than that in matched paracancerous tissues; and suggested that HCC patients with a relatively high cir-ITCH expression have a better prognosis. Collectively, the results revealed that cir-ITCH has an inhibitory action on HCC and may be a hopeful biomarker for the assessment of susceptibility and prognosis in patients with HCC [80]. Modulating function of hsa_circ_0015756 on hepatoblastoma cell by acting as a miR-1250-3p sponge has also been found based on circRNAs microarray analysis. Silencing hsa_circ_0015756 reduces viability, proliferation, and invasiveness of hepatoblastoma cells [81]. Fu et al. conducted a series of experiments to prove the point that the expression level of hsa_circ_0005986 is related to the tumor size, BCLC staging, and microvascular infiltration. The underlying mechanism of hsa_circ_0005986-regulated carcinogenesis of HCC is by modulating Notch1 expression through interaction with miR-129-5p [82]. They further showed the low expression of hsa_circ_0004018 is associated with AFP level in serum, diameter, and differentiation of tumor and Barcelona clinical stage in HCC. Remarkably, hsa_circ_0004018 has the specific expression characteristics indicative of different HCC stage in various chronic liver diseases [83]. Based on all these findings, there are reasons to believe that circRNAs combined with traditional biomarkers can make a more accurate diagnostic method for hepatic cancer.
3.4 Gynecologic Cancer
Gynecologic cancer is closely related to women mainly including breast cancer, endometrial cancer, cervical cancer, and ovarian cancer. The incidence of gynecological tumors has increased year by year, which causes serious harm to women’s physical and mental health. Breast cancer has the highest incidence of women malignancy tumors in the world, which is also the biggest factor leading to the death of female cancer patients [84]. Breast cancer accounts for 23% of the global new female cancer cases and accounts for 14% of the total number of global cancer deaths [84, 85]. Ovarian cancer represents the highest mortality rate among gynecologic cancer with no less than 204,000 new cases and 125,000 deaths cases per year [86]. Only a small proportion of patients can be diagnosed in the early stage due to its concealed characteristics and lack of effective early diagnosis. In fact, more than 70% of the patients are diagnosed in advanced stage; and their 5-year survival rate is lower than 30% [87]. Endometrial cancer originates from the endometrial epithelium. Its mortality is second only to ovarian cancer. It accounts for 7% of all malignant tumors in women, and the incidence of malignant tumors in female reproductive system is as high as 20–30% [88]. Although early-stage endometrial cancer has a good prognosis, the median survival time for advanced or recurrent cases does not exceed 1 year [86]. Clinical diagnosis of gynecologic malignancies is mainly by physical signs, tumor markers, B-ultrasonography, computed tomography (CT), and magnetic resonance imaging (MRI). Common gynecological tumor markers include cancer markers such as cancer antigen 125 and 19-9 (CA-125 and CA19-9), but their specificity is doubtful. Researches have been trying to find high-specificity diagnostic markers among circRNAs, which is of great significance for timely treatment of gynecological cancer, reduction of metastasis, and improvement of prognosis.
3.4.1 Breast Cancer
As previously mentioned, Cdr1as can indirectly regulate the expression of miR-7 target gene, while Cdr1as/miR-7 can affect tumorigenesis and development of tumor through multiple pathways. Early studies have showed that the expression level of endogenous miR-7 is negatively correlated with Pak1 and is positively correlated with homeodomain transcription factor HOXD10. In the transformation process of breast cancer from high invasion phenotype to low invasion phenotype, Pak1 protein levels gradually increase, while miR-7 and HOXD10 gradually decrease. In highly invasive breast cancer cells, miR-7 can inhibit their proliferation activity, invasiveness, and tumorigenic potential. These indicate that the miR-7/Pak1 pathway may play an important role in the development of breast cancer. Therefore, Cdr1as is considered also involved in the regulation of breast cancer [89]. A total of 1155 differentially expressed circRNAs were screened out in 51 breast cancer patients by using the whole genome transcript profile technique, of which 715 were upregulated and 440 were downregulated. The expression levels of hsa_circ_103110, hsa_circ_104689, and hsa_circ_104821 were elevated in breast cancer tissues, while the expression levels of hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 were downregulated among the selected circRNAs. Further investigation of the circRNAs targeting complementary miRNAs response elements revealed that progesterone receptor (PR)-negative was related to the upregulation of hsa_circ_104689 and hsa_circ_104821 and the downregulation of hsa_circ_406697. The diagnostic accuracy of hsa_circ_100219 was the highest with the AUC of 0.78 (95% CI: 0.69–0.88). Combined hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 had a higher judgment value for judging breast cancer (AUC: 0.82, 95% CI: 0.73–0.90) [90]. With the purpose to investigate the expression profile and possible regulatory mechanisms of oncogenic circRNAs in breast cancer, Liang et al. used circRNA microarray to screen abnormally expressed circRNAs in breast cancer tissues and found that circ-ABCB10 was highly expressed in breast cancer tissues. The authors then verify the result of the chip by using a large amount of samples. The loss-of-function experiments in vitro demonstrated that subtraction of circ-ABCB10 level in breast cancer cells can inhibit cell proliferation and promote apoptosis. The bioinformatics technique was used to predict the existence of complementary sequences in circ-ABCB10 and miR-1271, which was then verified by luciferase reporter assays. Finally, it was confirmed in breast cancer cells that the inhibition of miR-1271 can restore function of circ-ABCB10, demonstrating the spongy effect of circ-ABCB10 on miR-1271 [91]. In a study investigating whether hypoxia regulates proliferation through circRNAs, a hypoxic model in breast cancer cells was established. The increased expression of circ-DENND4C was detected under hypoxic conditions, whereas knockdown of HIF1α could reduce the expression of circ-DENND4C. This confirmed the correlation between circ-DENND4C and HIF1α in the hypoxic model. It was further found that knockdown circ-DENND4C can inhibit the abnormal proliferation of breast cancer cells in anoxic environment, indicating that circ-DENND4C has the function of promoting breast cancer cell proliferation under hypoxic conditions. Finally, the expression level of circ-DENND4C was found related to the tumor volume, and the larger tumors contained more circ-DENND4C [92]. As a noteworthy downregulated circRNA in breast cancer cells, when the expression of circ_000911 is enhanced, the proliferation, migration, and invasion ability of breast cancer cells are all inhibited, and meanwhile the cell apoptosis accelerates. The miR-449a was identified as a related miRNA to circ_000911 by using a biotin-labeled probe method. Overexpression of cir_000911 in breast cancer could increase the expression of Notch1, which is a functional target of miR-449a. Signal transduction reporter array and western blot analysis confirmed that NF-κB signaling transduction is a functional target of the circ_000911/miR-449a pathway [93].
3.4.2 Reproductive System Tumors
By RNA sequencing of primary ovarian cancer, peritoneal metastases, and lymph node metastases in three patients with ovarian cancer, circRNAs with significant differences in expression in epithelial ovarian cancer were found, including many new genes such as HIPK2/3 and ZKSCAN1. The number of differentially expressed circRNAs is much higher than that of the corresponding linear mRNA in metastatic lesions. In addition, various cancer-associated signaling pathways including NF-κB, PI3K/Akt, and TGF-β have the opposite expression trends in circRNAs and linear mRNAs. Consensus of circRNAs expression provides new candidates for cancer treatment and prognosis [94]. Endometrial cancer (EC) and cervical cancer also belong to the female reproductive system malignancy. Lately, researchers used RNA sequencing technology to identify EC-specific circular transcriptomes. By comparison, the overall abundance of circRNAs in EC (14,707) was lower than that in normal endometrium (21,340). On this basis the researchers identified 120 differentially expressed circRNAs between EC tissues and normal endometrial tissues by collecting and analyzing samples from 6 EC patients, in which unique hotspot genes, such as cancer-specific ESR1 circular isoforms, were regarded with the value of EC diagnosis and progress detection. The circular isoform of DNAH14 may be involved in the regulation of tumor-associated pathways. The DMD and DMBT1 genes undergone significant changes during generation of the circular transcript, suggesting that they may be involved in the pathological changes of EC [95]. The work of Abdelmohsen’s team demonstrated an example of a functional model of a protein regulation through circRNAs endogenous binding in cervical cancer. They used the RIP assay to identify circRNAs interacting with HuR in Hela cells; and the most obviously changed candidate was circPABPN1(hsa_circ_0031288). Excess circ-PABPN1 can prevent the binding of HuR to linear PABPN1 mRNA and hence inhibits the translation of HuR [96].
3.5 Other Cancer
Some circRNAs also have the potential of biological markers in other tumors. For instance, the expression level of has_circ_104912 is significantly downregulated in laryngeal squamous cell carcinoma, while the expression level of has_circ_100855 is significantly increased [97]. circ_100290 can function in oral squamous cell carcinoma as a molecular sponge of miR-29 family [98]; high expression of circ-TTBK2 in glioma tissue may promote the development of glioma [99].
Acute pregranulocyte leukemia chromosomal translocation t(15;17)(q24;q21) leads to the formation of a key oncogenic fusion protein PML-RARα, and the circRNA, f-circRNA, forms after the translocation of this chromosome can also be carcinogenic [100]. Circ-TRIM24 (hsa_circ_0082582) and circ-FAM169A (hsa_circ_0007158) are significantly downregulated in bladder cancer tissues, while circ-BC048201 (hsa_circ_0061265), circ-PTK2 (hsa_circ_0005273), circ-ZFR (hsa_circ_0072088), and circ-TCF25 (hsa_circ_0041103) are significantly upregulated, and the circTCF25-miR-103a-3p/miR-107-CDK6 pathway is suggested as an important regulatory axis in bladder cancer [101]. 23 high- and 48 low-expression circRNAs and their corresponding binding sites for 354 bindable miRNA sequences are identified in basal cell carcinoma [102]. The phenomenon of abnormal expression of these circRNAs in various tumor tissues indicates that circRNAs have an inseparable relationship with the occurrence and development of tumors.
4 Conclusion and Prospect
CircRNAs is a type of closed-circular RNA molecule widely distributed in the transcriptome; they participate in the regulation of numerous biological activities in organisms. CircRNAs are not easily degraded by nucleases and are more stable than linear RNAs, which provide a foundation for them to be novel biomarkers for tumor diagnosis. The regulatory expression mechanisms of circRNAs are diverse. They can serve as “miRNA sponges” to perform posttranscriptional regulation by competitively binding to miRNAs, interact with snRNP or RNA polymerase II in the nucleus to regulate transcription, or bind to transcription factors and competitively regulate classic RNA splicing. CircRNAs accumulate in cells and release into exosomes and plasma. The amount of circRNAs released into exosomes from tumor tissue is three times more than that in tumor tissues [24].
CircRNAs play an irreplaceable regulatory role in the complex life process. Their abnormal expression can induce or impede the occurrence and development of cancer. They are promising biomarkers and even therapeutic targets for cancer. With the continuous development of high-throughput sequencing and bioinformatics technologies, the formation and function of circRNAs and their relationship with cancer have gradually attracted widespread attention in the scientific community and become a hotspot for researches of clinical disease after miRNAs and long noncoding RNAs (LncRNAs). For the moment, Circbase, Circ2Traits, CircNet, and other databases have included information of more than 100,000 circRNAs, which can assist us to predict the regulatory relationship of circRNA-miRNA-mRNA. This tool is extremely convenient for us to study circRNA systematically. At the same time, methods for constructing or interfering with circRNAs have emerged and matured, making it possible to artificially regulate the expression of intracellular circRNAs and being helpful to further explore the role of circRNAs in tumor cells.
By summarizing the researches on the relationship between circRNAs and cancer in recent years, we find that many circRNAs have abnormal expression in the tumor tissue/blood/exosome of tumor patients, among which “star molecules” such as circ-ITCH and Cdr1as have differential expression and play a regulatory role in different types of cancer. Cdr1as has the potential for being a biomarker for gastric cancer, hepatic cancer, colorectal cancer, cervical cancer, and so on. Hsa_circ_0000064 and hsa_circ_0013958 are related to tumor lymph node metastasis and TNM staging in lung cancer. Circ-100876 is associated with the prognosis of lung cancer patients. The reduction of has_circ_002059 in gastric cancer has a predictive role in distant metastasis and TNM staging. The combination of hsa_circ_0000096 and hsa_circ_002509 exhibit a high diagnostic value for gastric cancer. Circ-BANP is a potential diagnostic marker for colorectal cancer, and the elevation of has_circ_001569 in colorectal cancer is proven helpful to the diagnosis and staging of disease. The rise of hsa_circ_0067934 in esophageal cancer suggests the progression of tumor staging. The same phenomenon is found in other cancers. In certain kind of cancer, there are many circRNAs with different expression changes; on the other hand, the same circRNA can correspond to different types of cancer. The summary of cancer-related circRNAs that have been uncovered is shown in Table 14.1.
With the continuous expansion of existing researches, the relationship between circRNAs and cancer networks have become increasingly clear. However, our knowledge on circRNAs is only the tip of the iceberg. There is still a long way to go to uncover the mysterious veil of circRNAs function. We have reason to believe that with the discovery of more and more circRNAs, the diagnosis, progression, and prognosis evaluation system of various cancer based on circRNAs will be mature, which is of great significance for cancer treatment.
References
Sanger HL, Klotz G, Riesner D et al (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73(11):3852–3856
Sorber K, Dimon MT, Derisi JL (2011) RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res 39(9):3820–3835
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333
Hsiao KY, Sun HS, Tsai SJ (2017) Circular RNA – New member of noncoding RNA with novel functions. Exp Biol Med 242(11):1136
Zhang Y, Zhang XO, Chen T et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806
Li Z, Huang C, Bao C et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256
Rybakwolf A, Stottmeister C, Glažar P et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870
Guo JU, Agarwal V, Guo H et al (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7):409
Ferlay J, Soerjomataram I, Dikshit R et al (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5):E359–E386
Dusek L, Muzik J, Maluskova D et al (2014) Epidemiology of screening-targeted cancers according to new data of the Czech National Cancer Registry. Klin Onkol 27(Suppl 2):19–39
Rosenberg AR, Kroon L, Chen L et al (2015) Insurance status and risk of cancer mortality among adolescents and young adults. Cancer 121(8):1279
Torre LA, Bray F, Siegel RL et al (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108
Kirwan A, Utratna M, O'Dwyer ME et al (2015) Glycosylation-based serum biomarkers for cancer diagnostics and prognostics. Biomed Res Int 2015:490531
Huang G, Li S, Yang N et al (2017) Recent progress in circular RNAs in human cancers. Cancer Lett 404:8
Xin Z, Ma Q, Ren S et al (2017) The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics 16(2):80
Boutros PC (2015) The path to routine use of genomic biomarkers in the cancer clinic. Genome Res 25(10):1508–1513
Kalia M (2015) Biomarkers for personalized oncology: recent advances and future challenges. Metabolism 64(3 Suppl 1):S16–S21
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9(9):22–32
Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4(9):721
Rupaimoole R, Calin GA, Lopez-Berestein G et al (2016) miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov 6(3):235–246
Leva GD, Garofalo M, Croce CM (2014) microRNAs in cancer. Annu Rev Pathol 9(1):287
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16(3):203
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Li Y, Zheng Q, Bao C et al (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna-a Publication of the Rna. Society 19(2):141–157
Schwanhäusser B, Busse D, Li N et al (2013) Corrigendum: global quantification of mammalian gene expression control. Nature 495(7439):126–127
Szabo L, Morey R, Palpant NJ et al (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16(1):1–26
Rybak-Wolf A, Stottmeister C, Glažar P et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870
Li Z, Huang C, Bao C et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Ashwal-Fluss R, Meyer M, Pamudurti NR et al (2014) circRNA biogenesis competes with Pre-mRNA Splicing. Mol Cell 56(1):55–66
Conn S, Pillman K, Toubia J et al (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160(6):1125–1134
Dudekula DB, Panda AC, Grammatikakis I et al (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Howlader N, Noone AM, Krapcho M, et al (2012) Previous version: SEER cancer statistics review,1975–2009(Vintage 2009 Populations). National Cancer Institute. http://seer.cancer.gov/csr/1975_2009_pops09/
Ferlay J, Soerjomataram I, Dikshit R et al (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5):E359
Institute NC (2015) Cancer of the pancreas – SEER stat fact sheets. http://seer.cancer.gov/statfacts/html
Maemondo M, Inoue A, Kobayashi K et al (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362(25):2380
Wang BY, Huang JY, Cheng CY et al (2013) Lung cancer and prognosis in taiwan: a population-based cancer registry. J Thorac Oncol 8(9):1128–1135
Luo YH, Zhu XZ, Huang KW et al (2017) Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother 96:892–898
Yao JT, Zhao SH, Liu QP et al (2017) Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 213(5):453–456
Fang T, Yun W, Zhe X et al (2017) Circular RNA CircHIPK3 promotes NCI-H1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1. Chin J Lung Cancer 20(7):459
Zhu X, Wang X, Wei S et al (2017) hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 284(14):2170
Zhao J, Li L, Wang Q et al (2017) CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem 44(6):2138–2146
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in cancer. Cancer Res 73(18):5609
Wan L, Zhang L, Fan K et al (2016) Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-Catenin pathway. Biomed Res Int 2016(1):1579490
Tian F, Yu CT, Ye WD et al (2017) Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem Biophys Res Commun 493(3):1260
Aoyama T, Nishikawa K, Fujitani K et al (2016) Early results of a randomized two-by-two factorial phase II trial comparing neoadjuvant chemotherapy with two and four courses of cisplatin/S-1 and docetaxel/cisplatin/S-1 as neoadjuvant chemotherapy for locally advanced gastric cancer. Ann Oncol 28(8):1876–1881
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115
Acebron SP, Niehrs C (2016) β-Catenin-independent roles of Wnt/LRP6 signaling. Trends Cell Biol 26(12):956–967
Sui W, Shi Z, Xue W et al (2017) Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep 37(3):1804–1814
Chen J (2017) 250P Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 388:208–219
Li P, Chen H, Chen S et al (2017) Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer 116(5):626–633
Peifei L, Shengcan C, Huilin C et al (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132
Zhang Y, Li J, Yu J et al (2017) Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget 8(14):22936–22943
Li WH, Song YC, Zhang H et al (2017) Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers 2017:4587698
Huang YS, Jie N, Zou KJ et al (2017) Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep 16(3):2469–2476
Shao Y, Li J, Lu R et al (2017) Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med 6(6):1173–1180
Chen S, Li T, Zhao Q et al (2017) Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 466:167–171
Bachmayrheyda A, Reiner AT, Auer K et al (2015) Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 5(8057):8057
Zhu M, Xu Y, Chen Y et al (2017) Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother 88:138
Guo J, Jin L, Zhu C et al (2016) Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther 9:7451–7458
Xie H, Ren X, Xin S et al (2016) Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 7(18):26680
Hsiao KY, Lin YC, Gupta SK et al (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77(9):2339
Huang G, Zhu H, Shi Y et al (2015) cir-ITCHPlays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One 10(6):e0131225
Su H, Lin F, Xia D et al (2016) Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 14(1):225
Li F, Zhang L, Li W et al (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 6(8):6001
Xia W, Qiu M, Chen R et al (2016) Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 6:35576
Qu S, Song W, Yang X et al (2015) Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genomics Data 5(C):385
Li H, Hao X, Wang H et al (2016) Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem 40(6):1334
Wang FS, Fan JG, Zhang Z et al (2014) The global burden of liver disease: the major impact of China. Hepatology 60(6):2099
Nguyen VTT, Law MG, Dore GJ (2009) Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat 16(7):453–463
Zhang JY (2007) Mini-array of multiple tumor-associated antigens to enhance autoantibody detection for immunodiagnosis of hepatocellular carcinoma. Autoimmun Rev 6(3):143
Samuel M, Chow PK, Chan SYE et al (2009) Neoadjuvant and adjuvant therapy for surgical resection of hepatocellular carcinoma. Cochrane Database Syst Rev 1(1):CD001199
Crissien AM, Frenette C (2014) Current management of hepatocellular carcinoma. Gastroenterol Hepatol 10(3):153
Qin M, Liu G, Huo X et al (2016) Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 16(1):161–169
Shang X, Li G, Liu H et al (2016) Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine 95(22):e3811
Lei Y, Gong X, Lei S et al (2016) The circular RNA Cdr1as Act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 11(7):e0158347
Xu L, Zhang M, Zheng X et al (2017) The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol 143(1):1–11
Huang XY, Huang ZL, Xu YH et al (2017) Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep 7(1):5428
Ren S, Xin Z, Xu Y et al (2017) Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle 16(22):2204–2211
Guo W, Zhang J, Zhang D et al (2017) Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget 8(29):48169–48177
Liu BH, Zhang BB, Liu XQ et al (2018) Expression profiling identifies circular RNA signature in hepatoblastoma. Cell Physiol Biochem 45(2):706
Fu L, Chen Q, Yao T et al (2017) Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 8(27):43878–43888
Fu L, Yao T, Chen Q et al (2017) Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget 8(35):58405–58416
Center MM (2011) Global cancer statistics. CA Cancer J Clin 61(2):69
Goto N, Hiyoshi H, Ito I et al (2014) Identification of a novel compound that suppresses breast cancer invasiveness by inhibiting transforming growth factor-β signaling via estrogen receptor α. J Cancer 5(5):336–343
Grushko TA, Filiaci VL, Mundt AJ et al (2008) An exploratory analysis of HER-2 amplification and overexpression in advanced endometrial carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol 108(1):3–9
Wang H, Mu X, Zhou S et al (2014) NEDD9 overexpression is associated with the progression of and an unfavorable prognosis in epithelial ovarian cancer. Hum Pathol 45(2):401–408
Tangjitgamol S, Khunnarong J, Srijaipracharoen S (2014) Medical morbidities in endometrial cancer patients. Int J Gynecol Cancer 24(9):1623–1627
Reddy SD, Ohshiro K, Rayala SK et al (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 68(20):8195
Lü L, Jian S, Shi P et al (2017) Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget 8(27):44096–44107
Liang HF, Zhang XZ, Liu BG et al (2017) Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7(7):1566–1576
Liang G, Liu Z, Tan L et al (2017) HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res 37(8):4337–4343
Wang H, Xiao Y, Wu L et al (2018) Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol 52(3):743–754
Ahmed I, Karedath T, Andrews SS et al (2016) Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 7(24):36366–36381
Chen BJ, Byrne FL, Takenaka K et al (2018) Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget 9(5):5786–5796
Abdelmohsen K, Panda AC, Munk R et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14(3):361–369
Xuan L, Qu L, Han Z et al (2016) Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 8(2):932
Chen L, Zhang S, Wu J et al (2017) circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene 36(32):4551–4561
Zheng J, Liu X, Xue Y et al (2017) TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol 10(1):52
Guarnerio J, Bezzi M, Jeong JC et al (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165(2):289–302
Zhong Z, Lv M, Chen J (2016) Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 6:30919
Sand M, Bechara FG, Sand D et al (2016) Circular RNA expression in basal cell carcinoma. Epigenomics 8(5):619–632
Yang H, Luo J, Liu Z et al (2015) MicroRNA-138 regulates DNA damage response in small cell lung cancer cells by directly targeting H2AX. Cancer Investig 33(4):126
Liu YC, Li JR, Sun CH et al (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44(D1):D209
Peifei L, Shengcan C, Huilin C et al (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136
Wang X, Zhang Y, Huang L et al (2015) Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 8(12):16020–16025
Huang G, Hua Z, Shi Y et al (2015) cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One 10(6):e0131225
Mcglynn LM, Mccluney S, Jamieson NB et al (2015) SIRT3 & SIRT7: potential novel biomarkers for determining outcome in pancreatic cancer patients. PLoS One 10(6):e0131344
Yeh YM, Chuang CM, Chao KC et al (2013) MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int J Cancer 133(4):867–878
Yu SY, Wang YP, Chang JY et al (2014) Increased expression of MCM5 is significantly associated with aggressive progression anda poor prognosis of oral squamous cell carcinoma. J Oral Pathol Med 43(5):344–349
Sand M, Bechara FG, Gambichler T et al (2016) Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 83(3):210–218
Wang Q, Tang H, Yin S et al (2013) Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep 29(5):2046–2052
Jin H, Jin X, Zhang H et al (2017) Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget 8(15):25571–25581
Wang K, Sun Y, Tao W et al (2017) Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett 394:1–12
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (81670571 and 81370559 to C. Yang; 81400635 to F. Wang), Joint Projects in Major Diseases funding from Shanghai Municipal Commission of Health and Family Planning (2014ZYJB0201 to C. Yang), Joint Projects for Novel Frontier Technology in Shanghai Municipal Hospital from Shanghai Municipal Commission of Health and Family Planning (SHDC12014122 to C. Yang), Shanghai Medical Guide Project from Shanghai Science and Technology Committee (14411971500 to F. Wang), grants from Chinese Foundation for Hepatitis Prevention and Control (TQGB20140141 to F. Wang), and funds from Shanghai Innovation Program (12431901002 to C. Yang).
Competing Financial Interests
The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Xia, L., Song, M., Sun, M., Wang, F., Yang, C. (2018). Circular RNAs as Biomarkers for Cancer. In: Xiao, J. (eds) Circular RNAs. Advances in Experimental Medicine and Biology, vol 1087. Springer, Singapore. https://doi.org/10.1007/978-981-13-1426-1_14
Download citation
DOI: https://doi.org/10.1007/978-981-13-1426-1_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1425-4
Online ISBN: 978-981-13-1426-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)