Abstract
During the process of beer fermentation, higher alcohols and esters are two important substances influencing beer flavor. Deletion of BAT2 gene coding amino acid transaminase can effectively reduce production of higher alcohols. On the other hand, alcohol acetyltransferase (AATase) encoded by the ATF2 gene is one of the most important enzymes for acetate ester synthesis. The objective of this study is to construct engineered brewer’s yeast strains for moderate production of acetate esters and less yield of higher alcohols. The industrial beer yeast strain S5 was selected as the parental strain, by means of overexpressing ATF2 and knocking out BAT2 to appropriately increase the concentration of acetate and reduce the concentration of higher alcohols. The engineered strain S5-L, featuring partial BAT2 allelic genes replaced by the constructed ATF2 overexpression cassette, was obtained. The fermentation results indicated that the engineered strain S5-L had a moderate accrete of ethyl acetate compared with the parental strain. The concentration of ethyl acetate produced by the engineered strains S5-L increased to 7.6 mg L−1, about 1.28-fold higher than that produced by the parental S5 cells. The concentration of higher alcohols produced by the engineered strains S5-L reduced to 51.49 mg L−1, about 84.77% of the parental S5 cells. Isobutanol and propanol reduced to 8.30 and 9.72 mg L−1, which was 73.13% and 79.47% of that of the parental strain S5, respectively. Research on the beer yeast genes ATF2 and BAT2 influencing the generation amount of higher alcohol and volatile esters lays a foundation to improve the flavor and quality of beer.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Saccharomyces cerevisiae
- Ethyl acetate
- Higher alcohols
- Alcohol acetyl transferase
- Amino acid transaminase
1 Introduction
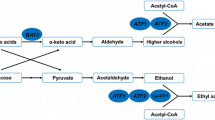
In the yeast fermentation process, the by-products, carbon dioxide and alcohol form a unique beer flavor. The by-products include higher alcohols, esters, phenolic compounds and so on [1]. Among them, esters and higher alcohols are two of the most important groups of volatile flavor compounds. As we all known, beer alcohols and esters are generated in the main fermentation stage. However, levels of higher alcohols in the fermentation solution are often too high for favorable beer development, while beer yeast strains display low capacity for ester production. Thus, development of methods by which to decrease the generation of higher alcohols and increase the production of aromatic esters in beers, particularly through the use of industrial brewer’s yeast, is of great importance.
Alcohol acetyltransferases (AATases), which catalyze the transformation of alcohols and acetyl-coenzyme A into acetate esters, are key enzymes involved in ester synthesis [2,3,4]. Three types of AATases (namely, AATase, AATase I, and AATase II) have been studied, and they are encoded by the ATF1, Lg-ATF1, and ATF2 genes respectively [5,6,7,8,9]. Several researchers have reported that, compared to ATF1, overexpression of ATF2 caused slight increase in the process of synthesis of ester. Similarly, Lg-ATF1 has a very limited role in the synthesis of volatile esters [10,11,12]. In the previous study, the ester production was observed to increase significantly when overexpressed ATF1 in beer yeast, which leaded to the disharmony of beer flavor [13]. Overexpression of ATF2 in industrial brewer’s yeast is observed for appropriately increasing the production of ester, thus beer flavor behave more harmonious.
Higher alcohols, also known as fusel alcohols, include propyl alcohol, isoamyl alcohol, isobutyl alcohol, active amyl alcohol and so on. About 80% outputs of higher alcohols are formed during the primary fermentation. Higher alcohols are one of the inherent flavor ingredients of beer, and the coordination between all kinds of higher alcohols can make beer palate soft and unique taste [14]. In the amino acid catabolism pathway, knocking out the amino acid transaminase (BCAT), which is encoded by BAT1 and BAT2 gene, can reduce the concentration of isobutanol and isoamyl alcohol [15]. The product of the BAT2 gene has been previously reported to play an important role in the production of higher alcohols than the BAT1 gene [16].
In this article, the brewing yeast strain with part BAT2 allele insteaded by ATF2 gene was constructed to improve a moderate amount of acetate content and reduce higher alcohol content. Our data show that ATF2 overexpression and BAT2 deletion can improve the acetate ester content in beer while significantly reducing its isoamyl alcohol content. The results in this article are useful in future developments in the beer industry.
2 Materials and Methods
2.1 Strains and Media
Escherichia coli DH5α, the parental strain Saccharomyces cerevisiae S5, and plasmid pUC-BBAK, pUC-PIA2K were obtained from the Microbiological Culture Collection Center of Tianjin Industrial Microbiology Key Laboratory, Tianjin University of Science and Technology, People’s Republic of China.
Plasmid pUC-BBAK contained two homology DNA fragments of upstream and downstream of BAT2 gene and loxP-kanMX-loxP gene disruption cassette, named BA, BB and K, respectively. Plasmid pUC-PIA2K was used in the preparation of the PGK P -PGK T expression cassette and the ATF2 expression genes. The ATF2 gene was connected between the promoter and the terminator, the direction is consistent with them.
E. coli DH5α used for the preparation and construction of the plasmid, was grown at 37 °C in Luria-Bertani medium (1% Bacto-tryptone, 0.5% yeast extract, and 0.5% NaCl). Ampicillin was added to the medium at a final concentration of 100 μg mL−1 to select positive E. coli transformants. The parental strain S. cerevisiae S5 was usually cultured in YPD medium (1% yeast extract, 2% Bacto-peptone, and 2% glucose). G418 was added to the medium at a final concentration of 100 μg mL−1 to select positive yeast transformants. The optimum growth temperature of S. cerevisiae was 30 °C. During fermentation, the yeast cells were cultured in wort medium. Wort medium was prepared from crushed malt and distilled water according to the ratio of material to water 1:4, and saccharification was performed according to certain routes with sugar meter adjusted to 10 °Brix. All of the solid media used in this study contained 2% agar.
2.2 DNA Manipulation
The DNA operation in this study was performed according to the standard procedure described by Ausubel [17]. In-Fusion DNA ligase, TaKaRa LA Taq DNA polymerase, 5000 DNA Marker, 15,000 DNA Marker and restriction enzymes were used for DNA manipulation, and these reagents were purchased from the TaKaRa Biotechnology.
2.3 Plasmid Construction
The polymerase chain reaction (PCR) primers used in this work are listed in Table 1. A 3379-bp PGKp-ATF2-PGKt fragment was amplified via PCR from the plasmid pUC-PIA2K, which contained ATF2 gene (1608-bp) under the control of phosphoglycerate kinase I gene promoter and terminator (1771-bp). A 5309 bp BB-pUC19-BA-KanMX fragment was amplified via PCR from the plasmid pUC-BBAK, which contained the homology arms BA (488-bp) and BB (522-bp) and loxP-kanMX-loxP fragment (1613-bp). The recombinant plasmid pUC-PABBK was obtained from the ligation of PGKp-ATF2-PGKt fragment and BB-pUC19-BA-KanMX fragment reacted for 30 min with the use of In-Fusion DNA ligase under 50 °C.
2.4 Yeast Transformation and Screening
Transformation fragment PABBK was obtained by PCR amplification from the constructed plasmid pUC-PABBK, and then transformed into yeast genome through the method of LiAc transformation. G418-resistant transformants were selected and identified via PCR.
2.5 Fermentation Method
First of all, the yeast cells were cultured in tube containing 7 mL wort medium and static cultured for 24 h in 30 °C incubator. The culture was added to the 150 mL triangle bottle containing 45 mL wort medium with 10% inoculation quantity, and static cultured for 24 h in 16 °C incubator. Then 15 mL of the culture was transferred into 250 mL triangle bottle containing 135 mL wort medium, and static cultured 7–9 days in 10 °C incubator. Weight lost of Carbon dioxide was detected every 12 h, until the data is less than 0.2 g.
2.6 Gas Chromatography (GC) Analysis
After fermentation, samples were filtered and distilled from the wort medium, then used for GC analysis. Analysis was performed on an Agilent 7890C GC system. Capillary column was 30 m × 320 μm × 0.5 μm and column temperature was 75 °C. The temperature of the flame ionization detector (FID) was adjusted to 230 °C, the injector temperature was 200 °C, and the split ratio was 20:1. Nitrogen was used as the carrier gas, and the injection volume was 1.0 μL. Butyl acetate was used as the internal standard. A specific amount of each of the analytes was measured and used as a standard for machine calibration. Ethyl acetate, amyl acetate, isoamyl acetate, isobutanol and isoamyl alcohol were purchased from Merck.
2.7 Other Basic Performance Test
Weight lost of Carbon dioxide, residual sugar, alcohol degree, appearance fermentation degree and real fermentation degree were referred from People’s Republic of China country.
3 Results and Discussions
3.1 Construction of Recombinant Plasmid pUC-PABBK
The construction process of the recombinant plasmid pUC-PABBK was showed in Fig. 1.
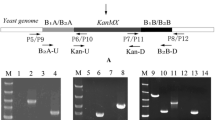
Plasmids pUC-BBAK and pUC-PIA2K were used as templates, and BB-pUC19-BA-KanMX (5310-bp) and PGKP-ATF2-PGKT (3379-bp) fragments were obtained by PCR amplification with primers BB-U, BK-D and PGK-U, PGK-D, respectively. Data was shown in Fig. 2. The two fragments were connected with In-Fusion DNA ligase. For PCR verification, the 6005-bp size of a fragment (Fig. 3a) would be gotten with the primers BA-U and BB-D. For enzyme digestion verification, the 8691-bp size of a fragment (Fig. 3b) would be gotten using the restriction enzyme Nco I.
3.2 Construction of Engineered Brewer’s Yeast Strains
The transformation fragment PABBK was amplified via PCR from the plasmid pUC-PABBK and integrated into the homologous genome of the S5 strain via LiAc transformation (Fig. 4). The resulting transformants were screened on YPD plates containing 0.50 mg mL−1 G418 [18]. The strain S5-L was selected as the correct recombinant after PCR analysis using the primer pairs U-①, D-① and U-②, D-②, separately (Fig. 5).
lanes 1, 2 were PCR amplification results from the recombinant (S5-L) genome and the parental strain (S5) genome using the primer pairs U-①, D-①; M 5000 DNA marker; lanes 3, 4 were PCR amplification results from the parental strain (S5) genome and recombinant (S5-L) genome using the primer pairs U-②, D-②
3.3 Basic Fermentation Performance of S5 and S5-L
The fermentation performance of S5 and S5-L were detected. The results (Table 2) showed that there was no significant difference in the basic fermentation performances of the engineered strains S5-L and the parental strain S5.
3.4 Production of Volatile Flavor Compounds During Fermentation
After fermentation, the esters and higher alcohols in parental strain and engineering strain were determined by GC analysis. The results showed that the content of acetate esters in engineering strain S5-L was improved than that of the parental strain S5. The content of ethyl acetate in engineering strain S5-L reached 7.60 mg L−1, which was 1.28-fold of that of the parental strain S5. Isoamyl acetate in engineering strain and parental strain was not detected. It was probably the result of that the content of isoamyl acetate was lower than the GC detection. After fermentation, the content of isoamyl alcohol in the engineering strain S5-L was reduced to 51.49 mg L−1, which was 84.77% of that of the parental strain S5. Isobutanol and propanol were reduced to 8.30 and 9.72 mg L−1, which was 73.13% and 79.47% of that of the parental strain S5 (Table 3), respectively.
4 Conclusion
In this work, with the action of PGK promoter, ATF2 gene, which encodes the AATase II, was overexpressed to increase acetate content. On the other hand, BAT2 gene, which encodes the amino acid transaminase, was knocked out to decrease the content of higher alcohols. The results of the final fermentation showed that ethyl acetate content was moderate increased while higher alcohols concentrations were effectively reduced. In this paper, the ester content has been improved, but there’s still a lot of work to do in order to achieve better results.
References
Styger G, Prior B, Bauer FF (2011) Wine flavor and aroma. J Ind Microbiol Biotechnol 38:1145–1159
NordstrÖm K (1963) Formation of ethyl acetate in fermentation with brewer’s yeast IV: metabolism of acetyl coenzyme A. J Inst Brew 69:142–153
NordstrÖm K (1964) Formation of esters from alcohols by brewer’s yeast. J Inst Brew 70:328–336
NordstrÖm K (1962) Formation of ethyl acetate in fermentation with brewer’s yeast III: participation of coenzyme A. J Inst Brew 68:398–407
Fujii T, Nagasawa N, Iwamatsu A, Bogaki T, Tamai Y, Hamachi M (1994) Molecular cloning, sequence analysis and expression of the yeast alcohol acetyltransferase gene. Appl Environ Microbiol 60:2786–2792
Fujii T, Yoshimoto H, Nagasawa N, Bogaki T, Tamai Y, Hamachi M (1996) Nucleotide sequence of alcohol acetyltransferase genes from lager brewing yeast, Saccharomyces carlsbergensis. Yeast 12:593–598
Yoshimoto H, Momma T, Fujiwara D, Sone H, Kaneko Y, Tamai T (1998) Characterization of the ATF1 and Lg-ATF1 genes encoding alcohol acetyltransferases in the bottom fermenting yeast Saccharomyces pastorianus. J Ferment Bioeng 86:15–20
Yoshimoto H, Fujiwara D, Momma T, Tanaka K, Sone H, Nagasawa N, Tamai T (1999) Isolation and characterization of the ATF2 gene encoding alcohol acetyltransferase II in the bottom fermenting yeast Saccharomyces pastorianus. Yeast 15:409–417
Lilly M, Bauer FF, Lambrechts MG, Swiegers JH, Cozzolino D, Pretorius IS (2006) The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 23(9):641–659
Nagasawa N, Bogaki T, Iwamatsu A, Hamachi M, Kumagai C (1998) Cloning and nucleotide sequence of the alcohol acetyltransferase II gene (ATF2) from Saccharomyces cerevisiae Kyokai No. 7. Biosci Biotechnol Biochem 62:1852–1857
Lilly M, Lambrechts MG, Pretorius IS (2000) Effect of increased yeast alcohol acetyltransferase activity on flavour profiles of wine and distillates. Appl Environ Microbiol 66:744–753
Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP (2003) Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl environ microbiol 69(9):5228–5237
Cui-Ying Z, Yu-Lan L, Ya-Nan Q, Jian-Wei Z, Long-Hai D, Xue L, Dong-Guang X (2013) Increased esters and decreased higher alcohols production by engineered brewer’s yeast strains. Eur Food Res Technol 236:1009–1014
Stewart GG (2005) Esters: the most important group of flavor-active beer compounds. Proc Congr Eur Brew Conv 30:100–101
Yoshimoto H, Fukushige T, Yonezawa T et al (2002) Genetic and physiological analysis of branched-chain alcohols and isoamyl acetate production in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 59(4):501–508
Eden A, Van Nedervelde L, Drukker M et al (2001) Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol 55(3):296–300
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current protocols in molecular biology. Wiley, New York
Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16(5–6):339–346
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Liu, X., Xu, J., Pi, L., Zhang, C., Xiao, D. (2018). Effects of ATF2 Overexpression with BAT2 Deletion on the Higher Alcohols and Esters in Beer Yeast. In: Liu, H., Song, C., Ram, A. (eds) Advances in Applied Biotechnology. ICAB 2016. Lecture Notes in Electrical Engineering, vol 444. Springer, Singapore. https://doi.org/10.1007/978-981-10-4801-2_8
Download citation
DOI: https://doi.org/10.1007/978-981-10-4801-2_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4800-5
Online ISBN: 978-981-10-4801-2
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)