Abstract
Atopic dermatitis (AD) is a chronic inflammatory skin disease in which there are considerable genetic contributions. Genome-wide association studies (GWASs) provide an unbiased method to identify the genetic factors of human diseases and phenotypes comprehensively. Although it is well known that loss-of-function mutations in FLG are the most significant genetic risk factor for AD, recent GWASs, immunochip analyses, and meta-analyses of GWASs have identified a number of loci associated with AD. Candidate genes identified by GWASs of AD are involved in skin barrier functions and innate and adaptive immune responses. Those findings imply a substantial overlap of genetic components with other autoimmune and inflammatory diseases. Genetic variants may influence molecular phenotypes, including RNA expression and stability, transcription factor binding, DNA methylation, histone modifications, and protein levels. Understanding the functional links between susceptibility variants and phenotypic traits is crucial to improve our knowledge of AD. Further interdisciplinary research is necessary for translation of the genetics of AD into clinical practice.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Atopic dermatitis (AD) is a chronic inflammatory skin disease in which there is a considerable genetic component [1]. Genome-wide association studies (GWASs) provide an unbiased method to identify the genetic factors of human diseases and phenotypes comprehensively [2]. Imputation is a statistical method used to infer untyped genotypes by employing a reference panel of extensively genotyped individuals [3]. Imputation is useful to combine data from GWASs performed using different platforms for meta-analysis, provides a high-resolution overview of association results, and increases the statistical power to identify associated loci. Recent developments in high-throughput genotyping and imputation technologies have made it possible to identify disease susceptibility loci convincingly. Although it is well known that loss-of-function mutations in FLG (encoding filaggrin) are the most significant genetic risk factor for AD [4], recent GWASs, immunochip analyses, and meta-analyses of GWASs have identified a number of loci associated with AD (Table 5.1
) and have improved our understanding of its pathogenesis.
2 Genome-Wide Association Studies of Atopic Dermatitis in European and Chinese Populations
The first GWAS for AD was reported in 2009. The study analyzed 939 cases and 975 controls as well as 270 complete nuclear families with two affected siblings [5]. Genetic variants that showed associations with AD in both discovery set were examined in two independent replication populations totaling 2637 cases and 3957 controls. The GWAS found that rs7927894 located 38 kb downstream of C11orf30 (11q13.5) was associated with AD (P = 7.6 × 10−10). The locus had been reported to be a susceptibility locus for Crohn’s disease, which shares many pathophysiological features with AD such as recurrent inflammation of the epithelium, defective barrier function, and dysfunction of innate immune responses against infections.
In a Chinese Han population, a GWAS of AD was conducted using 1012 cases and 1362 controls followed by a replication set of 3624 cases and 12,197 controls [6]. It also conducted a replication study using 1806 cases and 3256 controls from Germany. The GWAS identified novel two loci, TMEM232/SLC25A46 (5q22.1) and TNFRSF6B/ZGPAT (20q13.33), and found an association of a common variant, rs3126085 in the FLG locus, with AD at genome-wide significance levels. The association at the TNFRSF6B/ZGPAT locus was replicated in the German sample. TNFRSF6B (also called DCR3) encodes a TNF receptor superfamily gene that plays a suppressive role in FasL- and LIGHT-mediated cell death. Interestingly, LIGHT is a target for airway remodeling in asthma [14] and binds the herpes virus entry mediator (TNFRSF14) [15].
A genome-wide association meta-analysis using 5606 cases and 20,565 controls from 16 population-based studies was reported in 2011 [7]. The study was followed by one with 5419 cases and 19,833 controls from 14 studies that finally identified a total of three novel loci that reached genome-wide significance, OVOL1 (11q13.1), ACTL9 (19p13.2), and KIF3A (5q31.1). Interestingly OVOL1 and ACTL9 are implicated in epidermal proliferation and differentiation, and KIF3A is located in the cytokine cluster region at 5q31.1 containing IL13.
3 Genome-Wide Association Studies of Atopic Dermatitis in the Japanese Population
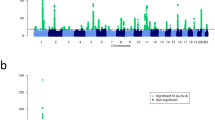
In 2012, a GWAS of AD in the Japanese population using a total of 1472 cases and 7971 controls was published, followed by a replication study with an additional 1856 cases and 7021 controls [8]. The study investigated 606,164 SNP loci and identified a total of 8 novel susceptibility loci for AD: IL1RL1/IL18R1/IL18RAP (2q12), the HLA region (6p21.3), OR10A3/NLRP10 (11p15.4), GLB1 (3p21.33), CCDC80 (3q13.2), CARD11 (7p22), ZNF365 (10q21.2), and CYP24A1/PFDN4 (20q13) (Fig. 5.1). The study also replicated the associations of the FLG, C11orf30, OVOL1, TNFRSF6B/ZGPAT, TMEM232/SLC25A46, ACTL9, and KIF3A/IL13 loci in prior GWASs for AD. All cases were recruited from hospitals in Japan, and the mean ages of the cases in the discovery and replication study were 28.6 and 33.0, respectively. Candidate genes identified by the GWAS suggested important roles for skin barrier functions, adaptive immune responses, IL-1 family signaling, regulatory T cells, and the vitamin D pathway in the pathogenesis of AD.
3.1 IL1RL1/IL18R1/IL18RAP
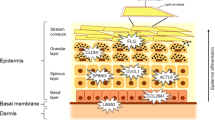
The 2q12 locus contains genes IL1RL1, IL18R1, and IL18RAP, which encode the receptors of IL-1 family cytokines that play crucial roles in the skin. IL1RL1, a component of the IL-33 receptor, is highly expressed in Th2 cells and mast cells. The levels of IL-33 expression are increased in inflamed skin tissue of subjects with AD, and it has been suggested that IL-33 produced in the damaged tissues of AD induces Th2-type inflammation [16].
3.2 The HLA Region
The HLA region contains hundreds of immune system genes, and the locus often shows the strongest association for most autoimmune diseases. Generally, seropositive diseases are associated with HLA class II, and seronegative diseases are associated with HLA class I [17]. The most significant association with AD in the GWAS was observed at rs176095 in the HLA class III region (P = 8.38 × 10−20). After logistic regression analysis of the HLA region, there were two independent association signals in the HLA class I and III regions. The HLA region shows significant associations with asthma [18] and peanuts allergy [19]. Recent studies have reported the involvement of autoimmunity in the pathophysiology of AD, and IgE antibodies against keratinocytes and endothelial cells have been identified in severe AD. However, further studies are necessary to elucidate the involvement of autoimmunity in chronic inflammation in subjects with AD.
3.3 OR10A3/NLRP10
The region at 11p15.4 contains two genes, NLRP10 and OR10A3. OR10A3 encodes an olfactory receptor family gene. NALP proteins sense both microbial and danger signals, and NLRP10 encodes a NALP family protein that lacks the leucine-rich repeat region. NLRP10 is highly expressed in the skin and plays a role in immune responses against invasive bacteria. A recent study using a mouse model of contact hypersensitivity has shown that NLRP10 is involved in T cell-mediated skin inflammation and plays a role in epidermal keratinocytes [20].
3.4 GLB1
GLB1 encodes β-galactosidase-1, but the involvement of the enzyme in allergic inflammation remains unclear. The 3p21.33 locus is located adjacent to the CCR4 gene, which encodes a chemokine receptor for CCL22 and CCL17 (also called TARC). Interestingly, TSLP derived from keratinocytes induces dendritic cells to produce TARC, and skin-specific recruitment of T cells during inflammation is mediated by CCR4. TARC is produced by endothelial cells, keratinocytes, and fibroblasts and plays an important role in AD, bullous pemphigoid, and mycosis fungoides [21]. Serum TARC levels are associated with the disease activity of AD [21].
3.5 CCDC80
The 3q13.2 locus includes CCDC80, which encodes a protein associated with the induction of peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα. A recent study has shown that PPARγ agonists suppress the maturation and migration of dendritic cells and decrease TSLP expression in the skin in a mouse model of AD [22]. C/EBPα is expressed in basal keratinocytes and upregulated in terminal differentiation of keratinocytes [23].
3.6 CARD11
The 3p21.33 locus contains CARD11, which encodes CARMA1. CARMA1 regulates T cell receptor/NF-kB signaling and controls completion of Th17 differentiation [24]. Interestingly, mice homozygous for the CARMA1 mutation develop AD with high levels of serum IgE. Sézary syndrome is an erythrodermic form of cutaneous T cell lymphoma characterized by scaling and itching [25]. Sézary cells, circulating atypical T lymphocytes, show a Th2 cytokine profile, activating CCR4 and CARD11 mutations in nearly one-third of patients. Overlap of the target genes would be helpful to unveil details of shared etiologies.
3.7 ZNF365
The 10q21.2 locus includes three genes, ZNF365, ADO, and EGR2. The most associated variant, rs10995251, is located within ZNF365. Although the region shows a suggestive association with AD in the Chinese population (P = 1.1 × 10−7), it is associated with AD at the genome-wide significance level in the Japanese population (P = 5.9 × 10−20). The locus is also associated with Vogt-Koyanagi-Harada syndrome, which is a multisystem autoimmune disorder affecting pigmented tissues in the ocular, integumentary, auditory, and central nervous systems [26]. Regulatory T cells control the maintenance of immune tolerance and protect the body from harmful immune responses to antigens. It is recognized that dysregulation of Tregs is responsible for the development of a number of immune-mediated diseases. A recent study has shown that induced Tregs characteristically express both LAG3 and EGR2 [27], and EGR2 is involved in the negative regulation of T cell proliferation and inflammation [28].
3.8 CYP24A1/PFDN4
The 20q13 locus contains CYP24A1 and PFDN4. PFDN4 encodes a subunit of a molecular chaperone complex, prefoldin. CYP24A1 encodes 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) 24-hydroxylase, a mitochondrial cytochrome P450 superfamily enzyme that initiates the degradation of the physiologically active form of vitamin D3 by hydroxylation of the side chain [29]. The active form of vitamin D3, 1,25(OH)2D3 is synthesized in the skin systemically after its exposure to sunlight. Since local 1,25(OH)2D3 synthesis activates innate immune responses, sun exposure is an important environmental factor in immune function. A recent study has shown an association between vitamin D deficiency and the severity of AD [30].
4 Immunochip Analysis for Atopic Dermatitis
The Immunochip array is an Illumina Infinium SNP array that was designed by researchers investigating 11 distinct inflammatory and autoimmune diseases in 2009. The chip covers a total of 195,806 SNPs and 718 small insertion-deletions, and the top 2000 associated variants for each disease are included. High-density genotyping study using the Immunochip array identified a total of four novel susceptibility loci for AD in 2013: IL2/IL21 (4q27), PRR5L (11p13), CLEC16A/DEXI (16p13.13), and ZNF652 (17q21.32) [9]. A total of 2425 German subjects with AD and 5449 controls were assessed, and a validation study with 7196 cases and 15,480 controls was conducted using populations from Germany, Ireland, Japan, and China. The IL2 locus is located near the IL21 gene. IL-2 is required for T cell activation and proliferation and regulates the proliferation and survival of regulatory T cells. Cyclosporin A is an inhibitor of calcineurin, which suppresses IL-2 production [31]. Interestingly, recent studies have shown associations of IL2RA and IL21 with immune-mediated diseases [17]. CLEC16A encodes a member of the C-type lectin domain containing family, and CLEC16A variants are associated with multiple immune-mediated diseases such as celiac disease, Crohn’s disease, and alopecia areata [32]. A recent study has shown that CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection and reactivity [32]. In the Japanese population, a total of three loci, PRR5L, CLEC16A/DEXI, and ZNF652, were replicated.
5 Recent Genome-Wide Association Studies of Atopic Dermatitis in Other Populations
A recent study focused on a total of 318 markers associated with any inflammatory trait obtained from a public GWAS database and assessed associations with AD in a three-step approach using 7130 AD cases and 9253 controls [10]. A functional nonsynonymous variant in IL6R (rs2228145, Asp358Ala), which determines the balance between the membrane bound (IL-6R) and soluble forms (sIL-6R), was significantly associated with AD. Interestingly the study also identified increased serum levels of sIL-6R and their influence on the prognosis and persistence of AD.
A GWAS of recalcitrant AD, which is defined as moderate-to-severe AD with allergic sensitization, in Korean children was reported in 2015 [11]. Genetic variants on 13q21.31 were associated with recalcitrant AD at genome-wide significance levels. The closest gene, protocadherin 9 (PCDH9), is located more than 1 Mb from those related variants. In the GWAS, four loci, NBAS (2p24.3), THEMIS (6q22.33), GATA3 (10p14), and SCAPER (15q24.3), showed P values <1 × 10−6.
A GWAS for AD using an imputed data set consisting of more than 1.6 million variants was reported in 2015 [12]. The GWAS assessed 924 tertiary care cases and 5506 population-based controls, followed by an independent replication of 1383 cases and 1728 controls in the German population. Finally, two novel susceptibility loci were identified in the combined analysis: XIRP2 (2q24.3) and DMRTA1 (9p21.3). However, the functions of XIRP2 and DMRTA1 remain unknown.
6 Multi-ancestry Genome-Wide Association Study for Atopic Dermatitis
GWAS meta-analyses for common diseases using large sample sizes have recently been conducted. Collaborative studies consisting of a number of cohorts ensure sufficient statistical power and reveal disease susceptibility loci. A multi-ancestry genome-wide association study for AD using 21,000 cases and 95,000 controls of European, African, Japanese, and Latino ancestry was reported in 2015 [13]. A replication study of the GWAS was conducted using 32,059 cases and 228,628 controls from 18 studies, and a total of 11 novel loci of AD were finally identified.
Among the 11 novel loci, MIR5708/ZBTB10 (8q21.13) was associated with asthma, and ETS1 (11q24.3), IL15RA/IL2RA (10p15.1), MIR5708/ZBTB10 (8q21.13), and LINC00299 (2p25.1) were associated with self-reported allergy [33]. The study indicated that 15 of the 36 reported psoriasis-associated variants were associated with AD (P < 0.05), and 10 variants showed the same direction of association. The GWAS revealed that two loci, CCDC80/CD200R1L (3q13.2) and OR10A3/NLRP10 (11p15.4), might be Japanese-specific signals.
Furthermore, the functional annotations of the associated variants were reviewed in ENCODE consortium [34] and Roadmap Epigenomics Consortium data [35], and their expression quantitative trait loci (eQTL) effects were evaluated in the MuTHER database [36]. The AD associations of DNase I hypersensitivity regions were strongly enriched compared with the rest of the genome, particularly in Th0, Th1, and Th17 cells, at genome-wide significance levels. Interestingly, the most significant cis eQTLs were identified at 2p13.3 (rs4852714 and rs6723629) and eQTLs for CD207 (langerin) in the skin. rs4852714 is also located in open chromatin regions with histone marks, suggesting roles for promoter or enhancer activity in lymphoblastoid cell lines. Langerin is highly expressed in Langerhans cells and Langerin+ dermal dendritic cells and has antiviral and antifungal effects [37].
The majority of the novel loci in this study harbor genes with functional annotations involved in autoimmunity. PPP2R3C regulates B cell maturation and survival, and dysregulation of PPP2R3C leads to autoimmunity in mice. The most significantly associated variant at 5p13.2 (rs10214237) is located 4 kb downstream of IL7R, which is in strong linkage disequilibrium with an IL7R missense variant, rs6897932 (r 2 = 0.94). It has already been reported that the risk allele enhances IL-7 bioavailability [38], and IL-7 transgenic mice show the AD-like features of severe dermatitis with pruritus and elevated serum IgE [39]. Hyper IgE syndrome (HIES) is characterized by AD with high serum IgE levels, and those patients show impaired Th17 function and suffer from recurrent staphylococcal infections. A recent study has shown that dominant negative mutations at the DNA binding site of the STAT3 gene cause HIES [40]. In this GWAS, common variants at the STAT3 locus (17q21.2) were associated with AD. Interestingly, a related genetic variant (rs5892724) at the MIR5708/ZBTB10 locus (8q21.13) is located in open chromatin and affects a STAT3 binding site [41]. ETS1, a plausible candidate gene at 11q24.3, plays roles in Th17 and B cell functions and in keratinocyte differentiation. A recent study has shown that ETS1 is a crucial regulator of ILC2 expansion and cytokine production [42]. Candidate genes at novel loci suggest roles for autoimmune regulation in the pathogenesis of AD, and the findings of this study imply a substantial overlap of genetic components with other autoimmune and inflammatory diseases.
7 Overlapping Loci Between Atopic Dermatitis and Other Diseases
7.1 Psoriasis
AD and psoriasis are common inflammatory diseases affecting the skin that show mutually exclusive clinical characteristics and different immune mechanisms. A recent study using GWAS data has demonstrated that AD and psoriasis have distinct genetic mechanisms in shared pathways involving immune responses and epidermal differentiation [43]. Intriguingly, the influence of genetic factors shows opposite effects in the pathways. Characterization of shared and opposing mechanisms will improve our knowledge of the pathogenesis of inflammatory skin diseases.
7.2 Atopic March
The term atopic march refers to the typical progression of allergic disorders that often begin in early childhood. In most cases, the first sign of the history is the development of AD in an infant. A recent multistage GWAS revealed seven loci involved in the atopic march [44]. The study assessed 2428 cases of infantile eczema followed by childhood asthma and 17,034 controls from 12 populations and identified seven loci: FLG (1q21.3), IL4/KIF3A (5q31.1), AP5B1/OVOL1 (11q13.1), C11orf30/LRRC32 (11q13.5), IKZF3 (17q21), EFHC1 (6p12.3), and TMTC2/SLC6A15 (17q21). EFHC1 and TMTC2/SLC6A15 are associated with the combined eczema plus asthma phenotype and had never been previously reported as susceptibility loci for allergic disease. The study also suggested a strong contribution of AD genes to the atopic march.
8 Epigenetic Analysis of Serum IgE Concentrations
IgE plays an important role in atopic inflammation. Cytosine methylation is generally associated with transcriptional silencing, and perturbations of DNA methylation patterns are often found in disease [45]. A recent study investigated epigenetic associations between serum IgE concentrations and DNA methylation by using Illumina HumanMethylation27 arrays, which assess CpG loci within proximal promoter regions of 14,475 genes [46]. The study identified associations between IgE and low methylation at a total of 36 loci with a meta-analysis false discovery rate <10−4. Annotated genes in several loci such as IL5RA, PGR2, PGR3, and GATA1 encode characteristic proteins of eosinophils. The most significant association was observed within an island of CpG, cg01998785, and adjacent to LPCAT2, which encodes lysophosphatidylcholine acetyltransferase 2. That enzyme functions in production of PAF in inflammatory cells. This study implies the presence of new therapeutically tractable pathways involving IgE production.
9 Perspectives
9.1 Rare Variant Association Studies
Since rare variants are not included in conventional arrays for GWAS, their contributions to AD susceptibility remain unclear. There are several methodologies for rare variant association studies: whole-genome sequencing, whole-exome sequencing, targeted sequencing of candidate genes, and exome arrays. Recent targeted gene sequencing studies have found rare coding genetic variants with strong effects on phenotype variation such as LDL and HDL cholesterol levels. However, a number of rare variant association studies have reported that most variants have modest-to-small effects on the variability of phenotypes [47].
9.2 Skin Microbiota and Immunity
Physical and immune skin barriers are maintained by interactions of keratinocytes, immune cells, and microbes under healthy conditions and also under wounding or infection [48]. Several potential mechanisms have been suggested for the influence of the skin microbiota on the initiation or amplification of skin diseases. Genetic components of patients may enhance sensing or translocation of the microbiota. Skin microbes such as S. aureus and C. albicans may contribute to tissue damage and inflammation in the context of infection. The antimicrobial function of keratinocytes, which are stimulated by IL-17 produced by skin commensal-specific T cells, could enhance skin immunity. Candidate genes identified in GWASs suggest roles for keratinocytes and Th17 cells in the pathogenesis of AD.
9.3 Functional Investigation of Susceptibility Loci
Genetic variants might influence molecular phenotypes, including RNA expression and stability, transcription factor binding, DNA methylation, histone modifications, and protein levels. The functional links between susceptibility variants and phenotypic traits are crucial to improve our knowledge of AD. To investigate how these genetic variants influence the pathophysiology of the disease is required for understanding the disease mechanism and utilization of the findings for prevention or treatment.
Expression quantitative trait loci are commonly used to investigate the effects of genetic variants on gene expression. To interpret susceptibility loci identified by GWAS, the Genotype-Tissue Expression (GTEx) project was launched to establish a database showing the relationship between genetic variants and gene expression in human tissues, including the skin (http://www.gtexportal.org/) [49].
The Encyclopedia of DNA Elements (ENCODE) project was launched in 2003 to identify functional elements in the human genome sequence. The NIH Roadmap Epigenomics Program, which builds on the ENCODE project, has shown the crucial role of epigenomic information for improving our knowledge of gene regulation, cellular differentiation, and human disease (http://www.roadmapepigenomics.org). A recent study has shown that trait- and disease-associated variants are enriched in tissue-specific epigenetic marks [50]. The database is a powerful tool to interpret the molecular mechanisms of human diseases.
Although recent studies identified interindividual variability of immune responses, those studies assessed only a few factors. To improve our understanding of the variability of immune responses in human pathologies more comprehensively, the Human Functional Genomics Project (HFGP) was launched in 2013 (http://www.humanfunctionalgenomics.org) [51]. The project combines “omics” techniques with rigorous phenotyping of immune responses in healthy and diseased subjects. The HFGP database will be a valuable tool for functional genomic studies of human immune-mediated disorders.
Conclusions
There are two major hypotheses regarding the mechanism of AD, abnormalities of the skin barrier and skin inflammation due to dysregulation of immune responses. Intriguingly, candidate genes of AD suggested by GWAS are involved in skin barrier functions and innate and adaptive immune responses. There are great differences in clinical phenotypes of AD among individuals. Since the definition of the phenotype is crucial for genetic studies, deep phenotyping and genotyping studies are needed, and further interdisciplinary research is necessary for translation of the genetics of AD into clinical practice.
References
Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387:1109–22. doi:10.1016/S0140-6736(15)00149-X.
Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi:10.1056/NEJMra0905980.
Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi:10.1038/nrg2796.
Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011;365:1315–27. doi:10.1056/NEJMra1011040.
Esparza-Gordillo J, Weidinger S, Fölster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi:10.1038/ng.347.
Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4. doi:10.1038/ng.851.
Paternoster L, Standl M, Chen CM, Ramasamy A, Bønnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2011;44:187–92. doi:10.1038/ng.1017.
Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi:10.1038/ng.2438.
Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodríguez E, Matanovic A, Marenholz I, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet. 2013;45:808–12. doi:10.1038/ng.2642.
Esparza-Gordillo J, Schaarschmidt H, Liang L, Cookson W, Bauerfeind A, Lee-Kirsch MA, et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J Allergy Clin Immunol. 2013;132:371–7. doi:10.1016/j.jaci.2013.01.057.
Kim KW, Myers RA, Lee JH, Igartua C, Lee KE, Kim YH, et al. Genome-wide association study of recalcitrant atopic dermatitis in Korean children. J Allergy Clin Immunol. 2015;136:678–684.e4. doi:10.1016/j.jaci.2015.03.030.
Schaarschmidt H, Ellinghaus D, Rodríguez E, Kretschmer A, Baurecht H, Lipinski S, et al. A genome-wide association study reveals 2 new susceptibility loci for atopic dermatitis. J Allergy Clin Immunol. 2015;136:802–6. doi:10.1016/j.jaci.2015.01.047.
Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi:10.1038/ng.3424.
Doherty TA, Soroosh P, Khorram N, Fukuyama S, Rosenthal P, Cho JY, et al. The tumor necrosis factor family member LIGHT is a target for asthmatic airway remodeling. Nat Med. 2011;17:596–603. doi:10.1038/nm.2356.
Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–36.
Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 2010;10:103–10. doi:10.1038/nri2692.
Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi:10.1038/nrg3502.
Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi:10.1038/ng.887.
Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun. 2015;6:6304. doi:10.1038/ncomms7304.
Damm A, Giebeler N, Zamek J, Zigrino P, Kufer TA. Epidermal NLRP10 contributes to contact hypersensitivity responses in mice. Eur J Immunol. 2016;46:1959–69. doi:10.1002/eji.201646401.
Saeki H, Tamaki K. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J Dermatol Sci. 2006;43:75–84.
Jung K, Tanaka A, Fujita H, Matsuda A, Oida K, Karasawa K, et al. Peroxisome proliferator-activated receptor γ-mediated suppression of dendritic cell function prevents the onset of atopic dermatitis in NC/Tnd mice. J Allergy Clin Immunol. 2011;127:420–429.e1–6. doi:10.1016/j.jaci.2010.10.043.
Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009;11:1181–90. doi:10.1038/ncb1960.
Molinero LL, Cubre A, Mora-Solano C, Wang Y, Alegre ML. T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc Natl Acad Sci U S A. 2012;109:18529–34. doi:10.1073/pnas.1204557109.
Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47:1426–34. doi:10.1038/ng.3444.
Hou S, Du L, Lei B, Pang CP, Zhang M, Zhuang W, et al. Genome-wide association analysis of Vogt-Koyanagi-Harada syndrome identifies two new susceptibility loci at 1p31.2 and 10q21.3. Nat Genet. 2014;46:1007–11. doi:10.1038/ng.3061.
Okamura T, Fujio K, Sumitomo S, Yamamoto K. Roles of LAG3 and EGR2 in regulatory T cells. Ann Rheum Dis. 2012;71:i96–100. doi:10.1136/annrheumdis-2011-200588.
Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 2005;6:472–80.
Hart PH, Gorman S, Finlay-Jones JJ. Modulation of the immune system by UV radiation: more than just the effects of vitamin D? Nat Rev Immunol. 2011;11:584–96. doi:10.1038/nri3045.
Peroni DG, Piacentini GL, Cametti E, Chinellato I, Boner AL. Correlation between serum 25-hydroxyvitamin D levels and severity of atopic dermatitis in children. Br J Dermatol. 2011;164:1078–82. doi:10.1111/j.1365-2133.2010.10147.x.
Haeck IM, Knol MJ, Ten Berge O, van Velsen SG, de Bruin-Weller MS, Bruijnzeel-Koomen CA. Enteric-coated mycophenolate sodium versus cyclosporin A as long-term treatment in adult patients with severe atopic dermatitis: a randomized controlled trial. J Am Acad Dermatol. 2011;64:1074–84. doi:10.1016/j.jaad.2010.04.027.
Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. 2015;42:942–52. doi:10.1016/j.immuni.2015.04.011.
Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–11. doi:10.1038/ng.2686.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi:10.1038/nature11247.
Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi:10.1038/nbt1010-1045.
Grundberg E, Small KS, Hedman ÅK, Nica AC, Buil A, Keildson S, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–9. doi:10.1038/ng.2394.
de Jong MA, Geijtenbeek TB. Langerhans cells in innate defense against pathogens. Trends Immunol. 2010;31:452–9. doi:10.1016/j.it.2010.08.002.
Lundström W, Highfill S, Walsh ST, Beq S, Morse E, Kockum I, et al. Soluble IL7Rα potentiates IL-7 bioactivity and promotes autoimmunity. Proc Natl Acad Sci U S A. 2013;110:E1761–70. doi:10.1073/pnas.1222303110.
Uehira M, Matsuda H, Nakamura A, Nishimoto H. Immunologic abnormalities exhibited in IL-7 transgenic mice with dermatitis. J Invest Dermatol. 1998;110:740–5.
Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62.
Stritesky GL, Jameson SC, Hogquist KA. Selection of self-reactive T cells in the thymus. Annu Rev Immunol. 2012;30:95–114. doi:10.1146/annurev-immunol-020711-075035.
Zook EC, Ramirez K, Guo X, van der Voort G, Sigvardsson M, Svensson EC, et al. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J Exp Med. 2016;213:687–96. doi:10.1084/jem.20150851.
Baurecht H, Hotze M, Brand S, Büning C, Cormican P, Corvin A, et al. Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet. 2015;96:104–20. doi:10.1016/j.ajhg.2014.12.004.
Marenholz I, Esparza-Gordillo J, Rüschendorf F, Bauerfeind A, Strachan DP, Spycher BD, et al. Meta-analysis identifies seven susceptibility loci involved in the atopic march. Nat Commun. 2015;6:8804. doi:10.1038/ncomms9804.
Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–6. doi:10.1038/nature14192.
Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–4. doi:10.1038/nature14125.
Auer PL, Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7:16. doi:10.1186/s13073-015-0138-2.
Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–9. doi:10.1126/science.1260144.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi:10.1038/ng.2653.
Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi:10.1038/nature14248.
Netea MG, Joosten LA, Li Y, Kumar V, Oosting M, Smeekens S, et al. Understanding human immune function using the resources from the Human Functional Genomics Project. Nat Med. 2016;22:831–3. doi:10.1038/nm.4140.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Tamari, M., Hirota, T. (2018). Genome-Wide Association Study for Atopic Dermatitis in the Japanese Population. In: Katayama, I., Murota, H., Satoh, T. (eds) Evolution of Atopic Dermatitis in the 21st Century. Springer, Singapore. https://doi.org/10.1007/978-981-10-5541-6_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-5541-6_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5540-9
Online ISBN: 978-981-10-5541-6
eBook Packages: MedicineMedicine (R0)