Abstract
All modern cells are delimited by cell membranes, which keep the integrity of the whole cell. The cell membrane is not only the boundary of the unit of life, but also a specific compartment that harbors many essential cellular activities. The unceasing progress of science and technology provides us a more comprehension and profound understanding of cell membranes. Here we summarize the major historical discoveries and theories that tackled the existence and structure of cell membranes, and the related techniques by which different membrane models have been proposed. We also discuss the strengths and weaknesses of the techniques and the suitability of different membrane models in various cell systems.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
2.1 Introduction
All cells are surrounded by a membrane layer called cell membrane or plasma membrane. Cell membranes consist of membrane lipid and proteins and are crucial for the life of the cell by defining the cell boundaries, therefore, maintaining the differences between the intracellular and extracellular environment. The cell membrane is not only the boundary of the unit of life, but also a specific compartment that harbors many essential cell functions including the provision of a stable internal environment, transportation of substances between cells and the environment, energy conversion, and signal transduction [1,2,3]. Due to its key role in a variety of activities, the cell membrane is one of the most attractive topics for multidisciplinary studies, including studies that combine chemistry, biology, and physics. For chemists, cell membranes are supermolecular structures that contain thousands of types of lipids, proteins, and saccharides [4]. Biologists consider cell membranes to be the first barrier to defend cells from external harms, and cell membranes are related to multiple functions and diseases [5]. From the physicists’ point of view, how the basic atoms and molecules can form a multi-functional and high-efficiency system is related to the origin of life [6].
After the basic composition of the cell membrane was discovered, scientists have paid more attention to the structure and function of the whole supermolecular complexes. The studying history of cell membrane structure can be traced back to the early twentieth century. Many scientists coming from different fields contributed to our understanding of membranes along the centuries. We can find that every breakthrough of membrane structure is always accompanied by the progress of science and technology. In this part, we will review the main discoveries that led to our current knowledge of biological membranes and the related techniques which were used to explore different membrane models. We will also discuss the strengths and weaknesses of the techniques and methods, and the suitability of different membrane models in various biological systems. This, we hope, will provide a more complete account of the discovery of membrane structure and open new perspectives for further uncovering the more refined and precise membrane structure at the sub-cellular and molecular level.
2.2 Membrane Models and Related Techniques
The properties and functions of the cell membrane are dependent on the structure of its own component molecules. The details of how these various kinds of lipids, proteins, and saccharides are organized to form a complex and ordered cell membrane are highly important. The investigation of this question requires a number of technological developments, which have been the basis of new understanding of cell membrane structure. Since last century, researchers have been working on the structure and function of cell membranes and proposed distinct membrane models to explain why cell membranes can play so many functions. Among them, the fluid mosaic model is one of the most classical models, which is widely accepted by biologists. Even so, the long history of studying cell membranes tells us that the membrane structure is far from completely understood. There are still open questions that remain to be resolved, such as the location and interaction of proteins in the membrane. Therefore, summarizing the development of cell membrane studies will help us more appropriately apply different membrane models to diverse disciplinary fields and discover novel models to explain what are happening or will happen in the membrane. Readers will find a timeline displaying the most dramatic contributions to membrane structure models up to the early twenty-first century in Fig. 2.1.
Summary of the main contributions related to cell membrane discovery between the first studies on cell membrane structures and the development of membrane models. The events are approximately stacked in chronological order from top to bottom. The orange boxes represent some major research influent to the history, and the green boxes reflect representative membrane models
2.2.1 Lipid Bilayer Model
From a historical perspective, monomolecular lipid films at the air–water interface (so-called Langmuir films) are the oldest membrane systems ever reported. The earliest record about this particular system (reported as oil films floating in water) came from Hammurabi period’s (eighteenth century BC) cuneiform-written Babylonian clay tablets [7]. Babylonians used it as one of their forms of divination, based on pouring oil on water and observing the shapes originated from the subsequent oil spreading. This application was also used in Greek, Chinese, and ancient Japanese cultures. The most often mentioned ancient application of lipid films is the calming effect of ripples on the water surface caused by spreading oil, which was already observed by sailors in ancient times and described by Plutarch, Aristotle, and Pliny. Pouring oil on stormy seas is believed to stem from the 429 to 651 AD period [7]. The scientific investigation of this phenomenon was initiated in 1744 by Benjamin Franklin, followed by contributions of Lord Rayleigh and Agnes Pockels. Until 1917, Langmuir presented the first evidence of the monomolecular nature of the lipid films by measuring the surface covered by lipid monolayers at the interface between water and air (Fig. 2.2) [7,8,9,10,11].
Langmuir films were used in a historically key experiment connected to our contemporary vision of a biological membrane. In 1925, two Dutch physicians, Gorter and Grendel explored the molecular structure of membranes [12]. They extracted the lipids from an erythrocyte sample; since these cells were known to lack internal membranes, they assumed that all lipids should come from the cell envelopes. Measuring the spread of lipids on water was done using a Langmuir’s trough. When Gorter and Grendel compared the surface covered by the lipids to the estimated sum of cell surfaces, they found a 2:1 ratio (Fig. 2.3) [11]. As a result, they concluded that cells were surrounded by a lipid membrane with two molecules thickness with hydrophobic components in the internal part of the membrane and the hydrophilic components in the external part [12].
Surface measurement of membrane lipid monolayers as a way to determine membrane structure. Summary of the method, consisting in the comparison between the area of lipids extracted from erythrocyte membranes and the estimated surface area of cells. The ratio of 2:1 indicated the lipid bilayer structure
This study has been commonly cited as the most conclusive argument in favor of the lipid bilayer nature of cell membranes. In spite of the elegance of this work, it is important to balance its contribution to the field because of its errors from both technical and theoretical grounds. First, the extraction technique employed could not isolate the totality of erythrocyte lipids from the samples [13]. Second, the equation that they used to estimate the surface of the erythrocytes also underestimated the cell area [13]. Some historical reviews on membrane discovery have argued that it was fortunate that the two errors neutralized each other in order to give credit to the lipid bilayer hypothesis that we now recognize in current membrane models. When calculating the surface covered by lipids, Gorter and Grendel measured the first detected pressure (i.e., the maximal continuous surface covered by an amount of lipid). A more accurate ratio for the lipid surface was measured at the collapse pressure in 1960s, which was 1.3:1 [13]. This ratio conformed to loose lipid bilayers, whereas the modern fluid mosaic model supports the idea that membrane lipids are tightly packed and the excess space is actually occupied by membrane proteins.
Disregarding their experimental mistakes, Gorter and Grendel’s work is the first documented evidence that cell lipids are arranged in a bilayer configuration. The contribution of lipid bilayer model can be helpful in the light of later progress in membrane studies. However, it should be noticed that at that time the presence of proteins as components of biological membranes was unknown.
2.2.2 Davson–Danielli Model
The formulation of the lipid bilayer model had opened the door to the molecular description of cell membrane structure. In an attempt to confirm or refute the lipid bilayer postulate, one of the first lines of research to be explored was the measurement of membrane thickness. In 1925, Fricke measured the static capacitance per surface unit with an estimation of the cell membrane dielectric constant. He extrapolated that the thickness of erythrocyte and yeast cell membranes was about 4 nm [14, 15]. This thickness supported the hypothesis of a lipid monolayer but not a bilayer. Although using the dielectric constant in these studies brought some questions, there were no enough tests to refute the estimation by Fricke at that time [16]. Until 1940, the invention of leptoscope provided a relatively accurate testing of measuring membrane thickness. It could measure very thin membranes by comparing the intensity of light reflected from a sample to the intensity of a membrane standard of known thickness [17]. The thickness it measured ranged from 8.6 to 23.2 nm. As a result, the lower value was compatible with the lipid bilayer, whereas the higher one supported the presence of supplementary superimposed layers [18]. Yet, the thickness of the membrane was only revealed after two decades when electron microscopy developed. The observation of membrane sections using the new instrument established the now accepted ~8 nm value for standard cell membranes [19].
Although the actual thickness of the membrane remained unclear during that time, studies in this field still took a huge step because the most influential membrane structure model introduced, namely the sandwich membrane model (Fig. 2.4). The genesis of this model followed previous studies on surface tension of various cells [20, 21]. As the surface tension values appeared to be much lower than would be expected for an oil–water interface, it was assumed that some substance was responsible for lowering the interfacial tensions in the surface of cells [21]. At this time, membranes were known to contain substantial quantities of proteins [22, 23] but little had been said about their position in cell membranes. Therefore, in 1935 Danielli and Davson proposed a sandwich membrane model (the Davson–Danielli model or the protein–lipid–protein model) [24]; in this model, the lipid layer was covered by a thin protein layer on both sides. This observation was significant progress in understanding better the compositional nature of biological membranes.
The Davson-Danielli model immediately became popular, and it dominated cell membrane studies for the following 30 years. There were some light polarization and X-ray diffraction studies to support the model. In order to provide insights into membrane structure, these methods required repeated structures, so the samples used were myelinized axons. These analyzes showed an alternation of protein and lipid layers in support of the sandwich model [25]. Unfortunately, we know now that this biological material is physiologically very specific and can hardly be compared to a regular cell membrane.
Despite the fact that the superposition of protein and lipid layers was characterized in this model, the authors were aware of its existent problems. In the surface tension experiments, they used triglyceride oils and other non-miscible lipids. These substances are very different from most natural amphipathic cell membrane components, and thus they are not suitable for a realistic comparison to cell membranes. Soon after the authors postulated the sandwich model, they added amphipathic cell membrane components such as fatty acids, cholesterol, or phospholipids to non-miscible mixtures, which significantly decreased the interfacial tension between water and highly hydrophobic substances [26]. Other contemporary debates were about membrane permeability. As the ion permeability developed and active transporters discovered, the authors acknowledged the three popular possibilities of that time: pores, simple diffusion, and the existence of some proteins that can span the membrane.
In summary, the Davson-Danielli model took into account the difference between the physical properties measured for natural membranes and membranes composed of pure lipids, for example, the low values of surface tension measured in the 1920s and 1930s in plasma membranes compared to lipid bilayers. However, although the researchers were conscious of the permeability of ions, sugars, and other hydrophilic solutes, the model they postulated implied that the protein layer did not interact with the hydrophobic parts of the lipid bilayer and that the protein layer formed by simple physical adsorption.
2.2.3 Unit Membrane Model
The direct characterization of membrane structure did not progress much until new techniques allowed dramatic discoveries. Electron microscopy emerged in the 1930s, however, sharper resolutions that allowed the direct observation of cell membranes was obtained till 1950s. Sjöstrand et al. demonstrated the “railroad track” structure of membrane cross sections after staining with heavy metals, which stained the phosphates of phospholipid headgroups and washed the proteins out [27]. The structure was known as two dense lines separated by a middle lighter space. Yet, which one among the whole structure or one of the dense parts should be considered as the quintessential cell membrane? In 1959, Robertson compared a collection of cross-section pictures and observed that the whole railroad track was consistent in a variety of cells [28] (Fig. 2.5). He thought that the railroad track fit the Davson–Danielli model and extended it as the unit membrane or the Davson–Danielli–Robertson (DDR) model. The model supported the concept of a “unit membrane” with a thickness of 6–8 nm where the proteins were coating the surface of the lipid bilayer in an extended β-sheet configuration, corresponding to the width of myelin sheaths observed in X-ray diffraction experiments. Robertson stressed two points in the model: first, that the three layers observed in the electron microscope cross-section shots were part of the same structure, the cell membrane, regardless of the other cell envelopes that might exist; and second, that this structure was universally shared among all biological membranes [29].
Unit membrane concept. Railroad track images from thin section electron microscopy of osmium-fixed cells: two dark lines separated by a clear region. Robertson summarized the available data and used many new examples to make the point that all cell membranes have a common structure. Reprinted by permission from Macmillan Publishers Ltd: Ref. [30], copyright 2003
The unit membrane model assigned the bilayer structure to all membrane systems, including cell membranes and cellular organelle membranes. However, many researchers called into questioning the idea that one unique membrane model could account for all biological membranes because it seemed contradictory to the large membrane diversity that was being discovered at that time [31, 32]. For example, it had been observed that the protein-lipid ratio from different membranes could vary from 1:4 to 5:1, suggesting that the amount of membrane proteins was not always enough to entirely cover the cell surface [31]. In addition, the lack of resolution prevented the observation of the railroad track in some bacteria, thus implying that the three-layer structure was either not always visible or not universal [33].
In 1966, observations made by Green and Benson challenged the unit membrane model [34, 35]. Working independently with inner mitochondria and chloroplast membranes, these authors demonstrated: first, that subunits not railroad tracks containing lipid and proteins can be separated from the whole membrane; and second, that these subunits can be reconstituted to regain activity. Based on these observations, a different model was postulated where lipids represent a solvent for embedded globular proteins, i.e., opposite to that postulated in the unit membrane model [35, 36]. Electron microscopy images from viral capsides also lent credit to the view that membranes could be made up of subunits [37]. From a historical perspective, it is noteworthy that these models emphasized the dominant role of proteins as the major structural components of membranes [38]. The so-called subunits that were being observed most likely corresponded to the functional proteins that dominated the mitochondrial and chloroplastic membranes, for example, ATPase or photosystems.
The unit membrane model was confronted and there was no consensus as to agreeing on a general model for biological membranes [36], i.e., a model that includes common relevant features existing in different biological membranes. In spite of the debates, the change in perspective that allowed the prevalence of the fluid mosaic model was highly impacted by the gathering of conclusive evidence from new techniques.
2.2.4 Fluid Mosaic Model
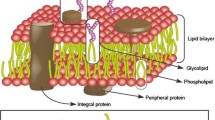
With the development of the freeze-fracture technique and immuno-electron microscopy, researchers identified antibody-recognizing isolated membrane proteins and proteins that spanned the lipid bilayer. Thus, Singer and Nicolson proposed their celebrated fluid mosaic model (FMM) of biological membranes in 1972 [39], and this model has since been a central paradigm in membrane science. The basics of the model have remained the same ever since the membrane is a lipid bilayer with hydrophilic parts on the sides and hydrophobic parts in the interior; proteins can interact with surface through transient polar contacts, but a lot of proteins are partially or totally embedded in the lipid bilayer where their hydrophobic parts also interact with the hydrophobic parts of lipids (Fig. 2.6).
Fluid mosaic membrane model as originally proposed in 1972. In this cross-sectional view, the solid bodies with stippled cut surfaces represent globular integral membrane proteins, which at long range are randomly distributed in the plane of the membrane. At short range, some may form specific aggregates. Reprinted from Ref. [40], Copyright 2014, with permission from Elsevier
Considering the big controversy going on at the beginning of the 1970s, the FMM incorporated for the first time many relevant experimental facts and the development of techniques: (1) the permeability and transport studies that predicted enzyme-like transmembrane proteins [41]; (2) the apparent lack and limited transverse mobility of lipids to make up complete bilayers [13], thus pointing out to the participation of proteins in the membrane plane; (3) electron microscopy pictures, including freeze-etching technique and immuno-EM studies that identified antibody-recognizing isolated membrane proteins [42]; (4) the stability of artificial lipid bilayers that supported them as suitable and sufficient components to make up structures similar in the biological membranes [43]; (5) the favorable conformations predicted for the membrane proteins, such as α-helical, globular, or membrane spanning [38].
As a key property, the Singer–Nicolson model on one hand assigned to the lipid bilayer component of membranes a certain degree of fluidity. The fluidity concept was meant to characterize the lipid bilayer as a kind of pseudo-two-dimensional liquid in which both lipids and membrane-associated proteins display sufficient lateral mobility to allow for function. On the other hand, it emphasized the mosaic nature of proteins, which could span the lipid bilayer. The overall random appearance of this lipid–protein fluid composite made the membrane look like a mosaic. The specific features and important contributions of the model are demonstrated in detail as follows.
First, the model recovered the lipid bilayer concept from Danielli and Davson as mentioned above. Membrane lipids are amphipathic. They possess both a hydrophobic and a hydrophilic moiety. Because of this amphipathic character, in an aqueous medium they can organize themselves on both sides of an imaginary plane, with the hydrophobic portions facing each other, and the polar moieties oriented to the outer, aqueous space.
Second, membrane proteins can be associated either to the lipid bilayer polar headgroups (peripheral proteins) or to the hydrophobic matrix (integral proteins). This idea of proteins embedded in a hydrophobic milieu had never been proposed in a clear and explicit way before. Moreover, the acknowledgment of the thermodynamic hydrophobic constraints improved our understanding of membrane proteins, for example, the first tridimensional structure of a transmembrane protein—the archaeal bacteriorhodopsin [44], the development of the patch-clamp technique which allowed the understanding of single ion channels [45, 46], and the late discovery of the aquaporins [47, 48].
Third, both lipids and proteins are in constant motion. In principle, three main modes of motion could be considered, rotational, translational, and transbilayer. Rotational motion occurs essentially around an axis perpendicular to the plane of the membrane. Both lipids and proteins rotate around their long axis. Protein rotation had been considered but not given much attention. It was experimentally demonstrated by Chapman and co-workers [49]. Translational diffusion occurs along the plane of the membrane, unhindered by diffusion barriers [50]. Transbilayer (or flip-flop) diffusion, though in theory possible, would not occur because of the energy barrier presented by the bilayer hydrophobic core to the polar groups of lipids and proteins.
Finally, the asymmetry of membranes has also proven to be a fruitful characteristic [51]. This is a direct consequence of the just mentioned lack of transbilayer motion. Asymmetry means that the two sides of a membrane are not identical. Lipids exhibit a relative asymmetry [52], i.e., the fraction of a given lipid in one of the monolayers is different from that in the other. A well-known example is phosphatidylserine, that is found almost entirely in the inner side of human red blood cell membranes. Conversely most, but not all, phosphatidylcholine occur in the outer monolayer [53]. Protein asymmetry is absolute, every single protein molecule in a membrane occurs with exactly the same orientation. Integral proteins are anchored to one or both side of the membrane immediately after synthesis and insertion into the bilayer.
Although FMM has been remarkably consistent with data collected on biological membranes since 1925, it was inevitable that the original model could not explain aspects of membrane dynamics. It had become obvious that the original model needed to be modified or augmented to reflect the emergence of new finds on extracellular and intracellular mechanisms that can affect the lateral distributions and movements of plasma membrane components [54, 55]. Thus, Nicolson elaborated the basic model in 1976 (Fig. 2.7) [55]. It added the interactions of extracellular matrix and membrane-associated cytoskeletal components with cell membranes and their potential influence on the mobility and distribution of transmembrane glycoproteins. Moreover, Nicolson pointed out the possibility that less mobile lipid–protein or lipid–lipid domains might exist in membranes as frozen or semi-frozen islands of less mobile lipids in a sea of fluid phospholipids. The revision of the FMM produced a few years after the original model was less homogeneous and contained additional information not included in the original model. However, this revised model retained all of the basic elements at the nanoscale level, and strictly speaking, it did not propose a new concept of membrane structure.
Modified schematic version of the fluid mosaic membrane model of biological membrane structure, as proposed in 1976. In this version, different snapshots in time are represented by the two panels. Some integral membrane glycoproteins are relatively free to diffuse laterally in the membrane plane within a fluid lipid region or domain, whereas others are “anchored” or relatively impeded by a cytoskeletal assemblage or an ordered or solid lipid phase. Reprinted from Ref. [40], Copyright 2014, with permission from Elsevier
The FMM successfully explains some characteristics of cell membranes, such as fluidity and protein mosaicism. However, techniques at that time had their limits, for example, immuno-EM only detected the separated proteins that labeled with antibodies in the membrane. Therefore, membrane domains and cell membrane-associated structures were not yet discovered, and some membrane functions, such as multiple protein-associated signal transduction and membrane endocytosis, were not explained clearly as well. Nonetheless, the FMM has proven its usefulness in describing basic cell membrane structure for over 40 years, and it remains the most explanatory hypothesis to understand biological membranes.
2.2.5 Lipid Raft Model
Approximately at the same time when the FMM was proposed, some lipid biophysicists reported experimental evidence that lipids could laterally segregate in membranes under certain temperature and compositional conditions and could form distinct lipid domains with particular structural characteristics [56,57,58]. Marcelja in 1976 [59] and Sackmann in 1984 [60] anticipated the possible role of different membrane regions induced by lipid–protein interactions as a physical basis for membrane-mediated processes. This discussion was also addressed by various researchers focused in the membrane biophysics research area [61,62,63]. Although novel experimental data and discussions were presented about the structure and dynamics of biological membranes, this view was largely ignored by most of life scientists. Until substantial evidence regarding protein clusters from biophysical studies emerged, such as centrifugation, detergent extraction, and cholesterol depletion by methyl-beta-cyclodextrin (MβCD) [64], Simons and Ikonen in 1997 postulated particular functional aspects of specialized domains called lipid rafts in biological membranes [65]. The lipid raft hypothesis proposes that the lipid bilayer is not a structurally passive solvent, but that the preferential association between sphingolipids, sterols, and specific proteins bestows cell membranes with lateral segregation potential [66] (Fig. 2.8).
Lipid raft domains in cell membranes. In this model, all membrane-associated proteins are clustered in protein islands (green lipids) that are surrounded by a sea of protein-free membrane (yellow lipids). The islands can be subdivided into raft and non-raft islands, which is also illustrated by their lipid composition (bright-green and dark-green lipids, respectively) and protein contents (gray and red proteins, respectively). The lipid raft domains are connected to the cytoskeleton (orange), most likely by actin. Reprinted with permission from Ref. [66]. Copyright (2006) National Academy of Sciences, U.S.A
The formation of lipid rafts is a dynamic and reversible process that confers functional signaling properties to cell membranes. As mentioned, lipid rafts are characterized by enrichments of cholesterol and sphingolipids, which are held together by hydrogen bonds, charge pairing, and hydrophobic and van der Waals forces [67,68,69]. Their constituents can quickly exchange with bulk membrane lipids as well as with lipids in other rafts [70]. Lipid rafts are generally nanosized domains (<300 nm diameter, most ~10–200 nm) that can contain integral and peripheral membrane proteins. These segregated domains are presumed to be behind a variety of membrane processes, such as formation of proteins clusters, signal transduction, endocytosis, cell polarization, and motility [2, 71,72,73].
The lipid raft model has long suffered controversy since there is little direct quantitative experimental evidence that such domains exist. The traditional method to study lipid rafts and their association with certain membrane proteins is based on the observation of the detergent-resistant membranes (DRMs), composed of mainly sphingomyelin, other saturated phospholipids, cholesterol, and GPI-anchored proteins [64, 71, 74]. However, this technique was severely criticized, because it utilized detergents that might induce phase separation and/or affect the partitioning of membrane proteins to a given phase [75,76,77]. Therefore, this disruptive measure tells us little about native membrane organization.
Since then, a variety of other techniques to study the phase behavior of model or native membranes in situ have been developed (Fig. 2.9). Conventional fluorescence microscopy has consistently failed to reveal large-scale, laterally segregated structures enriched in a major raft component, GPI-anchored proteins [72, 73]. One reason is its limited resolution, which is ~300 nm in lateral direction. Hence, the much smaller lipid rafts and/or extremely dynamic or simply compositional fluctuations could not be detected. In addition, light microscopy requires the use of contrast agents, fluorescent probes, because biological systems are poor in intrinsic fluorescent species. Probably, the most direct method to study lipid rafts is based on monitoring chemical composition of the cell membranes with mass spectrometry [78,79,80]. However, this method works under ultra-high vacuum and with freeze-dried samples. Atomic force microscopy (AFM) provides a nanoscopic resolution for imaging of membranes in aqueous phase [72, 81,82,83,84] and can clearly distinguish domains of liquid ordered phase due to their characteristic membrane width, being thicker by ~0.5 nm as compared to the liquid disordered phase. Nevertheless, AFM technique works well only in model or isolated native membranes immobilized on surfaces. Nuclear magnetic resonance provides direct information on the order of the lipid head groups and fatty acid chains, but it is limited to model membranes with simple lipid compositions [85,86,87].
Strengths and weaknesses of the most common methods for characterization of lipid rafts. Reprinted from Ref. [88], Copyright 2014, with permission from Elsevier
The recent development of super-resolution imaging further confirmed the existence of lipid rafts [89, 90]. One of the studies utilized stimulated emission depletion microscopy (STED)-fluorescence correlation spectroscopy as a main experimental technique [89]. It showed the presence of nanoscopic domains of sizes around 20 nm where plasma membrane proteins dwelled in periods of 10–20 ms. The study also demonstrated that these special domains owe in part their existence to sphingolipids in the cholesterol-enriched rafts.
The proposal of the lipid raft model has ascertained the structural and functional roles of cell membranes in signal transduction, protein-mediated endocytosis, and viral infection. However, membrane rafts remain one of the most controversial issues in biophysics, which is in part because the methods for their detection are still not perfect enough. In recent years, emerging super-resolution fluorescence microscopy, correlative AFM, and fluorescence microscopy have provided an unprecedented opportunity to study the cell membranes at the single-molecule level. We believe that the composition of lipid rafts and the interactions between multiple rafts will be revealed in the near future.
2.2.6 Modified Fluid Mosaic Model
More than forty years have passed since the fluid mosaic model proposed. During this period, myriads of experimental data have been produced that relate to the structure and dynamics of biomembranes. Although basic membrane structure of the FMM has been severely tested and found worthy, the FMM requires to be revised and modified, which would add the data published in the last forty years and some of the misconceptions inherent in the models of 1970s [39, 91].
In 2005, Engelman proposed an update of the Singer–Nicolson model [92]. He put forward the idea that “membranes are more mosaic than fluid.” Transmembrane proteins are so frequent in membranes that hardly any lipid molecule in the bilayer is left unperturbed (Fig. 2.10). Apart from the high density of transbilayer proteins in the membrane, he also pointed out three other features that are now considered to occur in most cell membranes. First, integral proteins exist voluminous extramembrane domains. This conclusion has been proved by the increasing data of the three-dimensional structures of many membrane proteins [93]. Second, contacts between proteins are more frequent. This phenomenon is observed in multiple events, for example, G-protein-mediated signaling, in which a receptor protein will physically interact with a G-protein that, in turn, will transiently bind and activate a cyclic ATPase, thus triggering a cellular response to the signal [94]. Third, the thickness of the lipid bilayer is uneven. Because the transmembrane portions of intrinsic membrane proteins exhibit a rough surface, and the whole domains have a noncritical size with respect to the lipids, membrane lipids are perturbed by the proteins. The lateral diffusion of lipids is hindered, lipid alkyl chains are disordered, and the bilayer thickness is irregular [95,96,97].
An amended and updated version of the fluid mosaic model with more mosaic proteins. Reprinted by permission from Macmillan Publishers Ltd: Ref. [92], copyright 2005
Engelman’s updated version of the FMM focuses more on the mosaic nature of transmembrane proteins. Without doubt, lipids have also been the key elements in the membrane. In 2008, Escribá et al. [98] published a review that shows how membrane lipids, and the structures they form, participate, and regulate numerous important cellular activities. In the paper, they discussed the influence of membrane asymmetry on the membrane lipid organization. The asymmetry of biological membranes includes lateral and cross-sectional asymmetry. The cross-sectional asymmetry reflects the lipid composition of each leaflet (Fig. 2.11). Membrane heterogeneity is further achieved by lateral asymmetry in which membrane regions (basal, lateral, apical), and different specialized membrane regions or microdomains (lipid rafts, caveolae, coated pits, synaptosomes, etc.) [99] extend the mosaic nature of membranes. The formation of these domains in part results from the nonideal mixing of lipids in membranes and in some cases, it is enhanced by the participation of certain cytoskeletal structures that underpin the lipid bilayer and that restrict the traffic of proteins and lipids.
Schematic illustration of a modification of the fluid mosaic membrane model. Different lipids are indicated in various colors forming specialized domains around integral membrane proteins and glycoproteins as well as being asymmetrically distributed across the membrane. Reproduced from Ref. [98] by permission of John Wiley & Sons Ltd
With the development of various techniques, our understanding of the structure and function of the cell membranes is more thorough and comprehensive. In 2014, Nicolson, one of the founders of the FFM, has also presented a modified FMM that takes into account many of the numerous contributions that have been made since the 1970s (Fig. 2.12) [40]. This model incorporates recent information on both membrane proteins and lipids, containing membrane domains, lipid rafts, and cytoskeletal fencing that were unknown in the 1970s. In this figure, the membrane has very crowded structure, with integral membrane proteins separated by only a few layers of phospholipid molecules. Although the dynamic properties of the membrane cannot be presented in a static medium, the author incorporated most of the important information into the model. In the figure, different types of interactions occur with integral membrane proteins and glycoproteins, membrane lipids, and membrane-associated cytoskeletal systems, and extracellular matrix components. These nonrandom interactions are important in controlling the mobilities, distributions, and aggregation states of membrane components.
An updated fluid mosaic membrane model that contains information on membrane domain structures and membrane-associated cytoskeletal and extracellular structures. Different integral proteins, glycoproteins, lipids, and oligosaccharides are represented by different colors, and where the membrane has been peeled-up to view the inner membrane surface cytoskeletal fencing. Reprinted from Ref. [40], Copyright 2014, with permission from Elsevier
In a word, the straightforward application of the Singer–Nicolson model as a frame of events is impossible without introducing new concepts. New concepts have the following attributes emphasizing the mosaic and dynamic properties of the cell membrane: (1) lipid domain structure, (2) cytoskeletal or other cytosolic interactions, or (3) homo- and hetero-associations with other integral proteins. Diffusion, intermolecular forces, and ever-changing membrane potential, and extracellular influences can dynamically generate and destroy supermolecular structures. Hence, the cell membrane model will undoubtedly become more and more complex, just as the original concepts of membrane structure have evolved into more and more complex structures.
2.3 Conclusions
In light of the long history of cell membrane studies, we must realize that the membrane structure is far from completely understood. Biological details are more complicated than the resolving power of a simple model, which describes generalized, uniform behavior of molecules in the membrane. Moreover, we should know that one model cannot fit every cellular membrane under all conditions. For example, the fluid mosaic model has been cited as the most successful general model of biological membranes; however, it does not provide adequate explanations for every cell membrane structure, such as lipid rafts. Therefore, the cell membrane structure needs to be refined and modified continually and considered on each specific case, such as different cell types and external environments.
Another fact of note is that every achievement made in the field of the cell membrane depends on the development of new techniques and experimental approaches. From traditional methods such as electron microscopy, X-ray crystallograph, fluorescence microscopy, nuclear magnetic resonance spectroscopy, and mass spectroscopy, to modern methods like atomic force microscopy (AFM), single molecular tracking, and super-resolution fluorescence microscopy (SRFM), all the techniques have provided and will provide the significant information of the membrane structure. Not only that, recent developed combined technologies, have also integrated the strengths of two kinds of instruments, which will offer complemented information and facilitate solving the problem from multiple angles. For example, the combined AFM and SRFM can directly visualize the whole cell membrane in situ (AFM) and have the potent ability to identify the local architecture of cell membranes (SRFM). Based on these new techniques, Wang et al. have proposed a semi-mosaic model for the red blood membrane and a protein layer–lipid–protein island (PLLPI) model for the nucleated cell membrane, which will be introduced in Chap. 3.
To thoroughly study the cell membrane, not only the techniques require faster development, questions regarding the membrane structure that were not involved before should also be focused on, including the accurate location of membrane proteins in the membrane, the membrane protein relationships, and the mechanism that underlies the formation of the protein pattern. These open questions will continue to intrigue investigators for some time.
References
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4):373–383
Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1(1):31–39
Glancy B, Balaban RS (2012) Role of Mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 51(14):2959–2973
Sacchettini JC, Baum LG, Brewer CF (2001) Multivalent protein-carbohydrate interactions. A new paradigm for supermolecular assembly and signal transduction. Biochemistry 40(10):3009–3015
Vandenabeele P, Galluzzi L, Vanden Berghe T et al (2010) Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11(10):700–714
Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409(6818):387–390
Dynarowicz-Latka P, Dhanabalan A, Oliveira ON (2001) Modern physicochemical research on Langmuir monolayers. Adv Coll Interface Sci 91(2):221–293
Langmuir I (1917) The constitution and fundamental properties of solids and liquids. II. liquids. J Am Chem Soc 39:1848–1906
Pockels A (1894) On the spreading of oil upon water. Nature 50:223–224
Rayleigh L (1899) The theory of anomalous dispersion. Phil Mag 48:151
Lombard J (2014) Once upon a time the cell membranes: 175 years of cell boundary research. Biol Direct 9:32
Gorter E, Grendel F (1925) On bimolecular layers of lipoids on the chromocytes of the blood. J Exp Med 41(4):439–443
Bar RS, Deamer DW, Cornwell DG (1966) Surface area of human erythrocyte lipids—reinvestigation of experiments on plasma membrane. Science 153(3739):1010–1012
Fricke H (1925) The electric capacity of suspensions with special reference to blood. J Gen Physiol 9(2):137–152
Fricke H, Curtis HJ (1934) Electric impedance of suspensions of yeast cells. Nature 134:102–103
Danielli JF (1935) The thickness of the wall of the red blood corpuscle. J Gen Physiol 19(1):19–22
Waugh DF, Schmitt FO (1940) Investigations of the thickness and ultrastructure of cellular membranes by the analytical leptoscope. Cold Spring Harb Symp Quant Biol 8:233–241
Davson H (1962) Growth of concept of paucimolecular membrane. Circulation 26(5):1022–1037
Robertson JD (1957) New observations on the ultrastructure of the membranes of frog peripheral nerve fibers. J Biophys Biochem Cytol 3(6):1043–1048
Cole KS (1932) Surface forces of the Arbacia egg. J Cell Comp Physiol 1(1):1–9
Danielli JF, Harvey EN (1935) The tension at the surface of mackerel egg oil, with remarks on the nature of the cell surface. J Cell Comp Physiol 5(4):483–494
Halliburton WD, Friend WM (1889) The stromata of the red corpuscles. J Physiol 10(6):532–549
Jorpes E (1932) The protein component of the erythrocyte membrane or stroma. Biochem J 26:1488–1503
Danielli JF, Davson H (1935) A contribution to the theory of permeability of thin films. J Cell Comp Physiol 5(4):495–508
Schmitt FO, Clark GL, Mrgudich JN (1934) X-ray diffraction studies on nerve. Science 80:567–568
Danielli JF (1937) The relations between surface pH, ion concentrations and interfacial tension. Proc R Soc Ser B-Biol Sci 122(827):155–174
Sjostrand FS, Andersson-Cedergren E, Dewey MM (1958) The ultrastructure of the intercalated discs of frog, mouse and guinea pig cardiac muscle. J Ultrastruct Res 1(3):271–287
Robertson JD (1959) The ultrastructure of cell membranes and their derivatives. Biochem Soc Symp 16:3–43
Robertson JD (1960) The molecular structure and contact relationships of cell membranes. Prog Biophys Mol Biol 10:343–418
Edidin M (2003) Timeline—Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol 4(5):414–418
Branton D, Park RB (1968) Papers on biological membrane structure. Bioscience. Little, Brown and Company, Boston
Korn ED (1966) Structure of biological membranes. Science 153(3743):1491–1498
Robinow CF (1962) On plasma membrane of some bacteria and fungi. Circulation 26(5):1092–1104
Benson AA (1966) On orientation of lipids in chloroplast and cell membranes. J Am Oil Chem Soc 43(5):265–270
Green DE, Perdue JF (1966) Membranes as expressions of repeating units. Proc Natl Acad Sci USA 55(5):1295–1302
Stoecken W, Engelman DM (1969) Current models for structure of biological membranes. J Cell Biol 42(3):613–646
Hechter O (1965) Role of water structure in molecular organization of cell membranes. Fed Proc 24(2S15):S91–102
Morange M (2013) What history tells us XXX. The emergence of the fluid mosaic model of membranes. J Biosci 38(1):3–7
Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175(4023):720–731
Nicolson GL (2014) The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim et Biophys Acta-Biomembr 1838(6):1451–1466
Mitchell P (1957) General theory of membrane transport from studies of bacteria. Nature 180(4577):134–136
Branton D (1966) Fracture faces of frozen membranes. Proc Natl Acad Sci USA 55(5):1048–1056
Bangham AD, Standish MM, Watkins JC (1965) Diffusion of univalent ions across lamellae of swollen phospholipids. J Mol Biol 13(1):238–252
Henderson R, Unwin PNT (1975) Three-dimensional model of purple membrane obtained by electron-microscopy. Nature 257(5521):28–32
Neher E, Sakmann B (1976) Single-channel currents recorded from membrane of denervated frog muscle-fibers. Nature 260(5554):799–802
Neher E (1992) Ion channels for communication between and within cells. Science 256(5056):498–502
Benga G, Popescu O, Pop VI et al (1986) Para-(chloromercuri) benzenesulfonate binding by membrane-proteins and the inhibition of water transport in human-erythrocytes. Biochemistry 25(7):1535–1538
Preston GM, Carroll TP, Guggino WB et al (1992) Appearance of water channels in Xenopus oocytes expressing red-cell CHIP28 protein. Science 256(5055):385–387
Razi Naqvi K, Gonzalez-Rodriguez J, Cherry RJ et al (1973) Spectroscopic technique for studying protein rotation in membranes. Nat: New Biol 245(147):249–251
Edidin M (1974) Rotational and translational diffusion in membranes. Ann Rev Biophys Bioeng 3:179–201
Rothman JE, Lenard J (1977) Membrane asymmetry. Science 195(4280):743–753
Daleke DL (2003) Regulation of transbilayer plasma membrane phospholipid asymmetry. J Lipid Res 44(2):233–242
Zwaal RFA, Comfurius P, Vandeenen LLM (1977) Membrane asymmetry and blood-coagulation. Nature 268(5618):358–360
Bretscher MS (1973) Membrane structure: some general principles. Science 181(4100):622–829
Nicolson GL (1976) Transmembrane control of receptors on normal and tumor-cells.1. Cytoplasmic influence over cell-surface components. Biochem Biophys Acta 457(1):57–108
Gebhardt C, Gruler H, Sackmann E (1977) Domain-structure and local curvature in lipid bilayers and biological-membranes. Zeitschrift Fur Naturforschung C-a J Biosci 32(7–8):581–596
Shimshick EJ, McConnell HM (1973) Lateral phase separation in phospholipid membranes. Biochemistry 12(12):2351–2360
Simons K, Van Meer G (1988) Lipid sorting in epithelial-cells. Biochemistry 27(17):6197–6202
Marcelja S (1976) Lipid-mediated protein interaction in membranes. Biochem Biophys Acta 455(1):1–7
Sackmann E (1984) Biological membranes. Physical basis for trigger processes and membrane structures. Academic Press, London
Opdenkamp JAF (1979) Lipid asymmetry in membranes. Annu Rev Biochem 48:47–71
Thompson TE, Tillack TW (1985) Organization of glycosphingolipids in bilayers and plasma-membranes of mammalian-cells. Ann Rev Biophys Biophys Chem 14:361–386
Vaz WLC, Almeida PFF (1993) Phase topology and percolation in multiphase lipid bilayers—is the biological membrane a domain mosaic. Curr Opin Struct Biol 3(4):482–488
Brown DA, Rose JK (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell-surface. Cell 68(3):533–544
Simons K, Ikonen E (1997) Functional rafts in cell membranes. Nature 387(6633):569–572
Lillemeier BF, Pfeiffer JR, Surviladze Z et al (2006) Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci USA 103(50):18992–18997
Ramstedt B, Slotte JP (2006) Sphingolipids and the formation of sterol-enriched ordered membrane domains. Biochim Et Biophys Acta-Biomembr 1758(12):1945–1956
van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9(2):112–124
Simons K, Gerl MJ (2010) Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol 11(10):688–699
Quinn PJ, Wolf C (2009) The liquid-ordered phase in membranes. Biochim Biophys Acta-Biomembr 1788(1):33–46
Brown DA, London E (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem 275(23):17221–17224
Jacobson K, Mouritsen OG, Anderson RGW (2007) Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 9(1):7–14
Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327(5961):46–50
Lingwood D, Simons K (2007) Detergent resistance as a tool in membrane research. Nat Protoc 2(9):2159–2165
Heerklotz H (2002) Triton promotes domain formation in lipid raft mixtures. Biophys J 83(5):2693–2701
Lichtenberg D, Goni FM, Heerklotz H (2005) Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 30(8):430–436
Sot J, Bagatolli LA, Goni FM et al (2006) Detergent-resistant, ceramide-enriched domains in sphingomyelin/ceramide bilayers. Biophys J 90(3):903–914
Boxer SG, Kraft ML, Weber PK (2009) Advances in imaging secondary ion mass spectrometry for biological samples. Ann Rev Biophys 38:53–74. doi:10.1146/annurev.biophys.050708.133634
Kraft ML, Weber PK, Longo ML et al (2006) Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science 313(5795):1948–1951
Lozano MM, Liu Z, Sunnick E et al (2013) Colocalization of the ganglioside G(M1) and cholesterol detected by secondary ion mass spectrometry. J Am Chem Soc 135(15):5620–5630
Chiantia S, Ries J, Kahya N et al (2006) Combined AFM and two-focus SFCS study of raft-exhibiting model membranes. ChemPhysChem 7(11):2409–2418
Giocondi MC, Vie V, Lesniewska E et al (2000) In situ imaging of detergent-resistant membranes by atomic force microscopy. J Struct Biol 131(1):38–43
Goksu EI, Vanegas JM, Blanchette CD et al (2009) AFM for structure and dynamics of biomembranes. Biochim Biophys Acta-Biomembr 1788(1):254–266
Shaw JE, Epand RF, Epand RM et al (2006) Correlated fluorescence-atomic force microscopy of membrane domains: structure of fluorescence probes determines lipid localization. Biophys J 90(6):2170–2178
Filippov A, Oradd G, Lindblom G (2003) The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J 84(5):3079–3086
Guo W, Kurze V, Huber T et al (2002) A solid-state NMR study of phospholipid-cholesterol interactions: sphingomyelin-cholesterol binary systems. Biophys J 83(3):1465–1478
Soni SP, LoCascio DS, Liu Y et al (2008) Docosahexaenoic acid enhances segregation of lipids between raft and nonraft domains: H-2-NMR study. Biophys J 95(1):203–214
Klymchenko AS, Kreder R (2014) Fluorescent probes for lipid rafts: from model membranes to living cells. Chem Biol 21(1):97–113
Eggeling C, Ringemann C, Medda R et al (2009) Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457(7233):1159–U1121
Wang Y, Gao J, Guo X et al (2014) Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res 24(8):959–976
Nicolson GL (1976) Transmembrane control of the receptors on normal and tumor cells: I. cytoplasmic influence over cell surface components. Biochem Biophys Acta 457(1):57–108
Engelman DM (2005) Membranes are more mosaic than fluid. Nature 438(7068):578–580
Abrahams JP, Leslie AGW, Lutter R et al (1994) Structure at 2.8 A resolution of F1-ATPase from bovine heart-mitochondria. Nature 370(6491):621–628
Golebiewska U, Scarlata S (2010) The effect of membrane domains on the G protein-phospholipase C beta signaling pathway. Crit Rev Biochem Mol Biol 45(2):97–105
Cortijo M, Alonso A, Gomezfernandez JC et al (1982) Intrinsic protein-lipid interactions—infrared spectroscopic studies of gramicidin-A, bacteriorhodopsin and Ca2+-ATPase in biomembranes and reconstituted systems. J Mol Biol 157(4):597–618
Arrondo JLR, Goni FM (1998) Infrared studies of protein-induced perturbation of lipids in lipoproteins and membranes. Chem Phys Lipid 96(1–2):53–68
Holt A, Killian JA (2010) Orientation and dynamics of transmembrane peptides: the power of simple models. Eur Biophys J Biophys Lett 39(4):609–621
Escriba PV, Gonzalez-Ros JM, Goni FM et al (2008) Membranes: a meeting point for lipids, proteins and therapies. J Cell Mol Med 12(3):829–875
Escriba PV (2006) Membrane-lipid therapy: a new approach in molecular medicine. Trends in Mol Med 12(1):34–43
Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2017YFA0505300), the National Natural Science Foundation of China (No. 21525314, 21703231, 21721003).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Gao, J., Wang, H. (2018). History and Traditional Techniques of Studying the Structure of Cell Membranes. In: Wang, H., Li, G. (eds) Membrane Biophysics. Springer, Singapore. https://doi.org/10.1007/978-981-10-6823-2_2
Download citation
DOI: https://doi.org/10.1007/978-981-10-6823-2_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-6822-5
Online ISBN: 978-981-10-6823-2
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)