Abstract
Microchip-mass spectrometry has been emerging as excellent analytical tool in the field of complex biological samples analysis. Microchip can play an important role, such as cell culture and sample preparation steps prior to mass spectral identification, which are beneficial from its ability to handle small sample quantities with the potential for high-throughput parallel analysis. Recent progress in chip-mass spectrometry including various approaches that combined microchip devices with electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) is described. Then the main applications of chip-mass spectrometry in proteomics and cell analysis during the recent years are reviewed. We also outlook their implications in the future. The goal of this chapter is to guide new and interested researchers to the important areas and strive toward full integration of microchip and mass spectrometry within biological research field.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Introduction
Mass spectrometry (MS), is the most used analytical tool today due to its high sensitivity and selectivity [1,2,3]. It has been developed as a powerful tool for the qualitative and quantitative detection of analytes ranging from inorganic elements to small organic molecules to macromolecules, such as proteins and nucleic acids. MS has been extensively applied to the study of drug cytotoxicity [4], cell—cell interaction [5, 6], and cellular metabolism [7]. MS has become increasingly important as an analytical tool for bioanalysis because of its sensitivity. MS allows the ionization of intact molecules to obtain a highly accurate molecular weight, making identification of molecules easier. As MS technology advanced, the advent of two “soft” ionization methods, namely the electrospray ionization (ESI) [8] and the matrix-assisted laser desorption/ionization (MALDI) [9, 10] revolutionized MS analysis. ESI produces highly charged ions directly from a liquid, thus facilitating online coupling of chromatographic separations to the mass spectrometer. However, the presence of high concentrations of salts in the samples can lead to severe ion suppression which makes analysis impossible. MALDI has a high tolerance to buffers and salts, but MALDI spectra tend to be very noisy in the mass range below 500 daltons, due to the presence of matrix ions. Therefore coupling mass spectrometry to separation technology was proposed. Despite many advantages in mass spectrometry, the direct application of MS detection to real complex sample is greatly limited. Here, gas chromatography (GC)-MS, liquid chromatography (LC)-MS and capillary electrophoresis (CE)-MS based on column separation have made greatly contribution for the separation and detection of molecules. Mass spectrometry based technology with higher throughput, more rapid and convenient analysis, and lower sample consumption are still in great demand to meet the new challenges in life science.
Miniaturized total analysis systems have attracted an increasing interest in analytical chemistry [11, 12]. There has been great progress in microfluidic devices, which have realized miniaturization of chemical analysis and integration of various chemical processes. This research field is called “micro total analysis systems (μ-TAS)” or “Lab-on-a-chip” [13,14,15,16]. Small fluidic channels, with sizes from tens to hundreds of μm which are fabricated using micro fabrication technology on a substrate of several cm have been utilized as a space in which reaction and analytical processes occur. The device is often called a “microchip.” By controlling the size, fluidics, surface properties, temperature of the micro scale channel, various chemical and analytical processes have been integrated and miniaturized. As a result, many advantages such as short analysis time, low consumption of sample and reagent materials, small waste volumes, effective reactions due to the large specific interfacial area, and small space requirements, have been achieved in this field. This strategy is effective not only from an industrial viewpoint but also widely applicable in various chemical fields, such as organic chemistry, physical chemistry, and cellular biochemistry. In the technology used for biological analysis, several microfluidic devices have been reported as using optical imaging [17], electrophoresis [18], and chemiluminescence [19, 20]. But these methods were limited in many aspects, for example, they lack the information for the characterization of chemical structure. An alternative strategy is to connect the microchip with mass spectrometer for signal analysis.
From 1997, Karger [21] and Ramsey [22] developed the microchip-mass spectrometry in modern biological analysis. A lot of microfluidic chip-mass spectrometry studies on the interface and applications were also reported [23, 24]. A successful research has been carried for peptide separation and identification by coupling a microchip with electrospray ionization quadrupole time-of-flight mass spectrometer [25, 26]. After these works, series dynamic analytical methods for cell culture and metabolism detection [27,28,29], cell-cell co-culture and their information exchange [30,31,32] have been developed, and realized their potential as an integrated platform for cellular biology analysis [27].
In our previous works, we developed integrated microfluidic devices containing multiple functional parts to directly couple with mass spectrometry through a silica-fused capillary for cells microenvironments analysis [4, 28,29,30]. Since 2008 we presents a newly analysis methods for cells study coupling microfluidic devices to ESI-MS and MALDI-MS [25, 31]. Attracted considerable research in recent years showing this method enormous potential for application in the biochemical analysis . That includes cell metabolism [27, 32,33,34], drug absorption [35, 36], drug permeability [37], cell–cell interactions [38,39,40,41], and cell secretion [6, 42]. Here, drug metabolism or cell secretion studies related functional parts, including cell culture, metabolism generation or cell secretion, sample pretreatment, and detection, were all integrated into the microfluidic devices. Compared with conventional methods, our methods offered several advantages, such as short analysis time (less than 10 min), low sample and reagent consumption (less than 100 mL). In order to detect rare samples, we also developed micro-sampling droplet combined with by paper spray ionization for online chemical monitoring of cell culture [7]. Our established platforms have the potential for drug screening and the diagnosis of specific disease with high throughput and low cost.
This chapter will provide a critical survey of recent advances and future trends of microchip-MS in biological sample analysis, with a special focus on various functional sample preparation and separation units integrated on microfluidic chips and the recent interface developments between chip and mass spectrometry . Then the biological applications of chip-MS, including proteomics and cell analysis, are summarized and discussed.
9.2 Mass Spectrometer Interface
Due to the recent development in microfluidic technologies, miniaturized microfluidic ion sources have received considerable attention in coupling with MS, and have gained a great achievement [32]. The interface between microfluidic chip and MS detector is a major challenge in the development of chip-MS technique. ESI can directly couple to microfluidic chips. The interface based on MALDI is typically performed off-line through deposition of the chip effluent onto a MALDI target or by directly ionizing from the chip.
9.2.1 ESI Interface
The major challenges for coupling of microfluidics to ESI-MS focus on the development of stable and effective interfaces . The existing chip-ESI MS interfaces are mainly the following three approaches: (1) spraying directly from the edge of a chip; (2) spraying from a chip-inserted fused-silica chip; (3) spraying from a microfluidic chip-integrated ESI-tip.
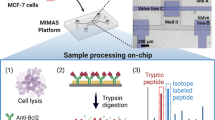
Several approaches for integrating a single electrospray emitter with different materials have been developed. For example, a robust PDMS microchip ESI-MS interface which containing an auxiliary channel to provide electrical contact was developed. Lin’s group developed integrated microfluidic devices containing multiple functional parts to directly couple with ESI-Q-TOF MS through a silica-fused capillary for biochemical analysis , which proposed a new tool for the cell analysis and opened a new research area in the life science. As shown in Fig. 9.1, drug metabolism or cell secretion studies related functional parts, including cell culture, metabolism generation or cell secretion, sample pretreatment, and detection, were all integrated into the microfluidic device. Compared with conventional methods, our methods offered several advantages, such as short analysis time (less than 10 min), low sample and reagent consumption (less than 100 μL). They established platform has the potential for drug screening and the diagnosis of specific disease with high throughput and low cost. Besides, Sun et al. [43] proposed a novel PDMS membrane-based microfluidic emitter for nano ESI-MS (Fig. 9.2a). This emitters were cutting into ∼100-μm-thick, and stable electrosprays could be achieved at low potentials (∼2 kV) and low flow rates (10 nL/min). This membrane-based ESI emitter enabled convenient integration with on-chip separations (e.g., microchip CE) for MS detection and multilayer PDMS microfluidic devices (e.g., pneumatic valve-integrated microchips). The integration of electrospray emitters on glass-based microfluidic chips is also of greatly interest due to important as the interface for chip to mass spectrometer [44]. Hoffman et al. [45] developed a glass-based Chip-MS platform with a monolithically integrated nanospray tip (Fig. 9.2b). The nanospray emitter was manufactured by computer numerical control (CNC) milling followed by heating and pulling. The electrospray performance of the integrated glass emitter was found to be comparable to commercially available nanospray needles . To improve the stability of the electrospray, the same research group optimized the manufacturing process including CNC milling and HF etching to generate a well-controlled opening of the fused pulled tips. These tips are of very small emitter dimensions (only a few microns) without dead-volume.
Microchip ES-MS interface. a Photographs of a PDMS-membrane-based microfluidic ESI emitter from top view and side view [43]. b Visualization of the integration nanospray emitter for electrospray process [45]. c Microchip dual ESI device with analyte emitter active and reference emitter active [53]. (Reprinted with permission from Ref. [43, 45, 53])
Recently, Single cell genomics has become a new frontier [46]. Proof-of-principle experiments have been performed to characterize peptides and metabolites in single cells using MALDI-MS [47] and ESI-MS [48]. However, limited sample processing strategies and low coverage of proteome and metabolome hindered their further developments. Thus, efficient sample processing in a single cell, high-throughput capabilities, as well as the improvement in detection sensitivity are urgently needed in single cell omics studies. The integration of multiple electrosprays simultaneously is a powerful method to improve MS sensitivity. Sequential multiple electrosprays using multichannel [49], multiple nozzles [50, 51], and gated multi-inlets [52] can realize high-throughput. Here, Mao et al. [53] reported a silicon-based monolithic multinozzle emitter array (MEA), it opens up the possibility of a fully integrated microfluidic system for ultrahigh-sensitivity and ultrahigh-throughput proteomics and metabolomics analysis (Fig. 9.2c). Digital microfluidic (DMF) device usually coupled with MALDI in an offline manner, however Baker et al. [54] realized the online coupling of digital microfluidic devices with electrospray ionization using an eductor, which consisted of a transfer capillary, a standard ESI needle, and a tapered gas nozzle. It has a good potential for rapid, versatile and high-throughput microfluidic analysis.
9.2.2 MALDI Interface
MALDI is another common soft ionization method, which was initially proposed in late 1980s [9, 10]. MALDI generally uses for large molecules analysis, such as proteins, peptides, and other biomolecules [43, 55]. Recently, high-density microarrays on microchip were directly adapted to the MALDI-MS analysis. Ability to obtain thousands of spectra from tissues or other samples by MALDI-MS makes it an optimal technology for high-throughput cell analysis and drug research [56,57,58,59]. On-line interfaces for MALDI-MS can be realized by delivering the matrix and analyte into the mass spectrometer mainly by the following three strategies, such as aerosol particles, capillaries, or mechanical means. However, the online coupling is challenge because a MALDI target is under vacuum while the operations on microfluidics are at atmospheric pressure. Microfluidic-based MALDI analysis is usually accomplished by dropping, spraying, or spotting the chip effluent onto a sample target and adding matrix. Spotting is the most widely used method for sample deposition, in which precise volume control of the droplets was very important. The most direct method for sample deposition is spotting with a robotic target spotter. The matrix and samples are online mixed before deposition or spotted onto a matrix-coated target. Using this approach, we developed inkjet automated single cells and matrices printing system for single cell MALDI analysis [60]. Single or several cells were introduced and printed onto ITO glass substrate by inkjet technology and phospholipids were detected by MALDI-MS cell analysis platform (Fig. 9.3a). A gold nanoparticles modified porous silicon chip based surface assisted laser desorption/ionization mass spectrometry (SALDI-MS) was developed to capture and analyze glutathione (GSH) in cells [61]. The silicon chip was array patterned for high throughput SALDI-MS detection and showed great potential for more efficient analysis of small thiol biomarkers in complex biological samples (Fig. 9.3b).
9.3 On-Chip Sample Pretreatment
The integration of sample pretreatment within microfluidic devices has become an important research area. In biological analysis, the analytes of interest are commonly at low abundance and mixed with complex interferences. Considering the limited volume of analysis system, pretreatment is roughly divided as preconcentration and separation. On-chip sample preconcentration and separation are especially needed during chip-MS analysis.
9.3.1 Sample Preconcentration
Before analytes are prepared on MS targets, samples usually need to go through preconcentration. Hence, preconcentration is necessary for effective MS analysis. For example, Lin group realized the extraction of cell metabolites from culture medium via microdialysis (Fig. 9.4a) [7]. A homemade microdialysis hollow fiber was immersed into cell culture medium, and as dialysate flowed into the fiber, glucose from culture media diffused through dialysis interface. The other end of the fiber came to paper spray ionization for MS detection. With the continuously flowing dialysate and generating droplets, on-line monitoring at glucose level in cell culture medium was achieved with the shortest time interval of 1.5 s. This work demonstrated a potential platform with label free MS detection for the monitoring of cellular culture system.
On-Chip sample preconcentration. a Microdialysis to extract glucose for paper spray ionization from cell culture medium [7]. b Schematic representation of the experimental setup. For the electrospray induction, high voltage (HV) is applied to the SPE-GEMS/MS microchip emitter, placed in front of the ion transfer capillary inlet of MS instrument. Meanwhile, for the fractionation of the analyte, enriched on the magnetic SPE sorbent, the eluent flow and composition are controlled by analytical LC pump and microsplitter valve [65]. (Reprinted with permission from Ref. [7, 65])
Novel paper spray cartridge with an integrated solid phase extraction (SPE) column was described by Zhang and Manicke [62]. The cartridge performs extraction and pre-concentration, as well as sample ionization by paper spray, from complex samples such as plasma. The cartridge allows for selective enrichment of target molecules from larger sample volumes and removal of the matrix, which significantly improved the signal intensity of target compounds in plasma samples by paper spray ionization .
Furthermore Hu’s group proposed a chip-based array magnetic solid phase microextraction (MSPME) system and on-line combined it with ICP-MS via microflow concentric nebulization for the determination of trace heavy metals in cells [63]. Wheeler group introduced a method to couple solid-phase microextraction (SPME) with HPLC-MS using digital microfluidics (DMF) [64]. In this technique, analytes in sample solution are extracted into a SPME fiber coating off-line, and then eluted into a droplet of desorption solvent on a digital microfluidic device, prior to analysis by HPLC-MS. The low volumes inherent to DMF allow for pre-concentration of analytes prior to analysis . The new SPME-DMF-HPLC-MS method was applied to the quantification of pg/mL-level free steroid hormones in urine. Girault group introduced an on-chip solid-phase extraction–gradient elution–tandem mass spectrometry (SPE-GEMS/MS), as shown in Fig. 9.4b [65]. This technique combines in a microfluidic format online sample preconcentration/purification on SPE sorbent with further fractionation and MS/MS analysis. Mikkonen et al. [66] used isoelectric focusing (IEF) to enrich samples in an open channel. After preconcentration, matrix was applied to the analytes by electrospray matrix deposition, resulting in effective crystallization in the channel for subsequent MALDI-MS detection. Then, they developed a novel method for preconcentration and purification of the Alzheimer’s disease related amyloid beta (Aβ) peptides by IEF in microchannels combined with their analysis by micropillar-matrix-assisted laser desorption ionization-time-of-flight-mass spectrometry (MALDI-TOF-MS) [67]. Yang and co-workers presented a microfluidic aptamer-based biosensor for detection of low-molecular-weight biomarkers in patient samples by using computer simulation and performing MALDI-MS analysis directly from the open microchannel [68]. Using a microfluidic device that integrates aptamer-based specific analyte extraction, isocratic elution, and detection by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry demonstrated rapid, sensitive and label-free detection of arginine vasopressin (AVP) in human plasma ultrafiltrate. This integrated on-chip sample processing enables the quantitative detection of low-abundance AVP by MALDI-TOF mass spectrometry in a rapid and label-free manner.
9.3.2 Sample Separation
On-chip separation technique was also developed on a chip-MS platform to achieve multi-component, high throughput analysis. Here, Ramsey’s group developed a microfabricated fluidic device for the automated real-time analysis of individual cells using capillary electrophoresis (CE) and electrospray ionization-mass spectrometry (ESI-MS) [69]. The microfluidic structure incorporates a means for rapid lysis of single cells within a free solution electrophoresis channel, where cellular constituents were separated, and an integrated electrospray emitter for ionization of separated components. The eluent was characterized using mass spectrometry . The dissociated heme group and the α and β subunits of hemoglobin from individual erythrocytes were detected as cells continuously flowed through the device. The average analysis throughput was approximately 12 cells per minute, demonstrating the potential of this method for high-throughput single cell analysis. Moreover, they utilized capillary electrophoresis–mass spectrometry (CE–MS) in an integrated microfluidic platform to analyze an intact, lysine-linked antibody drug conjugate (ADC) in order to assess post translational modifications and drug load variants [70].
Feasibility of multichannel parallel separation for sensitivity improvement in interfacing microchip electrophoresis hyphenated with inductively coupled plasma mass spectrometry (MCE-ICP-MS) was reported by Wang group [71]. By using 2–20 array lanes for parallel separation, the sensitivity of the MCE-ICP-MS system was proportionally improved by 2–20 folds. There are other work s concerning the application of MCE in metabolites probing with combination of electrochemical [72] or ESI-MS detection [73]. The sensitivity and quantitative ability of CE can be further strengthened via modification of sample introduction manner such as inkjet direct sampling (Fig. 9.5) [74]. About rapid liquid phase lithography prototyping creates the multifunctional device economically. Belder’s group presented a microfluidic chips containing porous polymer monolithic columns as a means to facilitate chemical transformations as well as both downstream chromatographic separation and mass spectrometric analysis [75].
9.4 Analytical Application
9.4.1 Proteomics
Proteomics has generated great research interest in elucidating cell function, to investigate the relationship between diseases state and clinical diagnostics [76]. Currently, one of the most successful analytical strategies of proteomics is 2D gel electrophoresis separation, in-gel digestion by proteases, and MS detection and analysis. However, many necessary sample-preparation steps in this process remain time consuming and labor intensive. Microfluidic systems have been proposed as the means of automating sequential sample-pretreatment steps and increasing sample throughput in parallel proteomics analysis [77]. Multiple processing steps can be seamlessly integrated on a single chip or multiple chips to allow fully automated sample processing, which can address the labor-intensity issues associated with proteomics analyses. Protein identification forms the basis for understanding protein functions and their roles in biological networks. Microfluidic-based approaches have been developed to perform various tasks that are essential for protein identification, such as enzymatic digestion [78,79,80,81,82], separation [83,84,85], and sample infusion [86]. Integrated microfluidic systems capable of performing multiple functions have also been reported for protein identification [87].
Wilson group developed a means to integrate hydrogen/deuterium exchange (HDX) labeling upstream of this system to study allosteric effects in enzyme inhibition [88]. As shown in Fig. 9.6a, a variable position micromixer was used to adjust the reaction time of the beta lactamase enzyme, TEM-1, with a covalent inhibitor clavulanate. This system was successfully applied to characterize structural changes and dynamics that occur upon acylation of TEM-1 by clavulanate. Both rapid structural changes at the active site and subtle changes in active-remote regions were identified.
MS-based investigation of protein–protein interactions usually involves the enrichment of a bait protein and its interacting partners followed by separation, digestion, and MS detection. Microfluidic-based approaches can greatly facilitate the identification of protein complexes and their related dynamic pathways. Recent developments of surface plasmon resonance (SPR) and MS have provided the feasibility of better integration that could be dedicated to the identification and characterization of protein–protein interactions [89]. Madeira et al. discoveedr protein–protein interactions using the surface plasmon resonance technique coupled to mass spectrometry (MS), as shown in Fig. 9.6b. A peptide or a protein is immobilized on a sensor chip and then exposed to brain extracts injected through the surface of the chip by a microfluidic system. The interactions between the immobilized ligand and the extracts can be monitored in real time.
9.4.2 Metabolomics
Cell metabolite analysis is of great interest to analytical chemistry and clinical medicine society, and some metabolites having been identified as important indicators of major diseases such as cancer and HIV. Recently, our group has published a series of works employing micro-SPE columns integrated to cell culture chip for establishment of drug cellular testing platform, study of cell-cell communication and drug metabolism. The device contained three components: One part composed of drug and culture medium injection entrance; the second part was used as cell culture and observation chamber for drug-induced cell apoptosis; the third part was composed of array of SPE micro columns for sample desalination. For examples, Gao et al. introduced a microfluidic platform to test drug permeability of intestine and coupled mass spectrometer for online curcumin detection [90]. Cell culture as well as sample desalting and extraction was integrated in the same chip by operating two independent channels separated by a semipermeable polycarbonate (PC) membrane , and integrating SPE micro-columns. Permeation of curcumin on microfluidic platform was characterized by HPLC and electrospray ionization quadrupole time-of-flight mass spectrometer (ESI-Q-TOF MS). Gao et al. developed an integrated chip-MS platform for high-throughput drug screening with an online ESI-Q-TOF MS. By using the technology combination, characterization of drug absorption and evaluation of cytotoxicity could be simultaneously realized (Fig. 9.7a) [35]. Chen et al. reported a microfluidic chip integrated ESI-Q-TOF MS platform based on a stable isotope labeling technology for qualitative and quantitative analysis of cell metabolism under antitumor drug genistein treatment [27]. They also reported a cell-compatible paper chip for in situ sensing of live cell components by PSI-MS, which allowed profiling the cellular lipids and quantitative measurement of drug metabolism with minimum sample pretreatment (Fig. 9.7b) [91]. Wei et al. fabricated a cell co-culture chip in which the secreted proteins were qualitatively and semi-quantitatively determined by a directly coupled mass spectrometer (Fig. 9.1b) [39]. PC12 cells and GH3 cells were co-cultured under various conditions to simulate the nervous system regulation of different organs. A SPE column was integrated to remove salts from the cells secretion prior to MS detection. A three-layer PDMS microfluidic device was fabricated to integrate valves for avoiding contamination between the cells co-culture zone and the pretreatment zone. The metabolites could be directly detected online with an ESI-Q-TOF MS after micro-SPE pretreatment. Zhang et al. constructed an in vitro liver model in a microchip to imitate and detect prodrug capecitabine (CAP) metabolism (Fig. 9.7c) [6]. CAP was metabolized into active metabolite in HepG2 cell and then transformed into final effective drug in tumor cells. SPE microcolumns were integrated into this device as pretreatment units prior to ESI-Q-TOF MS.
Cell metabolite analysis on chip-MS platform. a A schematic drawing shows the upstream concentration gradient generator, the downstream cell cultivation modules, and sample pretreatment module prior to ESI-Q-TOF MS detection. [35] b Schematic diagram of cell cultures on paper-based chip-MS for cell lipid analysis. [91] c Schematic of cell co-culture and on-chip preconcentration process on microchip. [5]. (Reprinted with permission from Ref. [5, 35, 91])
9.4.3 Glycomics
Protein glycosylation plays an important role in various biological processes, such as modification of protein function, regulation of protein–protein interactions, and control of turnover rates of proteins. Glycosylation is one of the most common post-translational protein modifications, having major roles in various biological processes [92,93,94,95,96,97]. Quantification is an important part of a glycomics study as in recent years; glycans have been identified as potential biomarkers for a number of diseases, including cancers, immune deficiencies, and cardiovascular diseases [98,99,100,101]. Accordingly, quantitation reliability is critical for analysis; meanwhile high-throughput and highly efficient quantitative methods are in high demand because of the vast number of samples required for quantitative glycomic analyses of clinical samples.
Glycans can be labeled with chromophores and detected and quantified by fluorescence spectroscopy. However, spectroscopy-based quantitation methods rely greatly on internal or external standards, and quantitation reliability decreases with increasing sample complexity. In recent years, mass spectrometry (MS) has become a dominant analytical instrument for glycomic research because of the ability to offer glycan identification, structure elucidation, and compositional quantitation through high resolution MS and triple quadrupole MS operated in selected reaction monitoring (SRM) or multiple reaction monitoring (MRM) modes [102, 103]. For the overall analysis of glycans or oligosaccharides, the competitive ionization in the ion source of a mass spectrometer is a significant barrier to observation of many trace-level analytes. The use of a separation technique prior to MS analysis is perhaps the most efficient approach to minimize the adverse effects from competitive ionization. The most reported separation methods are graphitized carbon chromatography [104], hydrophilic interaction chromatography, high performance anion-exchange chromatography, chip-based reverse-phase LC-MS. Here, Lebrilla’s group first used an Agilent HPLC-Chip/Time-of-Flight MS system for the separation of serum glycans, and then online detection by nano ESIMS [105]. The microfluidic chip consisted of an enrichment column, an analytical column packed both with porous graphitized carbon as the stationary phase, and an integrated nano-ESI emitter. By analyzing serum glycans in two groups of prostate cancer patients with different prognoses, they found a number of significant differences in N-glycan abundances. Lebrilla’s group has demonstrated the application of Chip-MS to glycomics by profiling N-glycans from human serum [106] and human milk glycome [107,108,109,110]. They have achieved [105] the comprehensive glycan profiling of human serum with the capability of isomer separation and quantitation by Chip LC-MS. In this study, a microfluid ic chip pa cked with graphiti zed carbon was used to chroma tographically separate the glycans from the serum sampl es of two groups of prostate cancer patients. More than 300 N-glycan species (including isomeric structures) were identified, corresponding to over 100 N-glycan compositions.
9.4.4 Single Cell Analysis
Nowadays, numerous methods have been developed to disperse and confine single cells in discrete compartments for further mass spectrometry (MS)-coupled analysis . The coupling of a microfluidic chip with MS has been emerging as a powerful analytical tool in cell-based assays [29, 111,112,113].
A stable isotope labeling assisted microfluidic chip-electrospray ionization MS platform was developed to qualitatively and quantitatively analyze cell metabolism [27]. Proteins secreted by co-cultured cells were qualitatively and semi-quantitatively determined by MS which was coupled with an integrated microfluidic device [39]. An electrochemical chip was coupled online to MS or LC–MS to generate phase I and phase II drug metabolites and to demonstrate protein modification by reactive metabolites . This new screening method gave the potential of this electrochemical chip as a complementary tool for a variety of drug metabolism studies in the early stages of drug discovery [113].
Recently, Liu’s group [114] developed a microfluidic platform deploying the double nano-electrode cell lysis technique for automated analysis of single cells with mass spectrometric detection. The proposed microfluidic chip features integration of a cell-sized high voltage zone for quick single cell lysis , a microfluidic channel for electrophoretic separation, and a nanoelectrospray emitter for ionization in MS detection. Built upon this platform, a microchip electrophoresis-mass spectrometric method (MCE-MS) has been developed for automated single cell analysis. In the method, cell introduction, cell lysis, and MCE-MS separation are computer controlled and integrated as a cycle into consecutive assays. Analysis of large numbers of individual PC-12 neuronal cells (both intact and exposed to 25 mM KCl) was carried out to determine intracellular levels of dopamine (DA) and glutamic acid (Glu) . To conduct the studies of cell metabolomics at single-cell resolution, MALDI-MS was used as an analytical detector on microplate chip. Amantonico et al. [47] validated a single-cell mass spectrometric method to evaluate the metabolic levels in the populations of unicellular organisms (Fig. 9.8a). Several cellular components (ADP, ATP, GTP, and UDP-Glucose) in single-cells of Closterium acerosum was easily analyzed by negative-mode MALDI-MS. Recently, Our group [115] reported a single-cell lipid profiling platform combining an inkjet nozzle cell manipulator with probe electrospray ionization mass spectrometry (PESI-MS) (Fig. 9.8b). Through inkjet sampling of a cell suspension, droplets with single cells were generated, precisely dripped onto a tungsten-made electrospray ionization needle, and immediately sprayed under a high-voltage electric field. Lipid fingerprints of single cells were obtained by a mass spectrometry (MS) detector.
9.5 Conclusion and Future Perspective
Coupling microfluidic chips to MS takes advantages of both technologies, addressing issues in conventional analytical methods such as efficiency, sensitivity and throughput. In this chapter, we documented the advances in coupling microfluidic chips to MS in the past decade, covering both innovations in Chip-MS interfaces and their applications in biological research and clinical diagnosis. ESI interfaces have evolved from off-the-edge spraying, inserted fused-silica capillary emitters to fully integrated micro fabricated emitters. There are remarkable advantages in integrated emitters such as easy fabrication and zero dead volume, and the number of corresponding publications has been rapidly increasing. In addition to a large number of innovations in Chip-MS interfaces, a wide range of applications has also been reported, including proteomics , glycemic, metabolic studies, cell analysis, and clinical diagnosis. Integration of multiple functions has been a major trend in coupling microfluidic chips with MS for automated sample preparation and MS detection with high sensitivity and throughput. Highly-automated, fully-integrated chip-MS would revolutionize the way that we do chemical and biological analysis today.
References
Bings NH, Skinner CD, Wang C, Colyer CL, Harrison DJ et al (2000) Coupling electrospray mass spectrometry to microfluidic devices with low dead volume connections. In: Harrison DJ, van den Berg, Albert (eds), Micro Total Analysis Systems ’98, Springer, Dordrecht 141–144
Astorga-Wells J, Jornvall H, Bergman T (2003) A microfluidic electrocapture device in sample preparation for protein analysis by MALDI mass spectrometry. Anal Chem 75:5213–5219
Grimm RL, Beauchamp JL (2003) Field-induced droplet ionization mass spectrometry. J Phys Chem B 107:14161–14163
Gao D, Li H, Wang N, Lin JM (2012) Evaluation of the absorption of methotrexate on cells and its cytotoxicity assay by using an integrated microfluidic device coupled to a mass spectrometer. Anal Chem 84:9230–9237
Ong TH, Kissick DJ, Jansson ET, Com TJ, Romanova EV et al (2015) Classification of large cellular populations and discovery of rare cells using single cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem 87:7036–7042
Zhang J, Wu J, Li HF, Chen QS, Lin JM (2015) An in vitro liver model on microfluidic device for analysis of capecitabine metabolite using mass spectrometer as detecto. Biosens Bioelectron 68:322–328
Liu W, Wang NJ, Lin XX, Ma Y, Lin JM (2014) Interfacing microsampling droplets and mass spectrometry by paper spray ionization for online chemical monitoring of cell culture. Anal Chem 86:7128–7134
Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246:64–71
Karas M, Hillenkamp F (1988) Laser desorption ionization of proteins with molecular massesexceeding 10,000 daltons. Anal Chem 60:2299–2301
Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y et al (1988) Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Sp 2:151–153
Janasek D, Franzke J, Manz A (2006) Scaling and the design of miniaturized chemical-analysis systems. Nature 442:374–380
Ríos A, Escarpa A, González MC, Crevillén AG (2006) Challenges of analytical microsystems. TrAC Trend Anal Chem 25:467–479
Reyes DR, Iossifidis D, Auroux PA, Manz A (2002) Micro total analysis systems. 1. introduction, theory, and technology. Anal Chem 74:2623–2636
Auroux PA, Iossifidis D, Reyes DR, Manz A (2002) Micro total analysis systems. 2. analytical standard operations and applications. Anal Chem 74:2637–2652
Vilkner T, Janasek D, Manz A (2004) Micro total analysis systems. 3. Recent Developments. Anal Chem 76:3373–3385
Dittrich P, Tachikawa K, Manz A (2006) Micro total analysis systems. Latest Advancements and Trends. Anal Chem 78:3887–3907
Liu W, Dechev N, Foulds IG, Burke R, Parameswaran A et al (2009) A novel permalloy based magnetic single cell micro array. Lab Chip 9:2381–2390
Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M et al (2010) Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science 329:533–538
Su R, Lin JM, Qu F, Gao Y, Chen Z et al (2004) Capillary electrophoresis microchip coupled with on-line chemiluminescence detection. Anal Chim Acta 508:11–15
Su R, Lin JM, Uchiyama K, Yamada M (2004) Integration of a flow-type chemiluminescence detector on a glass electrophoresis chip. Talanta 64:1024–1029
Koster S, Verpoorte E (2007) A decade of microfluidic analysis coupled with electrospray mass spectrometry: An overview. Lab Chip 7:1394–1412
Whitesides GM (2006) The origins and the future of microfluidics. Nature 442:368–373
Lee J, Soper SA, Murray KK (2009) Microfluidic chips for mass spectrometry-based proteomics. J Mass Spectrom 44:579–593
Meyvantsson I, Beebe DJ (2008) Cell culture models in microfluidic systems. Annu Rev Anal Chem 1:423–449
Li H, Liu J, Cai Z, Lin JM (2008) Coupling a microchip with electrospray ionization quadrupole time-of-flight mass spectrometer for peptide separation and identification. Electrophoresis 29:1889–1894
Zheng Y, Li H, Guo Z, Lin JM, Cai Z (2007) Chip-based CE coupled to a quadrupole TOF mass spectrometer for the analysis of a glycopeptide. Electrophoresis 28:1305–1311
Chen Q, Wu J, Zhang YD, Lin JM (2012) Qualitative and quantitative analysis of tumor cell metabolism via stable isotope labeling assisted microfluidic chip electrospray ionization mass spectrometry. Anal Chem 84:1695–1701
Gao D, Liu H, Jiang Y, Lin JM (2013) Recent advances in microfluidics combined with mass spectrometry: technologies and applications. Lab Chip 13:3309–3322
Li H, Zhang Y, Lin JM (2014) Recent advances in coupling techniques of microfluidic device-mass spectrometry for cell analysis. Scientia Sinica Chimica 44(5):777–783
Jie M, Mao S, Li H, Lin JM (2017) Multi-channel microfluidic chip-mass spectrometry platform for cell analysis. Chin Chem Lett. doi:10.1016/j.cclet.2017.05.024
Wei H, Li H, Lin JM A microfluidic device combined with ESI-Q-TOF mass spectrometer applied to analyze herbicides on a single C30 bead. J Chromatogr A 1216: 9134–9142
Mao S, Gao D, Liu W, Wei H, Lin JM (2012) Imitation of drug metabolism in human liver and cytotoxicity assay using a microfluidic device coupled to mass spectrometric detection. Lab Chip 12(1):219–226
Gao D, Wei H, Guo GS, Lin JM (2010) Microfluidic cell culture and metabolism detection with electrospray ionization quadrupole time-of-flight mass spectrometer. Anal Chem 82:5679–5685
Zhang J, Chen F, He Z, Ma Y, Uchiyama K, Lin JM (2016) A novel approach for precisely controlled multiple cell patterning in microfluidic chip by inkjet printing and the detection of drug metabolism and diffusion. Analyst 141:2940–2947
Gao D, Li H, Wang N, Lin JM (2012) Evaluation of the absorption of methotrexate on cells and its cytotoxicity assay by using an integrated microfluidic device coupled to mass spectrometer. Anal Chem 84(21):9230–9237
Liu W, Lin JM (2016) Online monitoring of lactate efflux by multi-channel microfluidic chip-mass spectrometry for rapid drug evaluation. ACS Sensor 1(4):344–347
Gao D, Liu H, Lin JM, Wang Y, Jiang Y (2013) Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip 13:978–985
Mao S, Zhang J, Li H, Lin JM (2013) A novel strategy for high throughput signaling molecule detection to study cell–to–cell communication. Anal Chem 85(2):868–876
Wei H, Li H, Mao S, Lin JM (2011) Cell signaling analysis by mass spectrometry under co-culture conditions on an integrated microfluidic device. Anal Chem 83(24):9306–9313
Wu J, Jie M, Dong X, Qi H, Lin JM (2016) Multi-channel cell co-culture for drug development based on glass microfluidic chip-mass spectrometry coupled platform. Rapid Commun Mass. SP 30:80–86
Jie M, Li H, Lin L, Zhang J, Lin JM (2016) Integrated microfluidic system for cell co-culture and simulation of drug metabolism. RSC Adv. 6:54564–54572
Wei H, Li H, Gao D, Lin JM (2010) Multi-channel microfluidic devices combined with electrospray ionization quadrupole time-of-flight mass spectrometry applied to the monitoring of glutamate release from neuronal cells. Analyst 135:2043–2050
Sun X, Kelly RT, Tang K, Smith RD (2011) Membrane-based emitter for coupling microfluidics with ultrasensitive nanoelectrospray ionization-mass spectrometry. Anal Chem 83:5797–5803
Yue GE, Roper MG, Jeffery ED, Easley CJ, Balchunas C et al (2005) Glass microfluidic devices with thin membrane voltage junctions for electrospray mass spectrometry. Lab Chip 5:619–627
Hoffmann P, Haeusig U, Schulze P, Belder D (2007) Microfluidic glass chips with an integrated nanospray emitter for coupling to a mass spectrometer. Angew Chem Int Edit 46:4913–4916
Wang D, Bodovitz S (2010) Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol 28:281–290
Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R (2010) Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal Chem 82:7394–7400
Lapainis T, Rubakhin SS, Sweedler JV (2009) Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal Chem 81:5858–5864
Xue Q, Foret F, Dunayevskiy YM, Zavracky PM, McGruer NE et al (1997) Multichannel microchip electrospray mass spectrometry. Anal Chem 69:426–430
Schultz GA, Corso TN, Prosser SJ, Zhang S (2000) A fully integrated monolithic microchip electrospray device for mass spectrometry. Anal Chem 72:4058–4063
Kim W, Guo M, Yang P, Wang D (2007) Microfabricated monolithic multinozzle emitters for nanoelectrospray mass spectrometry. Anal Chem 79:3703–3707
Moini M, Jiang L, Bootwala S (2011) High-throughput analysis using gated multi-inlet mass spectrometry. Rapid Commun Mass Sp 25:789–794
Mao P, Wang HT, Yang PD, Wang DJ (2011) Multinozzle emitter arrays for nanoelectrospray mass spectrometry. Anal Chem 83:6082–6089
Baker CA, Roper MG (2012) Online coupling of digital microfluidic devices with mass spectrometry detection using an eductor with electrospray ionization. Anal Chem 84:2955–2960
Ericson C, Phung QT, Horn DM, Peters EC, Fitchett JR et al (2003) An automated noncontact deposition interface for liquid chromatography matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem 75:2309–2315
Lee J, Soper SA, Murray KK (2009) Development of an efficient on-chip digestion system for protein analysis using MALDI-TOF MS. Analyst 134:2426–2433
Xie W, Gao D, Jin F, Jiang Y, Liu H (2015) Study of phospholipids in single cells using an integrated microfluidic device combined with matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem 87:7052–7059
Lazar IM, Kabulski JL (2013) Microfluidic LC device with orthogonal sample extraction for on-chip MALDI-MS detection. Lab Chip 13:2055–2065
Yang M, Nelson R, Ros A (2016) Toward analysis of proteins in single cells: a quantitative approach employing isobaric tags with MALDI mass spectrometry realized with a microfluidic platform. Anal Chem 88:6672–6679
Korenaga A, Chen F, Li H, Uchiyama K, Lin JM (2016) Inkjet automated single cells and matrices printing system for matrix-assisted laser desorption/ionization mass spectrometry. Talanta 162:474–478
Wu J, Jie M, Li H, He Z, Wang S (2017) Gold nanoparticles modified porous silicon chip for SALDI-MS determination of glutathione in cells. Talanta 168:222–229
Zhang C, Manicke NE (2015) Development of a paper spray mass spectrometry cartridge with integrated solid phase extraction for bioanalysis. Anal Chem 87:6212–6219
Wang H, Wu Z, Chen B, He M, Hu B (2015) Chip-based array magnetic solid phase microextraction on-line coupled with inductively coupled plasma mass spectrometry for the determination of trace heavy metals in cells. Analyst 140:5619–5626
Choi K, Boyacı E, Kim J, Seale B, Barrera-Arbelaez L et al (2016) A digital microfluidic interface between solid-phase microextraction and liquid chromatography–mass spectrometry. J Chromatogr A 1444:1–7
Gasilova N, Srzentić K, Qiao L, Liu B, Beck A et al (2016) On-chip mesoporous functionalized magnetic microspheres for protein sequencing by extended bottom-up mass spectrometry. Anal Chem 88:1775–1784
Mikkonen S, Rokhas MK, Jacksén J, Emmer Å (2012) Sample preconcentration in open microchannels combined with MALDI-MS. Electrophoresis 33:3343–3350
Mikkonen S, Jacksén J, Roeraade J, Thormann W, Emmer Å (2016) Microfluidic isoelectric focusing of amyloid beta peptides followed by micropillar-matrix-assisted laser desorption ionization-mass spectrometry. Anal Chem 88:10044–10051
Yang J, Zhu J, Pei R, Oliver JA, Landry DW et al (2016) Integrated microfluidic aptasensor for mass spectrometric detection of vasopressin in human plasma ultrafiltrate. Anal Methods 8:5190–5196
Mellors JS, Jorabchi K, Smith LM, Ramsey JM (2010) Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal Chem 82:967–973
Redman EA, Mellors JS, Starkey JA, Ramsey JM (2016) Characterization of intact antibody drug conjugate variants using microfluidic capillary electrophoresis–mass spectrometry. Anal Chem 88:2220–2226
Cheng H, Liu J, Xu Z, Wang Y, Ye M (2016) Improving sensitivity for microchip electrophoresis interfaced with inductively coupled plasma mass spectrometry using parallel multichannel separation. J Chromatogr A 1461:198–204
Saylor RA, Reid EA, Lunte SM (2015) Microchip electrophoresis with electrochemical detection for the determination of analytes in the dopamine metabolic pathway. Electrophoresis 36:1912–1919
Nordman N, Sikanen T, Moilanen ME, Aura S, Kotiaho T (2011) Rapid and sensitive drug metabolism studies by SU-8 microchip capillary electrophoresis-electrospray ionization mass spectrometry. J Chromatogr A 1218:739–745
Chen FM, Rang Y, Weng Y, Lin LY, Zeng HL et al (2015) Drop-by-drop chemical reaction and sample introduction for capillary electrophoresis. Analyst 140:3953–3959
Dietze C, Schulze S, Ohla S, Gilmore K, Seeberger PH et al (2016) Integrated on-chip mass spectrometry reaction monitoring in microfluidic devices containing porous polymer monolithic columns. Analyst 141:5412–5416
Haynes PA, Roberts TH (2007) Subcellular shotgun proteomics in plants: looking beyond the usual suspects. Proteomics 7:2963–2975
Chao TC, Hansmeier N (2013) Microfluidic devices for high-throughput proteome analyses. Proteomics 13:467–479
Liuni P, Rob T, Wilson DJ (2010) A microfluidic reactor for rapid, low pressure proteolysis with on-chip electrospray ionization. Rapid Commun Mass Sp 24:315–320
Fan HZ, Bao HM, Zhang LY, Chen G (2011) Immobilization of trypsin on poly(urea-formaldehyde)-coated fiberglass cores in microchip for highly efficient proteolysis. Proteomics 11:3420–3423
Ji J, Nie L, Qiao L, Li Y, Guo L et al (2012) Proteolysis in microfluidic droplets: An approach to interface protein separation and peptide mass spectrometry. Lab Chip 12:2625–2629
Lee J, Soper SA, Murray KK (2011) A solid-phase bioreactor with continuous sample deposition for matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry. Rapid Commun Mass Sp 25:693–699
Min Q, Zhang X, Wu R, Zou H, Zhu JJ (2011) A novel magnetic mesoporous silica packed S-shaped microfluidic reactor for online proteolysis of low-MW proteome. Chem Commun 47:10725–10727
Fritzsche S, Hoffmann P, Belder D (2010) Chip electrophoresis with mass spectrometric detection in record speed. Lab Chip 10:1227–1230
Nordman N, Sikanen T, Aura S, Tuomikoski S, Vuorensola K et al (2010) Feasibility of SU-8-based capillary electrophoresis-electrospray ionization mass spectrometry microfluidic chips for the analysis of human cell lysates. Electrophoresis 31:3745–3753
Chambers AG, Mellors JS, Henley WH, Ramsey JM (2011) Monolithic integration of two-dimensional liquid chromatography-capillary electrophoresis and electrospray ionization on a microfluidic device. Anal Chem 83:842–849
Sun X, Kelly RT, Tang K, Smith RD (2010) Ultrasensitive nanoelectrospray ionization-mass spectrometry using poly(dimethylsiloxane) microchips with monolithically integrated emitters. Analyst 135:2296–2302
Wang C, Jemere AB, Harrison DJ (2010) Multifunctional protein processing chip with integrated digestion, solid-phase extraction, separation and electrospray. Electrophoresis 31:3703–3710
Rob T, Gill PK, Golemi-Kotra D, Wilson DJ (2013) An electrospray ms-coupled microfluidic device for sub-second hydrogen/deuterium exchange pulselabelling reveals allosteric effects in enzyme inhibition. Lab Chip 13:2528–2532
Madeira A, Ohman E, Nilsson A, Sjogren B, Andren PE et al (2009) Coupling surface plasmon resonance to mass spectrometry to discover novel protein-protein interactions. Nature Protoc 4:1023–1037
Gao D, Liu H, Lin JM, Wang Y, Jiang Y (2012) Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip 13:978–985
Chen Q, He Z, Liu W, Lin X, Wu J, Li H, Lin JM (2015) Engineering cell-compatible paper chips for cell culturing, drug screening, and mass spectrometric sensing. Adv. Healthc. Mater. 4:2291–2296
Helenius A, Aebi M (2001) Intracellular functions of N-linked glycans. Science 291:2364–2369
Rudd PM, Woods RJ, Wormald MR, Opdenakker G, Downing AK et al (1995) The effects of variable glycosylation on the functional activities of ribonuclease, plasminogen and tissue-plasminogen activator. Biochim Biophys Acta 1248:1–10
Rudd PM, Wormald MR, Stanfield RL, Huang M, Mattsson N et al (1999) Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J Mol Biol 293:351–366
Varki A (1993) Biological roles of oligosaccharides - all of the theories are correct. Glycobiology 3:97–130
Iida J, Meijne AML, Knutson JR, Furcht LT, McCarthy JB (1996) Cell surface chondroitin sulfate proteoglycans in tumor cell adhesion, motility and invasion. Semin Cancer Biol 7:155–162
Reitter JN, Means RE, Desrosiers RC, Reitter JN, Means RE et al (1998) A role for carbohydrates in immune evasion in AIDS. Nat Med 4:679–684
Dennis JW, Granovsky M, Warren CE (1999) Protein glycosylation in development and disease. Bioessays News Rev Mol Cell Dev Biol 21:412–421
Lowe JB, Marth JD (2003) A genetic approach to mammalian glycan function. Annu Rev Biochem 72:643–691
Mechref Y, Hu Y, Garcia A, Zhou S, Desantos-Garcia JL et al (2012) Defining putative glycan cancer biomarkers by MS. Bioanalysis 4:2457–2469
Mechref Y, Hu Y, Garcia A, Hussein A (2012) Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis 33:1755–1767
Tao SJ, Huang YN, Boyes BE, Orlando R (2014) Liquid chromatography-selected reaction monitoring (LC-SRM) approach for the separation and quantitation of sialylated N-glycans linkage isomers. Anal Chem 86:10584–10590
Zhou SY, Hu YL, DeSantos-Garcia JL, Mechref Y (2015) Quantitation of permethylated n-glycans through multiple-reaction monitoring (MRM) LC-MS/MS. J Am Soc Mass Spectr 26:596–603
Dallas DC, Martin WF, Strum JS, Zivkovic AM, Smilowitz JT et al (2011) N-Linked Glycan Profiling of Mature Human Milk by High-Performance Microfluidic Chip Liquid Chromatography Time-of-Flight Tandem Mass Spectrometry. J Agr Food Chem 59:4255–4263
Hua S, An HJ, Ozcan S, Ro GS, Soares S et al (2011) Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 136:3663–3671
Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ et al (2009) Profile of native Nlinked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics 9:1939–1951
Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM et al (2012) Comparison of the human and bovine milk Nglycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res 11:2912–2924
Tao N, Wu S, Kim J, An HJ, Hinde K et al (2011) Evolutionary glycomics: Characterization of milk oligosaccharides in primates. J Proteome Res 10:1548–1557
Leoz MLAD, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M et al (2012) Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res 11:4662–4672
Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL et al (2012) Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res 11:6124–6133
Zhou H, Castro-Perez J, Lassman ME, Thomas T, Li W et al (2013) Measurement of apo(a) kinetics in humansubjects using a microfluidic device with tandem massspectrometry. Rapid Commun Mass Sp 27:1294–1302
Gallagher R, Dillon L, Grimsley A, Murphy J, Samuelsson K et al (2014) The application of a new microfluidic device for the simultaneous identification and quantitation of midazolam metabolites obtained from a single micro-litre of chimeric mice blood. Rapid Commun Mass Sp 28:1293–1302
Brink FTGVD, Büter L, Odijk M, Olthuis W, Karst U et al (2015) Mass spectrometric detection of short-lived drug metabolites generated in an electrochemical microfluidic chip. Anal Chem 87:1527–1535
Li X, Zhao S, Hu H, Liu YM (2016) A microchip electrophoresis-mass spectrometric platform with double cell lysis nano-electrodes for automated single cell analysis. J Chromatogr A 1451:156–163
Chen F, Lin L, Zhang J, He Z, Uchiyama K et al (2016) Single-cell analysis using drop-on-demand inkjet printing and probe electrospray ionization mass spectrometry. Anal Chem 88:4354–4360
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Lin, L., Lin, JM. (2018). Microfluidics-Mass Spectrometry for Cell Analysis. In: Lin, JM. (eds) Cell Analysis on Microfluidics. Integrated Analytical Systems. Springer, Singapore. https://doi.org/10.1007/978-981-10-5394-8_9
Download citation
DOI: https://doi.org/10.1007/978-981-10-5394-8_9
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5393-1
Online ISBN: 978-981-10-5394-8
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)