Abstract
Whilst antibodies remain the most popular and trusted choice for molecular recognition, they may still pose challenges for biosensing applications due to their high cost, low reproducibility and large size. One long championed alternative to antibodies are nucleic acid aptamers. Nucleic acid aptamers are single-stranded DNA or RNA sequences that can bind to a target with high affinity and specificity. Nucleic acid aptamers, due to their varied advantages, have been gaining significant importance in both diagnostic and theranostic applications. Among various diseases, early diagnosis of cancer is one of the biggest concerns for patients and healthcare professionals worldwide. For the case of cancer, it is crucial to be able to deliver treatment whilst monitoring therapy response in real time. This is required in order to prevent over- or under-dosing the patients whilst treatment occurs. One of the most commonly used techniques for cancer diagnosis is to detect biomarkers (cancer-related proteins, small molecules and cancer cells) found in body fluids specific for a particular cancer type. In this chapter, we present a discussion on the use of aptamer-based biosensors (termed as “aptasensors”) for cancer diagnosis. The development of aptamer-based biosensor devices is an interdisciplinary field and relies on very distinct aspects such as characterisation of bio-recognition probes with their respective analytes, immobilisation onto electrode surfaces, development of anti-fouling surface chemistries, sensor design and fabrication and microfluidics. Special attention is given to different types of surface chemistry used for the development of simple, sensitive and cost-effective aptasensors. Utilisation of aptamers is an encouraging tool for the development of point-of-care (PoC) biosensors for the detection of different types of cancer. In the view of unparalleled merits of aptamers, recent achievements and future perspectives of the applications of aptamers are also discussed.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
9.1 Introduction
Whilst antibodies remain the most popular and trusted choice for molecular recognition, they may still pose challenges for biosensing applications due to their high cost, low reproducibility and large size. One long championed alternative to antibodies are nucleic acid aptamers. Nucleic acid aptamers are single-stranded DNA or RNA sequences that can bind to a target with high affinity and specificity. Nucleic acid aptamers, due to their varied advantages, have been gaining significant importance in both diagnostic and theranostic applications. Among various diseases, early diagnosis of cancer is one of the biggest concerns for patients and healthcare professionals worldwide. For the case of cancer, it is crucial to be able to deliver treatment whilst monitoring therapy response in real time. This is required in order to prevent over- or under-dosing the patients whilst treatment occurs. One of the most commonly used techniques for cancer diagnosis is to detect biomarkers (cancer-related proteins, small molecules and cancer cells) found in body fluids specific for a particular cancer type. In this chapter, we present a discussion on the use of aptamer-based biosensors (termed as “aptasensors”) for cancer diagnosis. The development of aptamer-based biosensor devices is an interdisciplinary field and relies on very distinct aspects such as characterisation of bio-recognition probes with their respective analytes, immobilisation onto electrode surfaces, development of anti-fouling surface chemistries, sensor design and fabrication and microfluidics. Special attention is given to different types of surface chemistry used for the development of simple, sensitive and cost-effective aptasensors. Utilisation of aptamers is an encouraging tool for the development of point-of-care (PoC) biosensors for the detection of different types of cancer. In the view of unparalleled merits of aptamers, recent achievements and future perspectives of the applications of aptamers are also discussed.
9.1.1 Nucleic Acid Aptamers
Nucleic acid aptamers are short single-stranded DNA or RNA oligonucleotides, which have gained massive attention as bioreceptors in biosensing applications (aptasensors) or medical therapy (Hianik and Wang 2009; Iliuk et al. 2011). Their specificity is similar or higher than that of antibodies, with dissociation constants (K d) in the range of nanomolar (nM) to picomolar (pM) levels. An important advantage of their use in the development of biosensor devices is their high affinity and specificity to proteins and other molecules with low molecular weight, for example, toxins (Castillo et al. 2012; Wang et al. 2011). When compared with their biological counterparts, aptamers have several advantages; for instance, aptamers are more stable than antibodies, making them suitable for applications in special conditions such as high temperature or extreme pH.
Furthermore, aptasensors can be regenerated without loss of integrity and selectivity (Mairal et al. 2008; Tombelli et al. 2005). One very interesting property of an aptamer is the conformational change that it undergoes once bound to its target, a property which has been widely utilised within biosensing applications (Jolly et al. 2015a; Radi et al. 2006). The aptamers reported to date are known to form loops, stems, hairpins, triplexes or quadruplex structures. For instance, the DNA aptamer specific to prostate-specific antigen (PSA), a protein biomarker for prostate cancer, forms a stable single-loop configuration. On the other hand, the DNA aptamer specific to alpha-methylacyl-CoA racemase (AMACR), another protein biomarker for prostate cancer, forms multiple-loop structures (Savory et al. 2010; Yang et al. 2014). The formation of loops is due to the specific interactions between nucleotides, adenine and thymine or guanine and cytosine present in DNA aptamer chains. Quadruplexes are nucleic acid sequences that are rich in guanine and are able to form a four-stranded structure. The DNA aptamer specific to thrombin forms a quadruplex structure, which is further stabilised by the presence of a cation, especially potassium, which sits in a central channel between each pair of tetrads (Macaya et al. 1993).
Aptamers can be easily chemically modified with various functional groups, such as thiol, amine or azide as well as biotin groups. This modification allows the immobilisation of aptamers to various solid supports. For example, modification of aptamers with thiols allows their association on the gold surface using Au–S interactions (Jolly et al. 2015a) or modification with azido groups via click chemistry (Hayat et al. 2013). In another case, one end of DNA or RNA aptamers can be modified with biotin and binding of these biomolecules through solid support is realised via avidin, streptavidin or neutravidin bridges (Cavic and Thompson 2002; Centi et al. 2007; Liss et al. 2002; Ostatná et al. 2008).
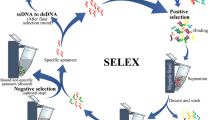
Aptamers were introduced in 1990 by two independent research groups: Tuerk et al. used the term SELEX (generalised scheme of systematic evolution of ligands by exponential enrichment) for selecting RNA ligands against T4 DNA polymerase; and Ellington et al. coined the term in vitro selection (Ellington and Szostak 1990; Robertson and Joyce 1990; Tuerk and Gold 1990). In contrast to antibodies that are obtained by molecular biology techniques, aptamers are prepared by a synthetic method using an in vitro selection procedure. Once an aptamer sequence is identified, it can be synthesised with high purity, reproducibility and relatively low cost. Although aptamers have a greater advantage over antibodies, they still possess a number of limitations, for example, degradation by nucleases (DNAse and RNAse) or protein fouling in serum due to DNA-binding proteins (Keum and Bermudez 2009; Sylvia et al. 1975).
A generalised SELEX method is presented in Fig. 9.1. The SELEX approach starts with a random library containing 1013–1016 ssDNA or RNA sequences (Iliuk et al. 2011; Song et al. 2008). The library is incubated with a target to initiate the first cycle of selection. This is typically followed by iterative cycles of absorption, recovery of bound DNA/RNA and amplification. Isolation of the bound DNA/RNA is the most critical step to ensure purity and selectivity. For example, the aptamer-target complex can be separated by filtration through nitrocellulose or by affinity chromatography from the unbound DNA/RNA sequences. The bound aptamers are then eluted and amplified by RT-PCR (for RNA libraries) or PCR (for DNA libraries) to generate new pools for the next selection cycle. An ideal aptamer selection procedure requires around 10–15 cycles.
9.1.2 Aptamer Assay Configuration
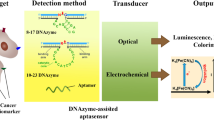
The first application of aptamers for biosensing was reported in 1996 where an optical aptasensor based on fluorescence-labelled aptamers was developed, for the detection of immunoglobulin G (IgG) (Davis et al. 1996). Since then, several aptasensors have been developed using different transduction techniques. Replacing the antibodies in a classical ELISA configuration has enabled the development of sophisticated assays, which are more robust, reproducible and economical. The high specificity of aptamers and the possibility of developing aptamers against different binding sites of target analyte offer high variability of assay configuration (Fig. 9.2) (Hianik and Wang 2009; Song et al. 2008). Like an antibody-based assay, aptamer-based assays can have different configurations to design the biorecognition events. Various assay configurations for aptamers have been reported. Nonetheless, the assay can be generalised into two categories: single-site binding and dual-site binding. The first simple, single-site binding assay consists of the attachment of aptamers onto a support and the interactions of aptamer/target can be directly monitored (Fig. 9.2a).
Examples of different assays based on aptamers. (a) Capture of the analyte by immobilised aptamers. (b) Sandwich-type assay with aptamers using two aptamers specific to two different sites of the analyte. (c) Capture of the analyte by the immobilised aptamers whilst a secondary antibody is used to detect in a modified ELISA format. (d) Capture of analyte by an immobilised antibody whilst aptamer is used as a secondary probe in a modified ELISA format
Although the single-site binding format is simple, fast and cheap, it may still impose problems with sensitivity and/or selectivity. Whereas, sandwich assay or dual-site binding assays consist of capturing the target by specific aptamer method followed by interactions with another aptamer or antibody (Fig. 9.2b–d). Again, a dual-site binding assay may enable detection of the target with high sensitivity and selectivity. However, such an assay involves several incubation steps, making it time consuming and expensive (Song et al. 2008).
9.2 Immobilisation Techniques
In order to fabricate a successful biosensor, surface engineering of the sensor transducer plays a key role. Surface chemistry has been demonstrated as a tool to engineer biosensor surfaces and is one of the most crucial aspects of biosensor construction. A proper control of the immobilisation step is essential in order to have good sensitivity, selectivity and stability of the biosensor (Campuzano et al. 2006). Broadly, immobilisation techniques could be divided into physical adsorption, covalent binding, affinity and entrapment. These techniques pose advantages and disadvantages, some of which are listed in Table 9.1.
A presentation on aptamer immobilisation strategy is presented based on the type of surface immobilisation coupled with the type of electrode surface. Briefly, we report the different types of aptasensors for the detection of cancer, based on physical adsorption, self-assembled monolayers (SAM) and polymer-based approach. Furthermore, with the advancements in the field of nanotechnology, exciting and powerful tools for the development of aptasensors have been developed. And therefore, this chapter further describes the different types of nanoparticles used for the fabrication of sensitive aptasensors for cancer diagnosis.
9.2.1 Physical Adsorption of Aptamers
Physical adsorption is a type of direct immobilisation technique of aptamers on a substrate via weak, liable bonds. The interactions involved are non-covalent such as van der Waals, hydrogen bonding, electrostatic and hydrophobic interactions. Although adsorption is one of the simplest and cost-effective methods to immobilise aptamers onto a surface of interest, it may result in a random orientation of the aptamer on the surface. Random orientation may lead to reduced activity along with reduced surface density. Furthermore, the interaction of aptamers via weak bonds could be easily broken, resulting in the loss of aptamers, limiting the immobilisation strategy. Nevertheless, Su et al. in 2007 reported the adsorption of low-molecular-weight ATP-binding DNA aptamers onto a microcrystalline cellulose membrane (Su et al. 2007). In 2015, Li et al. developed an optical aptasensor for the detection of multiple cancer in vitro and in vivo (Li et al. 2015). The study reports an aptasensor that binds specifically to the cell surface mucin 1 (MUC1) marker, which is overexpressed in a number of malignant tumours including prostate cancer and breast cancer (Gaidzik et al. 2013; Kimoto et al. 2013; Li et al. 2015). In 2013, Choi et al. developed a chemiluminescent-based aptasensor for the detection of PSA by immobilising DNA aptamers conjugated with fluorescent dye on magnetic Fe3O4 graphene oxide nanoparticles via π–π stacking. The aptamer labelled with a dye (Cy3) was immobilised on the surface of oxidised mesoporous carbon nanospheres via π–π stacking for an optical detection technique using fluorescence (Choi and Lee 2013).
To address the drawback of the random orientation of aptamers, Tzouvadaki et al. in 2016 reported the development of ultrasensitive memristive aptasensor based on a physical adsorption method for electrochemical detection of PSA. The immobilisation strategy was a combination of physical adsorption and affinity method where the silicon nanowires were first activated with hydroxyl groups by exposing the nanowires to piranha solution. First, a physically absorbed layer of biotin was prepared on the silicon nanowire. Later, the nanowire was incubated with streptavidin where it specifically binds to biotin on the surface. Finally, biotinylated aptamers were used to occupy the free spaces of streptavidin leading to the controlled orientation of DNA aptamers. The combined effect of memristor and immobilisation strategy led to detection down to attomolar levels (Tzouvadaki et al. 2016). On the other hand, Liu et al. reported a PSA aptasensor based on gold nanoparticles encapsulated by graphitized mesoporous carbon and bovine serum albumin (BSA) was used as a blocking molecule to reduce non-specific binding. Physical adsorption of nanoparticles on glassy carbon electrodes followed by affinity-based DNA aptamer immobilisation was used for the fabrication process. Differential pulse voltammetry (DPV) was used as a measurement technique with a limit of detection (LOD) of 7.58 pM (Liu et al. 2012).
Feng et al. in 2011 demonstrated a reusable graphene-based electrochemical aptasensor for the detection of nucleon on the cell surfaces of cancer cells using electrochemical impedance spectroscopy (EIS). The study reports the use of 3,4,9,10-perylene tetracarboxylic acid (PTCA), a water-soluble perylene derivative that can strongly bind on the graphene surfaces via hydrophobic and π–π interactions. Thereafter, amine terminated AS1411 aptamer was conjugated with free carboxylate groups of PTCA via ethyl(dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) chemistry. The aptasensor developed successfully differentiated cancer cells from non-cancerous cells (Feng et al. 2011).
9.2.2 Aptamers and Self-Assembled Monolayers
There are many techniques reported for aptamer immobilisation depending on the electrode surface and application. Specifically for gold electrode surfaces, self-assembled monolayer (SAM) is one of the most commonly reported techniques because it results in highly controlled density and thickness of the transducer surface (Hong et al. 1999). The key aspect to take into consideration for the immobilisation of oligonucleotide-based probes on gold surfaces is to have an optimum density (Keighley et al. 2008). Thiols, sulphides and disulphides have demonstrated very high affinity towards gold by forming a covalent bond between the sulphur and gold atoms, making alkane thiol-based SAMs very popular (Bain et al. 1989; Bain and Whitesides 1989; Love et al. 2005). For instance Su et al. reported a lab-on-paper electrochemical cyto-device to demonstrate detection of cancer cells together with multi-glycan profiling on living cancer cells. In such an approach thiolated DNA aptamers specific to K562 cancer cells were directly immobilized on a three-dimensional macroporous Au-paper electrode (Su et al. 2015). More recently, Rahi et al. have reported a relatively simple aptasensor for the detection of prostate cancer based on electrodeposited gold nanospheres using an arginine template to achieve a 50 pg/mL detection limit using differential pulse voltammetry technique. Again a thiolated anti-PSA DNA aptamer was used to bind to the gold surface followed by incubation with 6-mercapto hexanol (MCH) to cover the free gold spaces and to obtain a well-aligned monolayer (Rahi et al. 2016).
Sulphur in the proximity of gold undergoes the following reaction:
The sulphur–gold reaction is spontaneous with 80–90% coverage within the first few minutes, but a well-organised layer is formed in no less than 12–16 h (Schreiber 2000). The formation of well-organised SAMs depends not only on factors such as presence of contaminants and surface quality (roughness and purity), but also on the length of alkanethiols (Finklea 1996). The higher kinetics for longer alkyl thiols could be attributed to amplified van der Waals interactions (Darling et al. 2002). In 2005, Love et al. determined the maximum density of alkane thiol on gold surfaces to be 4.64 × 1014 molecules/cm2 with a gap of 4.99 Å between two adjacent molecules. The time-dependent well-organised SAM formation undergoes two main steps where there is spontaneous assembly within the first few minutes. In the early stages, alkane thiols lie flat on the gold electrode surface through physisorption, called the ‘lying-down’ phase (Camillone et al. 1994). Thereafter follows a chemisorption process where a crystalline or a semi-crystalline structure is formed due to van der Waals forces resulting in lateral movement until a tilt angle of about 30° between the hydrocarbon chains is achieved (Love et al. 2005). It is worth mentioning that the ability of the thiols to move laterally along the gold surface results in the formation of well-ordered layering and healing the defects (Ulman 1996).
In such a process, the terminal groups of the alkanethiol affect the SAM properties that define its interaction with biological molecules. Chun et al. reported an electrochemical aptasensor for the detection of HER2, which is a breast cancer biomarker. The group fabricated the aptasensor by using first a SAM layer of 3-mercaptopropanoic acid (MPA). Thereafter, the carboxylate group of MPA was used to covalently attach amine-terminated anti-HER2 DNA aptamer using EDC/NHS activation step (Chun et al. 2013). Another anti-HER2 aptasensor based on gold micro capacitor electrodes was reported to develop a label-free capacitive aptasensor for breast cancer biomarker (Qureshi et al. 2015). A similar surface chemistry was also reported to develop an aptasensor for the detection of human liver hepatocellular carcinoma cells using electrochemical methods (Kashefi-Kheyrabadi et al. 2014). In recent years, various nanomechanical systems have been developed (Eom et al. 2011). For instance, by monitoring the changing frequencies or deflection of microcantilevers, cantilever-based biosensors can be fabricated (Boisen et al. 2011). Such an approach was utilized to develop label-free microcantilever array sensor in static mode to detect lung cancer cells with a detection limit down to 300 cells/mL. Thiol chemistry was utilized to form a SAM on eight identical microcantilever gold surfaces, by using thiolated DNA aptamer specific to lung cancer cells, followed by backfilling with MCH (Chen et al. 2016).
Furthermore, the tailoring of the SAM layers has led to the establishment of mixed SAMs. Exploring different SAM layers, Jolly et al. demonstrated an optimised SAM of co-immobilised thiol-terminated sulfo-betaine and 11-mercaptoundecanoic acid (MUA) that can be used to create enhanced anti-fouling properties. Thereafter, carboxylate groups of MUA were used to bind an amine-terminated anti-PSA DNA aptamer using EDC/NHS activation step (Jolly et al. 2015b). In 2010 Wu et al. reported a ternary SAM on screen-printed gold electrodes as a potential anti-fouling SAM for amperometric detection of oligonucleotides. Ternary SAM here comprised of an optimised ratio of thiolated oligonucleotide probe and 1,6-hexanedithiol (HDT) and further passivated with 1-mercapto-6-hexanol (MCH). Based on similar surface chemistry with screen-printed electrodes, Miodek et al. demonstrated the development of an electrochemical aptasensor for the detection of thrombin (Miodek et al. 2015). Thrombin is a protein biomarker that plays an important role in cardiovascular diseases and inflammatory process and can indicate many pathological conditions, including cancer (Yeh et al. 2014). Furthermore, by tuning the composition of mixed SAMs, the amount of functional groups can be monitored for the efficient efficacy of biosensors. In fact, it is worth mentioning that by carefully selecting the spacer molecules, desired hydrophobicity or hydrophilicity as well as significant chemical reactivity could be achieved. Such a strategy could impede non-specific binding and therein improve the electrochemical signals (Herne and Tarlov 1997).
9.2.2.1 Silane-Based SAM and Aptamers
Deviating from metallic electrode surfaces for the development of aptasensors, non-metallic surfaces have also gained a lot of interest for the development of point-of-care devices. Different surface chemistries, mostly based on silane layers, have been demonstrated for the conjugation of bioreceptors. Sagiv (1980) demonstrated for the first time a chemisorbed monolayer using siloxane chemistry on a silicon surface. Since then, many reports have been published on bioconjugation techniques using different types of silane (Bañuls et al. 2013; Haensch et al. 2010). For instance, Sharma et al. demonstrated the development of electrochemical aptasensor by covalent attachment of amine-terminated DNA aptamers on 3-(2-aminoethylamino) propyl trimethoxysilane on indium tin oxide (ITO) surfaces via a glutaraldehyde linker. The developed aptasensor was used to detect lung cancer cells in the concentration of 103–107 cells/mL with a detection limit of 103 cells/mL within 60 s (Sharma et al. 2012). More recently, Pasquardini et al. developed an innovative single-photon avalanche diode (SPAD) system for the detection of vascular endothelial growth factor (VEGF), which is a circulating protein biomarker for cancer detection. The system employs the immobilisation of thiol-terminated anti-VEGF DNA aptamer via covalent bond on the SAM of 3-mercaptopropyltrimethoxysilane on silicon dioxide wafers. A secondary detector antibody labelled with horseradish peroxidase (HRP) was used to complete the sandwich assay with a demonstrated stability of the SAM up to 42 days (Pasquardini et al. 2015). A simple microfluidic assay was reported by Jolly et al. for an aptamer-based ELISA for both quantification and glycoprofiling of PSA for prostate cancer diagnosis. The group utilised an amine-terminated anti-PSA DNA aptamer immobilised in a microfluidic channel on a SAM of 3-glycidyloxypropyl) trimethoxysilane on glass surfaces. A secondary antibody or a lectin is used to quantify, by chemiluminescence, both the amount of PSA and its glycosylation levels (Jolly et al. 2016a).
9.2.3 Nanomaterials and Aptasensors
In the past decades, there has been a considerable interest in nanomaterials for their application in medicine and biology. Nanomaterials have been used as a physical approach to improving the pharmacokinetic and pharmacodynamic properties of different drug molecules, increasing their therapeutic benefit and, at the same time, minimizing side effects. Since then, the applications of these structures increased and nowadays have become one of the biggest research fields. Nanoparticles are particulate dispersions or solid particles with a size range from 1 to 100 nm. They are the fundamental components to fabricate nanostructures and were well described as smaller than the world of everyday objects described by Newton’s laws of motion, but bigger than an atom or a simple molecule that is governed by quantum mechanics (Horikoshi and Serpone 2013).
The major goal in designing nanoparticles is their controlled size and surface properties for specific applications. Once control parameters are achieved, nanoparticles can be introduced into medicine and biology in different ways, for example as drug delivery, fluorescent biological labels, probing DNA structure, detection of pathogens and proteins, tissue engineering, imaging contrast and, one of the newest applications, aptasensors. Due to all these possibilities, nanomaterial has been applied on biosensor development. The different structures and properties already improve the sensitivity lowering the LOD down to femtomolar levels and opening new possibilities for biosensing applications (Li et al. 2010). Using these nanomaterials, different signal transducers are employed for detection.
Given that a biosensor’s signal is generally proportional to the surface coverage, most methods of increasing the sensitivity of label-free biosensors revolve around surface modification to increase probe loading. An obvious method is simply multiplex detection with an array of sensors, but this often has the disadvantage of increasing the required sample volume and electronic complexity. Alternatively forming meso- and microporous surfaces with methods such as electrodeposition can provide increased surface area whilst still maintaining low sample volumes. One of the simplest ways to now increase surface area is to anchor nanoparticles to the surface. These nanoparticles may be formed from metals, oxides, semiconductors and conducting polymers, but it is the use of gold nanoparticles (AuNPs) which has attracted most attention for biosensing applications, in particular for biosensors based on optical and electrochemical transduction.
Nanomaterials are classified into different approaches, as based on the dimension or type of material that is produced (Hett 2004): in the first case, one-dimensional system such as thin film or monolayer, two-dimensional nanoparticles as carbon nanotubes (CNTs) single- or multi-walled carbon and three-dimensional nanoparticles as dendrimers and quantum dots (QDs). However, classifying them as a function of the material will be a better approach for discussing their application on biosensors.
Intentionally produced nanomaterials are divided into metallic (iron oxide, gold, silver), carbon structures (fullerene, carbon nanotubes), ceramic (silica, alumina), semiconductor (QDs), organic (protein based, DNA based, liposomes, polymers, dendrimers) and hybrid (magneto liposomes) (Estelrich et al. 2014). From all the possibilities, this topic will focus on nanomaterials that are commonly applied to aptasensors.
9.2.3.1 Carbon-Based Materials
These nanomaterials commonly take the form of hollow spheres or tubes with many potential applications, including improved films, surface coatings and applications in electronics (Wang et al. 2016). The biosensing applications are very wide, including aptasensors. For instance, Zhang et al. (2014) reported an electrochemical aptasensor for thrombin. Thrombin plays an important role in cardiovascular diseases and inflammatory process and can indicate many pathological conditions, including cancer (Yeh et al. 2014). In this study a glassy carbon electrode modified with a graphene and porphyrin nanocomposite was used to immobilise thrombin aptamer via aptamer/graphene π–π stacking interactions and aptamer/porphyrin π–π stacking simultaneously. The result displays a linear response with a LOD of 0.2 nM. This aptasensor benefits from the synergetic effects of graphene, a nanomaterial with high conductivity and high surface area, its ability to interact with porphyrin and the aptamer specificity (Zhang et al. 2014).
Recently, Nawaz et al. functionalized carbon nanotubes (CNTs) resulting in CNTs bearing benzoic acid and subsequently fabricated films on carbon. This assembly method offers an efficient protocol by using water as a solvent and one simple step for fabrication using a very small amount of CNTs. These modified electrodes were used to develop a DNA aptamer-based biosensor to detect mucin 1 (MUC1), a prevalent gene associated with breast cancer with a LOD of 0.02 U/mL (Nawaz et al. 2016).
Dendrimers are three-dimensional nano-sized polymers synthesized as spherical structures and the number of terminal groups increases exponentially as a function of the number of layers. Polyamidoamine dendrimers (PAMAM) are some of the most used and commercialised structures (Baker 2009). For instance Zhang et al. used gold electrode modified with PAMAM dendrimer to immobilise thrombin aptamer for the development of an EIS-based aptasensor (Zhang et al. 2009).
9.2.3.2 Metal Nanoparticles
Metal nanoparticles include noble metals, heavy metals, iron and metal oxides, such as titanium dioxide, that have unique physicochemical properties depending on their size and material, an easy and simple functionalisation, conductance and a high surface-to-volume ratio (Borghei et al. 2016). These properties provide an enhanced application for the commonly used biosensing techniques, particularly optical techniques.
Gold nanoparticles (AuNPs) should be highlighted as they are the most extensively studied nanomaterial and led to the development of numerous methods for molecular diagnostics, imaging, drug delivery and therapeutics because of their unique properties (Doria et al. 2012). For biosensing, AuNPs present excellent biocompatibility that allows using them for interfacing biological events. They can attach to biological probes, as described before, such as antibodies, enzyme, lectins, nucleic acids and glycans. AuNPs are already used as imaging and therapeutic agents (Borghei et al. 2016). In addition, because of their versatility in biological and medical applications, AuNPs have been investigated as signal enhancement probes for biosensors. Furthermore, AuNP conductivity permits direct electron transfer between many electroactive species and electrodes, which enables to use them for signal amplification, enhancing the analytical performance compared to other biosensor designs. Another important property is the high surface area. The diameter of AuNPs varies between 1 and 100 nm, offering a structure that increases the amount of biomolecules anchored on the surface, maintaining their bioactivity and increasing the sensitivity (Cao et al. 2011; Javier et al. 2008).
The application of AuNPs in medicine started almost two decades ago. In 2008, Javier et al. reported a platform for molecular specific using aptamer-based gold nanoparticles as contrast agents. They demonstrated an approach for prostate-specific membrane antigen (PSMA) detection obtaining reflectance images of cell lines treated with the anti-PSMA aptamer-gold conjugates (Javier et al. 2008). More recently, Borghei et al. reported a simple but highly sensitive colorimetric method based on aptamer/cell interaction for the detection of cancer cells. Cancer cells were able to uptake specific aptamers having affinity with receptors that are over-expressed in cancer cells. Such a process resulted in the removal of the aptamers from the solution, and leaving no free aptamers that can hybridise with complementary ssDNA/AuNP probes, leaving the solution red. Whereas in a negative control, with the absence of target cells or presence of normal cells, ssDNA/AuNP probes were able to hybridise with free aptamers and produced a purple solution. A linear response for MCF-7 cells was obtained with a LOD of 10 cells. This strategy can be extended to detect other receptors from different cancer cells (Borghei et al. 2016).
Quantum dots (QD) and magnetic beads (MB) are other nanoparticles very frequently reported for specific bio-application for molecular diagnostics and cancer applications. A QD is a semiconductor nanostructure that confines the motion of conduction in three spatial directions. QD has a discrete quantized energy spectrum; changing the size of QDs changes their optical properties and this property is the highlight of QDs. Due to these reasons, most of the transducers involved with QDs are optical. An example of QD application for biosensing is conjugate MUC1 aptamers with QDs to recognise MUC1 peptide. The strand includes additional bases capable of hybridisation with the complementary ssDNA sequence that carries the label. QD irradiation and FRET were observed for these MUC1-targeted probes down to 1 pM/μL (Singh et al. 2016).
MBs comprise a ferromagnetic elemental, alloy, oxide or composite structure. According to their material, they are divided into paramagnetic, antiferromagnetic and ferromagnetic (Xiao et al. 2016). As with QDs, optical biosensing is often developed for these nanomaterials due to their properties. Mostly, affinity-based method like avidin/biotin or streptavidin/biotin is used for the immobilisation of aptamers on magnetic beads. For instance, in 2006, Herr et al. developed an aptamer-conjugated MB for selective collection and detection of leukaemia cells. The study reports the conjugation of two types of aptamer-modified nanoparticles: aptamer-modified MB for target cell extraction, while aptamer-modified fluorescent nanoparticles for the detection using fluorescent imaging. By doing so, leukaemia cells were extracted from complex samples including whole-blood samples (Herr et al. 2006). In 2014, Hu et al. proposed a simple optical aptasensor for cancer biomarker AGR2 detection using UV–vis spectrometry. In this case, the aptasensor is sandwich-typed AuNPs/DNA/MBs where the aptamers/MBs target proteins and DNA probes on the AuNPs compete with proteins to hybridise with catchers. As a result, the increasing number of target proteins reduces the possibility of the sandwich structure formation with a picomolar range of detection limit (Hu et al. 2015). The sum of different properties allows an extraordinary performance of the aptasensor.
Researchers have also demonstrated the use of bivalent metal ions (M2+) for biosensing applications. Immobilisation of aptamers to the electrode surface can also be performed via histidine-tag/M2+/Nα,Nα-bis(carboxymethyl)-l-lysine hydrate (ANTA) chemistry. During this process, bivalent metal ions such as Cu2+ bound to ANTA anchored on the surface by a tetravalent chelation, leaving two available coordination sites for linking of histidine-modified aptamers. ANTA/Cu2+ form a stable complex and the binding of Cu2+ to the ANTA-modified self-assembled monolayer was studied by Stora et al., showing a dissociation constant of 5 nM obtained with impedimetric measurements (Stora et al. 1997). Because of the high affinity to the histidine sequence, the complex of ANTA with Cu2+ but also with other metal cations such as Ni2+, Zn2+ and Co2+ is widely used in protein purification methods and is known as ‘immobilised metal-ion affinity chromatography’ (IMAC) (Kronina et al. 1999; Ueda et al. 2003). The binding of histidine-modified molecules can be reversible; however, it needs a presence of highly concentrated (~0.2 μM) imidazole solution (Haddour et al. 2005).
Such strategy based on His-tag/M2+/ANTA chemistry has already been described in the case of immunosensors (Chebil et al. 2010) and aptasensors (Xu et al. 2013) and in both cases a wide linear response range was obtained. For example, the aptasensor based on ANTA/Cu2+ complex allowed to detect thrombin protein with a detection limit of 4.4 pM (Xu et al. 2013).
9.2.3.3 Assay Designs with Nanoparticles and Aptamers
The integration of nanoparticles with biosensors is also divided into immobilisation, amplification or both at the same time, as illustrated in Fig. 9.3. For example, AuNPs can be immobilised on different carbon-based matrices such as a carbon nanotube, graphene or graphene oxide, gold surfaces or other polymers (Sardar et al. 2009). This is an effective method to increase the amount of probes immobilised, especially thiol molecules which present high affinity to bind AuNPs. Moreover, due to the availability of high surface area of nanoparticles, the amount of biomolecules anchored is increased, consequently amplifying the signal.
Furthermore, metal NPs can be used to mediate reactions amplifying the signal. For example, capture probes can be first immobilised on a substrate. Thereafter, AuNPs modified with target probes are introduced for specific recognition, resulting in signal amplification. Using AuNPs as a well-established model, with the properties previously described and, additionally, due to their high absorption coefficient as well as the ability to enhance electromagnetic fields and fluorescence, allows exploring these particles in many different ways for biosensing purposes, especially in the development of optical, electrochemical and piezoelectric biosensors (Sardar et al. 2009). The different transducers can be applied to many biological probes, including aptamers.
For instance, Chai et al. in 2011 reported the use of SAMs to deposit AuNPs on electrode surfaces. The study reports a simple strategy to immobilise streptavidin-coated AuNPs on the surface of gold by employing a SAM of 1,3-propanedithiol. Thereafter, biotinylated DNA aptamer specific to PDGF-BB was immobilised using the conventional affinity interaction between streptavidin and biotin. Such a platform was used to capture the target PDGF-BB and finally a secondary label was used as a detector. The label comprised of AuNPs modified with N-(aminobutyl)-N-ethylisoluminol (ABEI) and DNA aptamers. The binding event of the sandwich assay was monitored by electrochemiluminescence method. The signal amplification by the AuNP was specific, simple and stable, with a detection limit as low as 2.7 × 10−14 M being achieved (Chai et al. 2011).
Other proteins, such as thrombin, have been extensively studied. Fang et al. proposed an electrochemiluminescence (ECL) aptasensor where aptamers labelled with AuNPs were first immobilised onto ITO electrode surface and, after catching the thrombin, signal aptamers tagged with ECL labels were attached to the assembled electrode surface. As a result, a sandwich-type assay was formed achieving 10 nM as LOD (Fang et al. 2008). Thrombin can also be detected using other approaches. Screen-printed electrodes (SPE) are economical electrochemical substrates with advantageous properties, such as disposability and simplicity and can be used for the rapid in situ analysis. Yeh et al. developed a system with a carbon SPE capture thrombin and an amplifier for recognising thrombin which is the multiple molecules of anti-thrombin antibody-modifying AuNPs. The electrochemical response presented a LOD of 1.5 pM (Yeh et al. 2014).
The whole cell can be captured by the aptasensor with good selectivity once they usually have specific proteins on the surface. Mucin 1 is a tumour maker protein in human breast carcinoma MCF-7 cells. An assay used aptamers to bind tumour markers on the surface of cancer cells, in this case, MCF-7 cells. Nanoporous materials modified with mucin 1 aptamers attach on the cell improving the biosensor performance and exhibiting a detection limit of 38 cells/mL (Yan et al. 2013).
Optical transducers are also applied in cancer diagnosis using aptasensors. Colorimetric detection is a signal transducer based on absorbance curves with low cost, fastness and a point-of-care compatible testing technique for cancer cells. Wang et al. recently designed a specific detection for MCF-7 cells with mucin 1 aptamer, the same samples previously described. But, in this case, the aptamer and PtAu nanoparticle with high catalytic activity were able to differentiate cancer cells from non-cancer cells and different cancer cell types with a LOD of 10 cells/mL in phosphate buffer solution and in the serum samples (Wang et al. 2015). Different nanoparticles can be used based on their properties. In some cases, nanoparticles can be combined. For example, Ye et al. published an interesting study with bimetallic nanoparticle (Cu–AuNP) combined with an iodide-catalysed system. The nanoparticle was modified with aptamers to human leukaemia CCRF-CEM cells, thus indirectly inducing the colorimetric signal variation of the system. The LOD of 5 cells in 100 μL shows a strategy that can be extended to other cancer cell assays (Ye et al. 2015).
Similar strategies have been developed for other optical transducers. Surface-enhanced Raman scattering (SERS) was applied to detect vascular endothelial growth factor (VEGF). A different strategy involves silver nanoparticle-ornamented gold nanoparticle pyramids (Ag–AuNPs) using an aptamer-based sensor that was ultrasensitive with a LOD of 22.6 aM. Limits lower than this have already been achieved with optical techniques (Zhao et al. 2015). Vance and Sandros presented an application of surface plasmon resonance imaging (SPRi) system to detect 7 zM (or 5 fg/mL) of C-reactive protein (CRP), in this case, combining aptamer-modified quantum dots (QDs) and microwave-assisted surface functionalisation (Vance and Sandros 2014).
Deviating from conventional nanoparticles, graphene oxide (GO) has also been used to develop aptasensors based on a fluorescence technique. By using the inherent capability of the ring in DNA guanine residues to absorb on the surface of graphene oxide by π–π interactions, He et al. demonstrated the development of an aptasensor that characterises epithelial malignancy by targeting MUC1. In such an approach, an anti-MUC1 labelled with a fluorescent dye (Cy5) was adsorbed on the GO resulting in close proximity of the dye to the surface. Consequently, a quenching effect is observed via energy transfer from dyes to GO. However in the presence of the target, the aptamer changes conformation resulting into increased distance between the dye and the GO, inducing the fluorescence restoration. The aptasensor was successfully tested in both buffer and blood serum with a detection limit of 28 nM (He et al. 2012). A similar strategy of using GO and the quenching effect was used for the detection of hepatocellular carcinoma, where the aptasensor was able to detect human liver cancer cell lines SMMC-7721 with a detection limit of 200 cells in 200 μL buffer (Xie et al. 2014).
9.2.4 Polymer-Based Aptasensors
The cost has always been a crucial factor in the development of novel biosensor devices for medical purposes. In order to reduce the cost of biosensor fabrication, the adoption of noble metals and their cleanroom processing are required to be kept at a minimum (Kiilerich-Pedersen et al. 2011). These factors have led to a shift from the use of gold and platinum to degradable polymer materials. As a result, the application of polymers is experiencing an increasing importance over traditional systems, especially within the area of immobilisation of aptamers for the detection of damages within target DNA (Liao et al. 2008; Radi 2011). Moreover, the development of novel polymeric materials led to a new trend within the area of biosensors for cancer diagnostics. The necessity for introducing the adoption of polymers aroused with the requirement of individualised and tailored methods of treatment for a more heterogeneous disease such as cancer (Luk and Zhang 2014). Polymer-based biosensors offer the ability of theranostic applications (emergence of therapy and diagnostics imaging into a single package), with their additional advantage of possessing excellent biocompatibility (Luk and Zhang 2014). These polymer-based nanomaterials demonstrate desirable biodegradability and structural versatility. Except at high concentrations, biopolymers are typically non-toxic and naturally degrade into safe materials (Clawson et al. 2011; Hu et al. 2010).
It is worth mentioning that depending on the type of polymer, different immobilisation strategies for aptamers have been reported. For example, a reagent-less electrochemical transduction-based aptasensor for the detection of PSA was developed: the study demonstrated the elimination of any redox labels by adopting a quinone-containing conducting polymer (Souada et al. 2015) on glassy carbon electrode surface. Short amine-terminated DNA aptamers have been first immobilised on the quinone-based conducting copolymer via EDC/NHS chemistry. When subjected to PSA, a strong current decrease (‘signal-off’) was generated due to heavier molecules of PSA compared to aptamer strands on the probe surface. This was next switched to a current drop (‘signal-on’) by the hybridisation of probe aptamer with its complementary strand DNA which breaks PSA–aptamer interactions. As a result, the developed switch signal system was able to detect PSA in ng/mL range and also evaluated the PSA–aptamer dissociation constant (K d), of ca. 2.6 nM. This dual-check system provided a full assurance of a perfectly specific recognition event (Souada et al. 2015).
There are many conducting polymers like polypyrrole (PPy), polythiophene and polyaniline that have been used for biosensor construction. Among them, PPy is one of the most extensively used conducting polymers in the design of bioanalytical sensors apart from polythiophene and polyaniline (Peng et al. 2009; Ramanavičius et al. 2006). This is due to its copious properties such as redox activity (Han et al. 2005), ion exchange and ion discrimination capacities (Johanson et al. 2005; Weidlich et al. 2005), strong absorptive properties (Azioune et al. 2005; Chehimi et al. 1999), catalytic activity (Khomenko et al. 2005) as well as biocompatibility (Wang et al. 2004). PPy as a polymer can be further characterised by its high electrical conductivity, hydrophilic character and high stability in water (Andrade 1985). In fact, its low oxidation potential enables a pyrrole polymer film to be grown from aqueous solutions which is compatible with most of the biological elements (Asavapiriyanont et al. 1984). Researchers have also worked on the modification of pyrrole monomers in order to provide enhanced anti-fouling properties. For example, in 2002 Rodrigez et al. synthesised pyrrole modified with biotin set apart by polyethylene glycol chain which prevented biosensor against non-specific interactions. More recently, Jolly et al. (2016c) reported the development of an aptasensor for AMACR detection using a voltammetry detection technique. In the study, a simple and efficient electrochemical patterning of amine-bearing PEG derivatives on PPy films has been developed. Such a method paved way to simple pyrrole monomer modification and demonstrated enhanced anti-fouling properties of PPy with PEG. His-tagged DNA aptamers specific to AMACR were then immobilised via coordination chemistry with copper ions. A very low detection limit of 1.4 fM was established in human plasma samples. The progression of a tumour and its transformation to different stages can be related to platelet-derived growth factor (PDGF), especially PDGF-B chain. Recently in 2016, Lee et al. fabricated an aptasensor based on multidimensional hybrid conductive nanoplate for the detection of the PDGF. PDGF has emerged as a critical cancer biomarker as it is associated with diverse cancers and other diseases (Lee et al. 2016; Shih et al. 2004). The sensor was fabricated by using multidimensional hybrid carboxylated polypyrrole plates (MHCPPs) that were functionalised with the PDGF-B-specific DNA aptamer. The vertical decoration of the polypyrrole nanosheets has managed to maximise the active surface area of the MHCPPs. The strategy demonstrated a dramatic increase in the interaction between the developed MHCPP-based sensor and the PDGF-BB analyte. Detection limit as low as 1.78 fM has been achieved for PDGF-B, by using field-effect transistor (FET)-type aptamer sensor (Lee et al. 2016).
Polymer-based approaches have also penetrated graphene-based FETs and are witnessing a rapid development growth and even considered as an alternative for post-silicon electronics. Kwon et al. demonstrated how to grow polypyrrole-converted nitrogen-doped few-layer graphene (PPy-NDFLG) on copper substrates which provided the recognition of the target molecules at really low concentrations of 100 fM. Such a process was carried out using a combined technique of chemical vapour deposition and vapour deposition polymerisation. The developed platform was used to immobilise amino-terminated anti-VEGF RNA aptamer using Schiff-base reaction via glutaraldehyde-conjugated 1,5-diaminonaphthalene (DAN) (Kwon et al. 2012).
Development of aptasensor based on single-polymer-based nanowire has also been reported. Huang et al. in 2011 reported a single-step electrochemical deposition of single-PPy nanowire between two gold electrode junctions in a patterned polymethylmethacrylate (PMMA) nanochannel. A different immobilisation strategy was utilised where a mixture of pyrrole monomer and aptamer was used, with the aptamers encapsulated within the polymer without any covalent bonding. By using microfluidic systems the group demonstrated the detection of immunoglobulin E (IgE) and mucin 1 (MUC1) with their specific aptamer. Mucin 1 is a protein biomarker which has been reported to be over-expressed in almost all human epithelial cell carcinomas like breast cancer, ovarian cancer and lung cancer (Croce et al. 2003; Hough et al. 2000; Maeshima et al. 1997). The detection of the protein immunoglobulin E (IgE) was achieved within a range from 0.01 to 100 nM and the aptasensor performed excellent sensitivity with a fast response and rapid stabilisation time (∼20 s). A detection limit of 2.66 nM was obtained for MUC1 using conductance measurements, which is significantly sensitive compared to commercially available MUC1 diagnosis assay (800 nM) (Huang et al. 2011).
Although conducting polymers have always been in the interest for the development of electrochemical biosensors, there are also reports on the use of non-conducting polymers such as chitosan. For instance, Tahmasebi et al. adopted carbon nanotube (CNT)-based polymer materials in order to provide a sensitive electrochemical aptasensor for the detection of PSA using EIS technique. The study demonstrates the benefits of nanomaterials composed of carboxylic acid-functionalised CNTs and chitosan for the immobilisation of PSA aptamer on probe surface with a LOD of 22 pM. The experimental results also proved the higher aptamer immobilising capability of chitosan-CNT composite when compared to CNTs or chitosan alone (Tahmasebi and Noorbakhsh 2016).
9.2.4.1 Molecular Imprinting and Aptamers
In response to the challenges faced by the conventional methods of protein imprinting for biosensor studies, molecular imprinting has adopted new approaches to overcome its limitations (Menger et al. 2016). One of the approaches is to incorporate aptamers within the molecularly imprinted polymers (MIPs). A study carried out on an aptamer-MIP hybrid receptor for the detection of prostate-specific antigen in 2016 reported much more sensitive recognition characteristics compared to that of the aptamer alone (Jolly et al. 2016b). The MIP cavity that had been developed was observed to act synergistically with the enclosed aptamer forming a hybrid receptor which provided much sensitive recognition characteristics (termed as ‘apta-MIP’). A simple strategy was demonstrated by using dopamine monomers and PSA-specific thiolated DNA aptamers. A thiolated anti-PSA DNA aptamer was complexed with PSA prior to being immobilised on the gold electrode surface. Thereafter, a controlled electropolymerisation of dopamine around the complex enabled the entrapment of the complex. The PSA was then removed from the template and the fabricated sensor was used to study the subsequent rebinding of PSA. The study has adopted EIS as a method of evaluation by looking into capacitance changes, where the apta-MIP sensor showed a detection limit of 1 pg/mL of PSA (Jolly et al. 2016b). Within the same year, another novel method of luminescent ‘double recognition’ for the detection of ENR (enrofloxacin) was developed. This method also had two stages of recognition. Firstly, ENR-specific biotinylated aptamers were immobilised on a surface of up-conversion nanoparticles (UCNPs) in order to capture and concentrate ENR as the initial imprinting recognition safeguard. This was then followed by the polymerisation of methacrylic acid monomers neighbouring the aptamers of ENR, which interacted with the residual groups of ENR by using MIP techniques as the second recognition safeguard. The sensor demonstrated detection and quantification limits of 0.04 and 0.12 ng/mL, indicating the feasibility of the method for the detection of ENRs in real samples (Liu et al. 2017).
The studies have shown that MIPs offer an exchange rate of the target aptamer that is significantly higher than that of the antibodies. However, the recent methods of using MIPs have been only developed for the detection of a limited range of proteins. As a result, the research in MIPs still requires improvement in terms of sensitivity and the application on real samples (Menger et al. 2016).
9.3 Outlook
Nucleic acid aptamers represent a challenging and fascinating venue and a possible replacement of antibodies for both therapeutics and diagnostics. Indeed early technology for the application of aptamers for sensing applications has its own limitations but it has continued for more than two decades with the integration of many modifications and advancements in the field. Aptamers, although they have been raised against a number of targets over the last two decades, are expected to see further improvement in their variety, affinity, diversity and half-lives. Therapeutic use of aptamers is a well-established field; for example, aptamer compound named Pegaptanib was approved as a drug for clinical use for endovascular age-related macular degeneration (Gragoudas et al. 2004). Conversely, in the field of diagnostics, especially diagnostic systems based on biomolecular binding events, are still under the dominancy of immunoassays. However, the diagnostic field is now showing that with the use of aptamers, some of the limits of current diagnostic tools can be circumvented, such as flexibility for signal transduction and detection (Liu et al. 2009). For instance, the ability of the aptamers to distinguish small differences between proteins sharing similar surface homology may allow aptamers to differentiate cells based on cancerous and non-cancerous. The use of natural and synthetic nucleotides is still developing and paving the way towards advanced biosensor development. Developments in biochemistry and molecular biology have led to a deeper understanding of the role of nucleic acids and showed that the functions they play are far greater than originally expected. This leads to a new world of biosensing applications, where nucleic acid-based biosensing approaches can have an unparalleled impact on clinical diagnosis, prognosis and monitoring. It can be seen from the plethora of reported literature how researchers from different fields are coming together to realise high-throughput aptamer-based biosensors for use with complex matrix samples such as clinical or environmental. The ease of manipulation of oligonucleotides, controlled surface chemistry approaches and ‘straightforward’ charge distribution make them optimal bioreceptors for biosensing applications. Moreover, the commercialisation of biosensors has been fuelled right after the first glucose test in a PoC format.
However, the process of commercialisation of aptamer-based biosensors for the detection of cancer biomarkers is still at an early stage (proof of concept) and requires further developments. There is still not a self-contained answer to why nucleic acid aptamers have not yet penetrated into the clinical laboratories (Baird 2010). Although there are several challenges that need to be addressed for real clinical application, one of the most important is related to the conformational change of aptamers. Most of the aptamers generated via SELEX is known to have well-known interaction between the aptamer and its target which is dependent on the conformation of aptamers. However, such a conformation is largely affected by chemical and physical environment. As a result, the aptamers selected through in vitro SELEX procedures might have decreased or completely lose their binding efficiency. Overall, the development of aptamer-based biosensors for biomarker detection is expected to attract increasing interest because of its ease of synthesis and the possibilities of multiple modifications. Furthermore, there are many successful reports that have been published for cancer diagnosis; however, most of them have been carried out either in ideal buffer solutions or in vitro cultures. There are very few reports that have shown the tests with real cultures or in animals.
These studies not only demonstrate the enormous potential, but also prove how they can be used for a wide range of other biomarkers for various diseases that exploit target/probe features similar to those of the systems that have been reported previously. It has become apparent that the aptasensor field has reached a new level of maturity where it has been employed to detect multiple biomarkers. Aptasensors have already shown their specificity and versatility to be applied to different surfaces and systems with different transducers. The use of nanoparticles combined to aptamers addressing the increase of sensibility opens the door to new detection levels not previously conceived. The future of these approaches aims to develop a multiplexing platform with the capacity to distinguish different biomarkers with ultra-low levels.
Nevertheless, the sensitivity of an aptasensor is affected not only by the surface chemistry used but also the analytical method used for the detection. Furthermore, the development of a multiplexed platform for parallel sensing of different biomarkers of cancer would help assist clinicians with deeper information on the pathological and physiological state of the patient (in particular the disease). Such a device could be represented as a point-of-use device that can provide a first assessment of the patient’s state which would accelerate the diagnosis and or prognosis speed. Not only limited to diagnosis, the device can be used for surveillance purposes in order to monitor patients at risk or those being treated (either post-surgery or during medication). In the near future, it is likely that cancer detection using aptamers will undoubtedly benefit from the integration of novel aptamers with miniaturised transducer platforms, and therefore in some regard revolutionise the cancer diagnostics on a global platform.
References
Andrade JD (ed) (1985) Surface and interfacial aspects of biomedical polymers: volume 1—surface chemistry and physics. Plenum Press, New York
Asavapiriyanont S, Chandler G, Gunawardena G, Pletcher D (1984) The electrodeposition of polypyrrole films from aqueous solutions. J Electroanal Chem Interfacial Electrochem 1:229–244
Azioune A, Siroti F, Tanguy J, Jouini M, Chehimi MM, Miksa B, Slomkowski S (2005) Interactions and conformational changes of human serum albumin at the surface of electrochemically synthesized thin polypyrrole films. Electrochim Acta 50:1661–1667
Bain CD, Whitesides GM (1989) Formation of monolayers by the coadsorption of thiols on gold: variation in the length of the alkyl chain. J Am Chem Soc 111:7164–7175
Bain CD, Troughton EB, Tao YT, Evall J, Whitesides GM, Nuzzo RG (1989) Formation of monolayer films by the spontaneous assembly of organic thiols from solution onto gold. J Am Chem Soc 111:321–335
Baird GS (2010) Where are all the aptamers? Am J Clin Pathol 134:529–531
Baker JR (2009) Dendrimer-based nanoparticles for cancer therapy. Hematology Am Soc Hematol Educ Program 2009:708–719
Bañuls MJ, Puchades R, Maquieira Á (2013) Chemical surface modifications for the development of silicon-based label-free integrated optical (IO) biosensors: a review. Anal Chim Acta 777:1–16
Boisen A, Dohn S, Keller SS, Schmid S, Tenje M (2011) Cantilever-like micromechanical sensors. Rep Prog Phys 74:036101
Borghei YS, Hosseini M, Dadmehr M, Hosseinkhani S, Ganjali MR, Sheikhnejad R (2016) Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal Chim Acta 904:92–97
Camillone N III, Eisenberger P, Leung T, Schwartz P, Scoles G, Poirier G, Tarlov M (1994) New monolayer phases of n-alkane thiols self-assembled on au (111): preparation, surface characterization, and imaging. J Chem Phys 101:11031–11036
Campuzano S, Pedrero M, Montemayor C, Fatás E, Pingarrón JM (2006) Characterization of alkanethiol-self-assembled monolayers-modified gold electrodes by electrochemical impedance spectroscopy. J Electroanal Chem 586:112–121
Cao X, Ye Y, Liu S (2011) Gold nanoparticle-based signal amplification for biosensing. Anal Biochem 417:1–16
Castillo G, Trnkova L, Hrdy R, Hianik T (2012) Impedimetric aptasensor for thrombin recognition based on CD support. Electroanalysis 24:1079–1087
Cavic BA, Thompson M (2002) Interfacial nucleic acid chemistry studied by acoustic shear wave propagation. Anal Chim Acta 469:101–113
Centi S, Tombelli S, Minunni M, Mascini M (2007) Aptamer-based detection of plasma proteins by an electrochemical assay coupled to magnetic beads. Anal Chem 79:1466–1473
Chai Y, Tian D, Gu J, Cui H (2011) A novel electrochemiluminescence aptasensor for protein based on a sensitive N-(aminobutyl)-N-ethylisoluminol-functionalized gold nanoprobe. Analyst 136:3244–3251
Chebil S, Hafaiedh I, Sauriat-Dorizon H, Jaffrezic-Renault N, Errachid A, Ali Z, Korri-Youssoufi H (2010) Electrochemical detection of d-dimer as deep vein thrombosis marker using single-chain d-dimer antibody immobilized on functionalized polypyrrole. Biosens Bioelectron 26:736–742
Chehimi MM, Abel ML, Perruchot C, Delamar M, Lascelles SF, Armes SP (1999) The determination of the surface energy of conducting polymers by inverse gas chromatography at infinite dilution. Synth Met 104:51–59
Chen X, Pan Y, Liu H, Bai X, Wang N, Zhang B (2016) Label-free detection of liver cancer cells by aptamer-based microcantilever biosensor. Biosens Bioelectron 79:353–358
Choi HK, Lee JH (2013) Role of magnetic Fe3O4 graphene oxide in chemiluminescent aptasensors capable of sensing tumor markers in human serum. Anal Methods 5:6964–6968
Chun L, Kim SE, Cho M, Choe WS, Nam J, Lee DW, Lee Y (2013) Electrochemical detection of HER2 using single stranded DNA aptamer modified gold nanoparticles electrode. Sensors Actuators B Chem 186:446–450
Clawson C, Ton L, Aryal S, Fu V, Esener S, Zhang L (2011) Synthesis and characterization of lipid–polymer hybrid nanoparticles with pH-triggered poly (ethylene glycol) shedding. Langmuir 27:10556–10561
Croce MV, Isla-Larrain MT, Demichelis SO, Segal-Eiras A, Gori JR, Price MR (2003) Tissue and serum MUC1 mucin detection in breast cancer patients. Breast Cancer Res Treat 81:195–207
Darling S, Rosenbaum A, Wang Y, Sibener S (2002) Coexistence of the (23×√3) au (111) reconstruction and a striped phase self-assembled monolayer. Langmuir 18:7462–7468
Davis KA, Abrams B, Lin Y, Jayasena SD (1996) Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res 24:702–706
Doria G, Conde J, Veigas B, Giestas L, Almeida C, Assunção M, Rosa J, Baptista PV (2012) Noble metal nanoparticles for biosensing applications. Sensors 12:1657–1687
Du M, Yang T, Zhao C, Jiao K (2012) Electrochemical logic aptasensor based on graphene. Sensors Actuators B Chem 169:255–260
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Eom K, Park HS, Yoon DS, Kwon T (2011) Nanomechanical resonators and their applications in biological/chemical detection: nanomechanics principles. Phys Rep 503:115–163
Estelrich J, Quesada-Pérez M, Forcada J, Callejas-Fernández J (2014) Introductory aspects of soft nanoparticles. In: Callejas-Fernández J, Estelrich J, Quesada-Pérez M, Forcada J (eds) Soft nanoparticles for biomedical applications. Royal Society of Chemistry, Cambridge, pp 1–18
Fang L, Lü Z, Wei H, Wang E (2008) An electrochemiluminescence aptasensor for detection of thrombin incorporating the capture aptamer labeled with gold nanoparticles immobilized onto the thio-silanized ITO electrode. Anal Chim Acta 628:80–86
Feng L, Chen Y, Ren J, Qu X (2011) A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials 32:2930–2937
Finklea H (1996) Electrochemistry of organized monolayers of thiols and related molecules on electrodes. Electroanal Chem 19:110–335
Gaidzik N, Westerlind U, Kunz H (2013) The development of synthetic antitumour vaccines from mucin glycopeptide antigens. Chem Soc Rev 42:4421–4442
Gragoudas ES, Adamis AP, Cunningham ET Jr, Feinsod M, Guyer DR (2004) Pegaptanib for neovascular age-related macular degeneration. N Engl J Med 351:2805–2816
Gulbakan B, Yasun E, Shukoor MI, Zhu Z, You M, Tan X, Sanchez H, Powell DH, Dai H, Tan W (2010) A dual platform for selective analyte enrichment and ionization in mass spectrometry using aptamer-conjugated graphene oxide. J Am Chem Soc 132:17408–17410
Haddour N, Cosnier S, Gondran C (2005) Electrogeneration of a poly (pyrrole)-NTA chelator film for a reversible oriented immobilization of histidine-tagged proteins. J Am Chem Soc 127:5752–5753
Haensch C, Hoeppener S, Schubert US (2010) Chemical modification of self-assembled silane based monolayers by surface reactions. Chem Soc Rev 39:2323–2334
Han DH, Lee HJ, Park SM (2005) Electrochemistry of conductive polymers XXXV: electrical and morphological characteristics of polypyrrole films prepared in aqueous media studied by current sensing atomic force microscopy. Electrochim Acta 50:3085–3092
Hayat A, Sassolas A, Marty JL, Radi AE (2013) Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 103:14–19
He Y, Lin Y, Tang H, Pang D (2012) A graphene oxide-based fluorescent aptasensor for the turn-on detection of epithelial tumor marker mucin 1. Nanoscale 4:2054–2059
Herne TM, Tarlov MJ (1997) Characterization of DNA probes immobilized on gold surfaces. J Am Chem Soc 119:8916–8920
Herr JK, Smith JE, Medley CD, Shangguan D, Tan W (2006) Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal Chem 78:2918–2924
Hett A (2004) Nanotechnology: small matter, many unknowns, Risk perception series. Swiss Reinsurance Company, Zürich
Hianik T, Wang J (2009) Electrochemical aptasensors–recent achievements and perspectives. Electroanalysis 21:1223–1235
Hong HS, Kim SJ, Lee KS (1999) Long-term oxidation characteristics of oxygen-added modified Zircaloy-4 in 360° C water. J Nucl Mater 273:177–181
Horikoshi S, Serpone N (2013) Microwaves in nanoparticle synthesis: fundamentals and applications. Wiley, New York
Hough CD, Sherman-Baust CA, Pizer ES, Montz F, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ (2000) Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 60:6281–6287
Hu CMJ, Kaushal S, Cao HST, Aryal S, Sartor M, Esener S, Bouvet M, Zhang L (2010) Half-antibody functionalized lipid−polymer hybrid nanoparticles for targeted drug delivery to carcinoembryonic antigen presenting pancreatic cancer cells. Mol Pharm 7:914–920
Hu Y, Li L, Guo L (2015) The sandwich-type aptasensor based on gold nanoparticles/DNA/magnetic beads for detection of cancer biomarker protein AGR2. Sensors Actuators B Chem 209:846–852
Huang J, Luo X, Lee I, Hu Y, Cui XT, Yun M (2011) Rapid real-time electrical detection of proteins using single conducting polymer nanowire-based microfluidic aptasensor. Biosens Bioelectron 30:306–309
Iliuk AB, Hu L, Tao WA (2011) Aptamer in bioanalytical applications. Anal Chem 83:4440–4452
Javier DJ, Nitin N, Levy M, Ellington A, Richards-Kortum R (2008) Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjug Chem 19:1309–1312
Johanson U, Marandi M, Tamm T, Tamm J (2005) Comparative study of the behavior of anions in polypyrrole films. Electrochim Acta 50:1523–1528
Jolly P, Formisano N, Estrela P (2015a) DNA aptamer-based detection of prostate cancer. Chem Pap 69:77–89
Jolly P, Formisano N, Tkáč J, Kasák P, Frost CG, Estrela P (2015b) Label-free impedimetric aptasensor with antifouling surface chemistry: a prostate specific antigen case study. Sensors Actuators B Chem 209:306–312
Jolly P, Damborsky P, Madaboosi N, Soares RR, Chu V, Conde JP, Katrlik J, Estrela P (2016a) DNA aptamer-based sandwich microfluidic assays for dual quantification and multi-glycan profiling of cancer biomarkers. Biosens Bioelectron 79:313–319
Jolly P, Tamboli V, Harniman RL, Estrela P, Allender CJ, Bowen JL (2016b) Aptamer–MIP hybrid receptor for highly sensitive electrochemical detection of prostate specific antigen. Biosens Bioelectron 75:188–195
Jolly P, Miodek A, Yang DK, Chen LC, Lloyd MD, Estrela P (2016c) Electro-engineered polymeric films for the development of sensitive aptasensors for prostate cancer marker detection. ACS Sens 1:1308–1314
Kashefi-Kheyrabadi L, Mehrgardi MA, Wiechec E, Turner AP, Tiwari A (2014) Ultrasensitive detection of human liver hepatocellular carcinoma cells using a label-free aptasensor. Anal Chem 86:4956–4960
Keighley SD, Li P, Estrela P, Migliorato P (2008) Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens Bioelectron 23:1291–1297
Keum JW, Bermudez H (2009) Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem Commun (45):7036–7038.
Khomenko V, Frackowiak E, Beguin F (2005) Determination of the specific capacitance of conducting polymer/nanotubes composite electrodes using different cell configurations. Electrochim Acta 50:2499–2506
Kiilerich-Pedersen K, Poulsen CR, Daprà J, Christiansen NO, Rozlosnik N (2011) Polymer based biosensors for pathogen diagnostics. In: Somerset V (ed) Environmental biosensors. INTECH, Rijeka, pp 193–212
Kimoto M, Yamashige R, Matsunaga KI, Yokoyama S, Hirao I (2013) Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol 31:453–457
Kronina VV, Wirth HJ, Hearn MT (1999) Characterisation by immobilised metal ion affinity chromatographic procedures of the binding behaviour of several synthetic peptides designed to have high affinity for Cu (II) ions. J Chromatogr A 852:261–272
Kwon OS, Park SJ, Hong JY, Han AR, Lee JS, Lee JS, JH O, Jang J (2012) Flexible FET-type VEGF aptasensor based on nitrogen-doped graphene converted from conducting polymer. ACS Nano 6:1486–1493
Lee J, Kim W, Cho S, Jun J, Cho KH, Jang J (2016) Multidimensional hybrid conductive nanoplate-based aptasensor for platelet-derived growth factor detection. J Mater Chem B 4:4447–4454
Li Y, Schluesener HJ, Xu S (2010) Gold nanoparticle-based biosensors. Gold Bull 43:29–41
Li Y, Deng L, Deng C, Nie Z, Yang M, Si S (2012) Simple and sensitive aptasensor based on quantum dot-coated silica nanospheres and the gold screen-printed electrode. Talanta 99:637–642
Li C, Meng Y, Wang S, Qian M, Wang J, Lu W, Huang R (2015) Mesoporous carbon nanospheres featured fluorescent aptasensor for multiple diagnosis of cancer in vitro and in vivo. ACS Nano 9:12096–12103
Liao W, Randall BA, Alba NA, Cui XT (2008) Conducting polymer-based impedimetric aptamer biosensor for in situ detection. Anal Bioanal Chem 392:861–864
Liss M, Petersen B, Wolf H, Prohaska E (2002) An aptamer-based quartz crystal protein biosensor. Anal Chem 74:4488–4495
Liu J, Cao Z, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109:1948–1998
Liu B, Lu L, Hua E, Jiang S, Xie G (2012) Detection of the human prostate-specific antigen using an aptasensor with gold nanoparticles encapsulated by graphitized mesoporous carbon. Microchim Acta 178:163–170
Liu X, Ren J, Su L, Gao X, Tang Y, Ma T, Zhu L, Li J (2017) Novel hybrid probe based on double recognition of aptamer-molecularly imprinted polymer grafted on upconversion nanoparticles for enrofloxacin sensing. Biosens Bioelectron 87:203–208
Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM (2005) Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev 105:1103–1170
Luk BT, Zhang L (2014) Current advances in polymer-based nanotheranostics for cancer treatment and diagnosis. ACS Appl Mater Interfaces 6:21859–21873
Ma W, Yin H, Xu L, Xu Z, Kuang H, Wang L, Xu C (2013) Femtogram ultrasensitive aptasensor for the detection of OchratoxinA. Biosens Bioelectron 42:545–549
Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J (1993) Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci 90:3745–3749
Maeshima A, Miyagi A, Hirai T, Nakajima T (1997) Mucin-producing adenocarcinoma of the lung, with special reference to goblet cell type adenocarcinoma: Immunohistochemical observation and Ki-ras gene mutation. Pathol Int 47:454–460
Mairal T, Özalp VC, Sánchez PL, Mir M, Katakis I, O’Sullivan CK (2008) Aptamers: molecular tools for analytical applications. Anal Bioanal Chem 390:989–1007
Menger M, Yarman A, Erdőssy J, Yildiz HB, Gyurcsányi RE, Scheller FW (2016) MIPs and aptamers for recognition of proteins in biomimetic sensing. Biosensors 6:35
Miodek A, Regan EM, Bhalla N, Hopkins NA, Goodchild SA, Estrela P (2015) Optimisation and characterisation of anti-fouling ternary SAM layers for impedance-based aptasensors. Sensors 15:25015–25032
Nawaz MAH, Rauf S, Catanante G, Nawaz MH, Nunes G, Marty JL, Hayat A (2016) One step assembly of thin films of carbon nanotubes on screen printed interface for electrochemical aptasensing of breast cancer biomarker. Sensors 16:1651
Ostatná V, Vaisocherová H, Homola J, Hianik T (2008) Effect of the immobilisation of DNA aptamers on the detection of thrombin by means of surface plasmon resonance. Anal Bioanal Chem 391:1861–1869
Pasquardini L, Pancheri L, Potrich C, Ferri A, Piemonte C, Lunelli L, Napione L, Comunanza V, Alvaro M, Vanzetti L (2015) SPAD aptasensor for the detection of circulating protein biomarkers. Biosens Bioelectron 68:500–507
Peng H, Zhang L, Soeller C, Travas-Sejdic J (2009) Conducting polymers for electrochemical DNA sensing. Biomaterials 30:2132–2148
Prabhakar N, Arora K, Singh SP, Singh H, Malhotra BD (2007) DNA entrapped polypyrrole–polyvinyl sulfonate film for application to electrochemical biosensor. Anal Biochem 366:71–79
Qureshi A, Gurbuz Y, Niazi JH (2015) Label-free capacitance based aptasensor platform for the detection of HER2/ErbB2 cancer biomarker in serum. Sensors Actuators B Chem 220:1145–1151
Radi AE (2011) Electrochemical aptamer-based biosensors: recent advances and perspectives. Int J Electrochem 2011:863196
Radi AE, Acero Sánchez JL, Baldrich E, O'Sullivan CK (2006) Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J Am Chem Soc 128:117–124
Rahi A, Sattarahmady N, Heli H (2016) Label-free electrochemical aptasensing of the human prostate-specific antigen using gold nanospears. Talanta 156:218–224
Ramanavičius A, Ramanavičienė A, Malinauskas A (2006) Electrochemical sensors based on conducting polymer—polypyrrole. Electrochim Acta 51:6025–6037
Robertson DL, Joyce GF (1990) Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344:467–468
Rodrıgez LMT, Billon M, Roget A, Bidan G (2002) Electrosynthesis of a biotinylated polypyrrole film and study of the avidin recognition by QCM. J Electroanal Chem 523:70–78
Sagiv J (1980) Organized monolayers by adsorption 1: formation and structure of oleophobic mixed monolayers on solid surfaces. J Am Chem Soc 102:92–98
Sardar R, Funston AM, Mulvaney P, Murray RW (2009) Gold nanoparticles: past, present, and future. Langmuir 25:13840–13851
Savory N, Abe K, Sode K, Ikebukuro K (2010) Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing. Biosens Bioelectron 26:1386–1391
Schreiber F (2000) Structure and growth of self-assembling monolayers. Prog Surf Sci 65:151–257
Sharma R, Agrawal VV, Sharma P, Varshney R, Sinha R, Malhotra B (2012) Aptamer based electrochemical sensor for detection of human lung adenocarcinoma A549 cells. J Phys Conf Ser 358:012001
Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC (2004) Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res 64:4783–4789
Singh S, Jha P, Singh V, Sinha K, Hussain S, Singh MK, Das P (2016) A quantum dot–MUC1 aptamer conjugate for targeted delivery of protoporphyrin IX and specific photokilling of cancer cells through ROS generation. Integr Biol 8:1040–1048
Song S, Wang L, Li J, Fan C, Zhao J (2008) Aptamer-based biosensors. Trends Anal Chem 27:108–117
Souada M, Piro B, Reisberg S, Anquetin G, Noël V, Pham M (2015) Label-free electrochemical detection of prostate-specific antigen based on nucleic acid aptamer. Biosens Bioelectron 68:49–54
Stora T, Hovius R, Dienes Z, Pachoud M, Vogel H (1997) Metal ion trace detection by a chelator-modified gold electrode: a comparison of surface to bulk affinity. Langmuir 13:5211–5214
Su S, Nutiu R, Filipe CD, Li Y, Pelton R (2007) Adsorption and covalent coupling of ATP-binding DNA aptamers onto cellulose. Langmuir 23:1300–1302
Su M, Ge L, Kong Q, Zheng X, Ge S, Li N, Yu J, Yan M (2015) Cyto-sensing in electrochemical lab-on-paper cyto-device for in-situ evaluation of multi-glycan expressions on cancer cells. Biosens Bioelectron 63:232–239
Sylvia P, Brehm SO, Hoch HJA (1975) DNA-binding proteins in human serum. Biochem Biophys Res Commun 63:24–31
Tahmasebi F, Noorbakhsh A (2016) Sensitive electrochemical prostate specific antigen aptasensor: effect of carboxylic acid functionalized carbon nanotube and glutaraldehyde linker. Electroanalysis 28:1134–1145
Tombelli S, Minunni M, Mascini M (2005) Analytical applications of aptamers. Biosens Bioelectron 20:2424–2434
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Tzouvadaki I, Jolly P, Lu X, Ingebrandt S, De Micheli G, Estrela P, Carrara S (2016) Label-free ultrasensitive memristive aptasensor. Nano Lett 16:4472–4476
Ueda E, Gout P, Morganti L (2003) Current and prospective applications of metal ion–protein binding. J Chromatogr A 988:1–23
Ulman A (1996) Formation and structure of self-assembled monolayers. Chem Rev 96:1533–1554
Vance SA, Sandros MG (2014) Zeptomole detection of C-reactive protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci Rep 4:5129
Wang X, Gu X, Yuan C, Chen S, Zhang P, Zhang T, Yao J, Chen F, Chen G (2004) Evaluation of biocompatibility of polypyrrole in vitro and in vivo. J Biomed Mater Res A 68:411–422
Wang L, Ma W, Chen W, Liu L, Ma W, Zhu Y, Xu L, Kuang H, Xu C (2011) An aptamer-based chromatographic strip assay for sensitive toxin semi-quantitative detection. Biosens Bioelectron 26:3059–3062
Wang K, Fan D, Liu Y, Wang E (2015) Highly sensitive and specific colorimetric detection of cancer cells via dual-aptamer target binding strategy. Biosens Bioelectron 73:1–6
Wang Z, Yu J, Gui R, Jin H, Xia Y (2016) Carbon nanomaterials-based electrochemical aptasensors. Biosens Bioelectron 79:136–149
Weidlich C, Mangold KM, Jüttner K (2005) EQCM study of the ion exchange behaviour of polypyrrole with different counterions in different electrolytes. Electrochim Acta 50:1547–1552
Wu J, Campuzano S, Halford C, Haake DA, Wang J (2010) Ternary surface monolayers for ultrasensitive (zeptomole) amperometric detection of nucleic acid hybridization without signal amplification. Anal Chem 82:8830–8837
Xiao D, Lu T, Zeng R, Bi Y (2016) Preparation and highlighted applications of magnetic microparticles and nanoparticles: a review on recent advances. Microchim Acta 183:2655–2675
Xie Q, Tan Y, Guo Q, Wang K, Yuan B, Wan J, Zhao X (2014) A fluorescent aptasensor for sensitive detection of human hepatocellular carcinoma SMMC-7721 cells based on graphene oxide. Anal Methods 6:6809–6814
Xu H, Gorgy K, Gondran C, Le Goff A, Spinelli N, Lopez C, Defrancq E, Cosnier S (2013) Label-free impedimetric thrombin sensor based on poly (pyrrole-nitrilotriacetic acid)-aptamer film. Biosens Bioelectron 41:90–95
Yan M, Sun G, Liu F, Lu J, Yu J, Song X (2013) An aptasensor for sensitive detection of human breast cancer cells by using porous GO/Au composites and porous PtFe alloy as effective sensing platform and signal amplification labels. Anal Chim Acta 798:33–39
Yang DK, Chen LC, Lee MY, Hsu CH, Chen CS (2014) Selection of aptamers for fluorescent detection of alpha-methylacyl-CoA racemase by single-bead SELEX. Biosens Bioelectron 62:106–112
Ye X, Shi H, He X, Wang K, He D, Yan LA, Xu F, Lei Y, Tang J, Yu Y (2015) Iodide-responsive Cu–Au nanoparticle-based colorimetric platform for ultrasensitive detection of target cancer cells. Anal Chem 87:7141–7147
Yeh FY, Liu TY, Tseng IH, Yang CW, LC L, Lin CS (2014) Gold nanoparticles conjugates-amplified aptamer immunosensing screen-printed carbon electrode strips for thrombin detection. Biosens Bioelectron 61:336–343
Zhang Z, Yang W, Wang J, Yang C, Yang F, Yang X (2009) A sensitive impedimetric thrombin aptasensor based on polyamidoamine dendrimer. Talanta 78:1240–1245
Zhang K, Xie M, Zhou B, Hua Y, Yan Z, Liu H, Guo LN, Wu B, Huang B (2013) A new strategy based on aptasensor to time-resolved fluorescence assay for adenosine deaminase activity. Biosens Bioelectron 41:123–128
Zhang H, Shuang S, Sun L, Chen A, Qin Y, Dong C (2014) Label-free aptasensor for thrombin using a glassy carbon electrode modified with a graphene-porphyrin composite. Microchim Acta 181:189–196
Zhao S, Ma W, Xu L, Wu X, Kuang H, Wang L, Xu C (2015) Ultrasensitive SERS detection of VEGF based on a self-assembled Ag ornamented–AU pyramid superstructure. Biosens Bioelectron 68:593–597
Zhu Y, Chandra P, Shim YB (2012) Ultrasensitive and selective electrochemical diagnosis of breast cancer based on a hydrazine–Au nanoparticle–aptamer bioconjugate. Anal Chem 85:1058–1064
Acknowledgements
PJ was funded by the European Commission FP7 Programme through the Marie Curie Initial Training Network PROSENSE (Grant No. 317420, 2012–2016). MRB was funded by FAPESP (process number 2013/26133-7). SU was funded by the UK EPSRC Centre for Doctoral Training in Sustainable Chemical Technologies. SKA was funded by the European Commission’s Horizon 2020 Programme through a Marie Skłodowska-Curie Individual Fellowship (Grant No. 655176). MM and PE acknowledge support from FAPESP and the University of Bath through the SPRINT program.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Jolly, P., Batistuti, M.R., Ustuner, S., Mulato, M., Arya, S.K., Estrela, P. (2017). Nucleic Acid-Based Aptasensors for Cancer Diagnostics: An Insight into Immobilisation Strategies. In: Chandra, P., Tan, Y., Singh, S. (eds) Next Generation Point-of-care Biomedical Sensors Technologies for Cancer Diagnosis. Springer, Singapore. https://doi.org/10.1007/978-981-10-4726-8_9
Download citation
DOI: https://doi.org/10.1007/978-981-10-4726-8_9
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4725-1
Online ISBN: 978-981-10-4726-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)