Abstract
There is a considerable market potential of extremophiles and their biomolecules; however, their high evolutionary rates and culturability limitations restrict their industrial exploration. The impressive development of “omics” technologies has allowed the culture-free technique, i.e., metagenomics as a valuable tool for mining the hidden information of the extremophiles and their biospheres. At present, environmental microbiomes are being studied using functional-based and sequence-based approaches. The growth of the extreme environmental metagenomics literature and projects is increased nearly tenfold in the last decade due to the advancement in the sequencing technologies. Here, in the context, we summarize all the aspects of extreme biosphere metagenomics with recent prospects and progress.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Extremophiles and Extreme Environments
These are various extreme habitats situated across the globe since the genesis of the earth. These extreme habitats harbor a rich microbial diversity. Certain old evidence of life like microfossils, stromatolites, microfibrous sedimentary rocks, and sedimentary carbon pool suggests that the microorganisms inhabit the earth since the archaean period, the time before 2.5 billion years (Stanley 2005). Such ancient microbial life had developed robust metabolic functions similar to many present-day living extremophiles thriving into extreme environments. The extremophiles hold secret survival “kits” to shelter at either single or multiple extreme conditions. Microbiologists are exploring their cellular properties like new gene pools, robust biomolecules, and metabolic uniqueness through the culturing methods since the discovery of extremophiles. However, despite the technological advancement for the investigation of extremophiles and their habitats, we have decoded the very limited information from the extreme biosphere (Rampelotto 2013).
Extreme territories support all the three taxonomic forms of life to flourish. However, the largest membership is represented by the Archaea followed by Bacteria and Eukaryotes. The ability of these extremophilic microorganisms to proliferate under extreme conditions is of immense importance for understanding microbial physiology and evolution. Extremophiles are best characterized according to their growth profiles, using marginal data, under certain culture conditions including salt concentration, temperature profile, pH scale, and growth under hydrostatic pressure (Mesbah and Wiegel 2008). The representative examples of extremophiles thriving in different extremities are thermophiles (45–60 °C temperature), hyperthermophiles (60–120 °C temperature), psychrophiles (below 0 °C temperature), acidophiles (below 4.0 pH), alkaliphiles (over 9.0 pH), piezophiles or barophiles (>0.5 MPa pressure), halophiles (>1 M NaCl concentration), and xerophiles (<0.85 water activity) (Horikoshi et al. 2010).
Culturability of the extreme habitats is very less due to the persistence of abiotic stresses and differences between natural environments and laboratory conditions. Additionally, culturability is a very complex physiological process that depends on various phenomena (Barer and Harwood 1999). Due to these limitations, the majority of such environment has remained unexplored. However, during the recent years, development and application of molecular techniques, such as PCR, cloning, and next-generation high-throughput sequencing, have proved quite valuable in judging the distribution and diversity of extreme habitats. Hence, due to the limitation of available culturing methods for the extremophiles, it is now being studied by the uncultivable approach referred as a metagenomics to translate the potentials of various extremophilic microorganisms. Furthermore, the holistic community can be deciphered through the metagenomics approach, whereas the traditional microbiology relies upon the cultivation of few clones or colonies. So, the metagenomics application provides the profiling of the microbial diversity of any extreme environment to analyze the entire community and can be delineated broadly as an environmental genomics, ecogenomics, or community genomics (Hugenholtz et al. 1998).
2 Metagenomics and Microbial Diversity
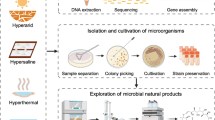
Development and discovery of various molecular biology techniques after the 1980s extend the genomic discipline toward its associated “omics” technologies. Metagenomics emerges in the ending of last century, which eliminates the culturability for mining the microbial information and revolutionizing microbial ecology. Metagenomics is the study of the collective forms of genomes directly isolated from the environmental sample for the comprehensive analysis of microbial diversity and ecology of a specific environment. Metagenomics studies provide the mechanism for analyzing previously unknown organisms, and at the same time, one can examine the diversity of organisms present in specific environments as well as analyze the complex interactions between members of a specific environment (Handelsman 2004). Metagenomics studies are conducted by two different approaches. One is function-based analysis, which deals with the total DNA extraction from environmental sample followed by cloning into suitable host and detection of expressed phenotypes in the host cells, whereas sequence-based analysis is mainly concerned with decoding of the extracted DNA and/or RNA using various sequencing platforms followed by assessment of taxonomic diversity. Both approaches are applicable to decipher hidden microbial gene pools and profiling of the microorganisms (Fig. 5.1). Early environmental gene sequencing is dealt with cloning of 16S rRNA gene to analyze the microbial taxonomic profile. Such work revealed the vast majority of microbial biodiversity that had been explored by cultivation-based methods (Hugenholtz et al. 1998). Slowly the shotgun Sanger sequencing or massively parallel pyrosequencing is applied to get largely unbiased samples of all genes from all the members of the sampled communities (Eisen 2007). The first metagenomics studies conducted using high-throughput sequencing by massively parallel 454 pyrosequencing transformed the studies of the microbial universe (Poinar et al. 2006; Edwards et al. 2006). Nowadays the various next-generation sequencing (NGS) platforms are utilized for the metagenomics studies, and continuous improvements in the existing sequencing technologies are often done by the original developers.
3 Environmental Metagenomics
According to the meeting report of Earth microbiome project, one quintillion (1030) microbial cells are present on the earth. Theoretical estimation of the average quantity of DNA in the microbial cell is 10 million base pairs. As yet, we have investigated hardly 1% of the total environmental DNA by global environmental DNA sequencing efforts (Gilbert et al. 2010). This statistics may also be far greater than actual analysis; so, the massive information of microbial life on the Earth is yet unknown and/or under-sampled. Hence, we are in the beginning stage in the study of the extreme environmental metagenomics.
Early environmental metagenomics projects are considered a key trigger to drive the field of extreme microbiomes. Environmental genomic studies of the Sargasso Sea (Venter et al. 2004) are a major breakthrough in the environmental metagenomics, which leads to developing the interest among the scientific communities to initiate and explore the microbial diversity of extreme habitats using metagenomics approach. The scientific literature on “extreme environmental metagenomics” available in public domains increased quickly in the last decade indicating the development of the field (Fig. 5.2). The rapid escalation of metagenomics projects in the various online databases indicated the quick growth of metagenomics field. Currently, more than 20% metagenomes submitted into public domains are derived from the various extreme biosphere including marine/ocean, abyssal plain, desert, hydrothermal vents, permafrost, glacier, salt marsh, thermal hot springs, geyser, soda lake, hypersaline lake, submarine volcano, black smoker, acid mine drainage, etc. (Table 5.1). Undoubtedly, it is due to the recent advances in high-throughput sequencing technologies with more sophisticated bioinformatics analysis pipeline making the metagenomics study very easy and rapid.
The recent identification of new gene pools and species of extremophiles from the extreme habitats geared up the exploration of microbial species for the industrial and biomedical potentials. At the beginning of metagenomics era, the giant vector, i.e., BAC, was used to construct the metagenomics library of the environmental DNA (Rondon et al. 2000) and function, and the sequence-based analysis was performed from each clone. Modern metagenomics approach makes it possible to know the physiology of the extremophiles, their role in the habitats and adaption to environmental pressures. So, the microbiome of extreme biosphere may help to establish the microbial community network structure, which is very useful to decode the microbial functionality, interaction, and community dynamics (Cowan et al. 2015). However, the various experimental challenges from sampling to sequencing should be addressed before conducting environmental metagenomics projects.
4 Challenges in Environmental DNA Extraction
The key challenges of conducting metagenomics studies include the sampling and transporting of the adequate intact environmental sample and extraction of the high-quality nucleic acid from the environmental sample. The stresses in the extreme site are the key hurdles for the extraction and the purification of the high-quality nucleic acids; so, the sample processing is prerequisite for environmental metagenomics project (Thomas et al. 2012). Isolation of poor-quality DNA may hamper the subsequent analysis, i.e., cloning and sequencing; so, the specific methods and protocol are needed to extract the high-quality and high molecular weight (HMW) community genomic DNA from the environmental sample. Various direct environmental DNA extraction methods including freezing-thawing, bead beating, and ultrasonication along with indirect extraction methods like PEG-NaCl-based and enzyme lysis with hot detergent treatment are being used for the extraction of environmental DNA and viable for the functional and structural profiling of the microorganisms (Delmont et al. 2011; Narayan et al. 2016). Based on direct and indirect extraction, nowadays commercial kits are also developed by many manufacturers to extract the nucleic acid in good quality and quantity from soil and water samples. However, the success of the DNA extraction depends on the microbial population and the physiological status of the cells.
Heterogeneous microbial communities exist in the environment, and all the microbial cells have the substantial structural variation in the cell wall and cell membrane. So the cellular dissimilarity in the microbial species restricts the selection of single universal cell lysis method to extract the nucleic acid. However, harsh treatment can be used but such conditions cause damage to DNA and mild treatment leads to less recovery. So, the combination of chemical, physical/mechanical, and biological cell lysis is best suitable for the extraction of high-quality environmental DNA (Bag et al. 2016). The environmental stresses have also increased the difficulties in extraction procedure as the extremities may give the adaptive and protective mechanisms to the microbial cell to survive in the extreme conditions, which may make the cell very resilient to lysis and consequently inadequate nucleic acid will be taken out that missing genomic contents of rare species. Hence, before starting on the extreme environmental metagenomics project, one should thoroughly study all the geological, biological, and cellular features to extract the high-quality HMW community genomic DNA (Table 5.2).
5 Sequencing Platforms
Metagenomics analysis using DNA sequencing technique is performed either through gene-targeted metagenomics (i.e., 16s rRNA or 18s rRNA) for taxonomic assessment or whole genome shotgun sequencing for structural and functional analysis (Ghelani et al. 2015; Dudhagara et al. 2015; Patel et al. 2015). Presently, various NGS platforms are effectively used for the metagenomics pipeline including the AB SOLiD System, 454 GS FLX, Illumina MiSeq, Roche-454, and Ion Torrent (Liu et al. 2012; Mardis 2013). All these sequencing techniques are not feasible in off-grid analysis and offer short read length, creating the difficulties in assembly process which consequently affects the downstream analysis including the taxonomic and functional profiling. Two modern sequencing platforms (1) PacBio sequencing and (2) Oxford nanopore sequencing recently emerge, which offer the advantages over the limitations of the above-discussed NGS techniques. Both provide longer reads mainly useful for the analysis of a diverse pool of microorganisms.
5.1 PacBio Sequencing
PacBio sequencing is a real-time sequencing developed by Pacific Biosciences, California, USA. The single-molecule real-time (SMRT) sequencing of single-stranded circular DNA is based on a template called SMRTbell, which is loaded into a chip referred as an SMRT cell (Travers et al. 2010). The key features of this sequencing method are the long read length up to 104 bp which makes it suitable for microbiome analysis by the full-length sequencing of target genes, i.e., 16S rRNA (~1500 bp) and 18S rRNA (~1800 bp) (Schadt et al. 2010). Longer read output is important to improve the contiguity in the assembly process. However, the higher error rate, high cost, and lower sequencing depth are major demerits of the technique (Rhoads and Au 2015). Recently, large contigs and minimizing the errors with >99% Q20 accuracy can be achieved using long read circular consensus sequencing (CCS) and place it comparatively affordable for the metagenomics analysis pipelines (Frank et al. 2016). So the aim of the metagenomics projects can be easily achieved by obtaining long contig sizes with negligible possibilities of misassemblies. PacBio sequencing is suggested for the analysis of microbial abundance and taxonomic assessment. Furthermore, the fusion assemblies using PacBio CCS and Illumina HiSeq contigs improve statistics of assembly, overall contig length and number.
5.2 Oxford Nanopore Sequencing
It is a very impressive fourth-generation sequencing method. It is based on the nanopore embedded in the membrane, which is kept at a certain voltage. When the ssDNA or ssRNA passes through the nanopore, the current level variation is detected resulting into decoding of nucleotide order (Ashkenasy et al. 2005). Oxford nanopore technologies have devised the portable MinION sequencer. This is very fast, is small in size, and produces the 200 kb long reads with high accuracy. Ultra-portability offers the in-field metagenomics analysis and hence overcomes the difficulties associated with the preservation and transportation of extreme environmental sample to the laboratory. Environmental samples from the glacier and hot springs are easily getting degraded in transit and biases acquired by taphonomic degradation during storage and subsequent extraction. The in situ microbial community analysis using portable nanopore sequencing methods improved agility to analyze environmental microbiomes (Edwards et al. 2016). However, the off-grid metagenomics analysis should be cross-validate before applying to the search the microbial life and extraterrestrial life in their habitation.
6 Future Prospects
Environmental microbiomics will bring new insights very shortly in the comprehensive determination of the microbial composition. The newest subdisciplines within the metagenomics field referred as metatranscriptomics and metaproteomics are also new hopes to offer the more resolution in structure and function of the microbial community. In the future, the single-step DNA extraction, rapid library preparation, and fast real-time in situ DNA and RNA sequencing will uplift the extreme biosphere microbiomics. The recent emergence of progressive miniaturization in the sophisticated tools and techniques will move the laboratory-dependent analysis toward in-field study to capture the more real microbial profile. Fusions of the sequencing and Raman spectroscopy, as well as mineralization of bioinformatics tools, are also the good hope in the future for search and analysis of microbial life. However, the universal standards of procedures and protocols should be established for uniformity research on environmental metagenomics, like human microbiome.
References
Ashkenasy N, Sánchez‐Quesada J, Bayley H, Ghadiri MR (2005) Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angew Chem Int Ed 117(9):1425–1428
Bag S, Saha B, Ojasvi Mehta DA, Kumar N, Dayal M, Pant A et al (2016) An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci Rep 6:26775
Barer MR, Harwood CR (1999) Bacterial viability and culturability. Adv Microb Physiol 41:93–137
Cowan DA, Ramond JB, Makhalanyane TP, De Maayer P (2015) Metagenomics of extreme environments. Curr Opin Microbiol 25:97–102
De Maayer P, Anderson D, Cary C, Cowan DA (2014) Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep 15(3):e201338170
Delmont TO, Robe P, Clark I, Simonet P, Vogel TM (2011) Metagenomic comparison of direct and indirect soil DNA extraction approaches. J Microbiol Methods 86(3):397–400
Dudhagara P, Ghelani A, Bhavsar S, Bhatt S (2015) Metagenomic data of fungal internal transcribed Spacer and 18S rRNA gene sequences from Lonar lake sediment, India. Data Brief 4:266–268
Edwards A, Debbonaire AR, Sattler B, Mur LA, Hodson AJ (2016) Extreme metagenomics using nanopore DNA sequencing: a field report from Svalbard, 78 N. bioRxiv 2016:073965
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM et al (2006) Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7(1):57
Eisen JA (2007) Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol 5(3):e82
Frank JA, Pan Y, Tooming-Klunderud A, Eijsink VG, McHardy AC, Nederbragt AJ, Pope PB (2016) Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci Rep 6:25373
Ghelani A, Patel R, Mangrola A, Dudhagara P (2015) Cultivation-independent comprehensive survey of bacterial diversity in Tulsi Shyam Hot Springs, India. Genomics Data 4:54–56
Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Brown CT et al (2010) Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci 3(3):243
Goordial J, Davila A, Lacelle D, Pollard W, Marinova MM, Greer CW et al (2016) Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J 10(7):1613–1624
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68(4):669–685
Horikoshi K, Grant WD (eds) (1998) Extremophiles: microbial life in extreme environments, vol 20. Wiley-Liss, Chichester
Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (eds) (2010) Extremophiles handbook. Springer Science & Business Media, Berlin
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180(18):4765–4774
Klingl A (2014) S-layer and cytoplasmic membrane–exceptions from the typical archaeal cell wall with a focus on double membranes. Front Microbiol 5:624
Koga Y (2012) Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012:789652
Kumar Gothwal R, Kumar Nigam V, Mohan MK, Sasmal D, Ghosh P (2007) Extraction of bulk DNA from Thar Desert soils for optimization of PCR-DGGE based microbial community analysis. Electron J Biotechnol 10(3):400–408
Kuske CR, Barns SM, Busch JD (1997) Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol 63(9):3614–3621
Liu L, Li Y, Li S, Hu N, He Y, Pong R et al (2012) Comparison of next-generation sequencing systems. Biomed Res Int 2012:251364
Mardis ER (2013) Next-generation sequencing platforms. Annu Rev Anal Chem 6:287–303
Mesbah NM, Wiegel J (2008) Life at extreme limits. Ann N Y Acad Sci 1125(1):44–57
Michels PC, Clark DS (1997) Pressure-enhanced activity and stability of a hyperthermophilic protease from a deep-sea methanogen. Appl Environ Microbiol 63(10):3985–3991
Morono Y, Terada T, Hoshino T, Inagaki F (2014) Hot-alkaline DNA extraction method for deep-subseafloor archaeal communities. Appl Environ Microbiol 80(6):1985–1994
Narayan A, Jain K, Shah AR, Madamwar D (2016) An efficient and cost-effective method for DNA extraction from athalassohaline soil using a newly formulated cell extraction buffer. 3 Biotech 6(1):1–7
Patel R, Mevada V, Prajapati D, Dudhagara P, Koringa P, Joshi CG (2015) Metagenomic sequence of saline desert microbiota from wild ass sanctuary, Little Rann of Kutch, Gujarat, India. Genomics Data 3:137–139
Poinar HN, Schwarz C, Qi J, Shapiro B, MacPhee RD, Buigues B et al (2006) Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science 311(5759):392–394
Purohit MK, Singh SP (2009) Assessment of various methods for extraction of metagenomic DNA from saline habitats of coastal Gujarat (India) to explore molecular diversity. Lett Appl Microbiol 49(3):338–344
Ramakrishnan V, Adams MWW (1995) Preparation of genomic DNA from sulfur-dependent hyperthermophilic Archaea. Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 95–96
Rampelotto PH (2013) Extremophiles and extreme environments. Life (Basel) 3(3):482–485
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13(5):278–289
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR et al (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66(6):2541–2547
Sagova-Mareckova M, Cermak L, Novotna J, Plhackova K, Forstova J, Kopecky J (2008) Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl Environ Microbiol 74(9):2902–2907
Schadt EE, Turner S, Kasarskis A (2010) A window into third-generation sequencing. Hum Mol Genet 19(R2):R227–R240
Stanley SM (2005) Earth system history. Macmillan, New York, NY, pp 302–323
Thomas T, Gilbert J, Meyer F (2012) Metagenomics-a guide from sampling to data analysis. Microb Inform Exp 2(1):3
Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW (2010) A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res 38(15):e159
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66–74
Verma D, Satyanarayana T (2011) An improved protocol for DNA extraction from alkaline soil and sediment samples for constructing metagenomic libraries. Appl Biochem Biotechnol 165(2):454–464
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Dudhagara, P., Kothari, R., Ghelani, A., Rank, J., Patel, R. (2017). Prospects and Progress in Extreme Biosphere Microbiome. In: Singh, R., Kothari, R., Koringa, P., Singh, S. (eds) Understanding Host-Microbiome Interactions - An Omics Approach. Springer, Singapore. https://doi.org/10.1007/978-981-10-5050-3_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-5050-3_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5049-7
Online ISBN: 978-981-10-5050-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)