Abstract
Genetic programmes that assist decision-making of a stem cell whether to self-renew or to differentiate into a committed cell type have been studied extensively over the past few decades. In the process of exploiting pluripotent nature of a stem cell, researchers across the globe channelized their efforts to derive target cell types from various sources of stem cells. The scientific know-how about cellular fate determining transcription factors (TFs) and the huge amount of information regarding the regulation of stem cell differentiation led researchers to come up with a highly attractive concept of cellular reprogramming. About three decades ago, a fascinating study revealed direct conversion of fibroblasts to muscle cells by overexpressing merely one transcription factor ‘MyoD’. Towards deciphering the underpinnings of cellular differentiation and self-renewal programmes, an offshoot of thought has emerged that advocated the interconversion within the somatic cell state. In present days the task of direct conversion, more popularly known as transdifferentiation, has been an excellent alternative approach to generate the cells of interest for clinical purpose.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Embryonic development is a highly orchestrated hierarchical process, where pluripotent stem cells are committed to germ layer-specific lineages. However, the potential to produce different cell types by stem cells is restricted progressively as development proceeds. Neural stem cells or mesenchymal stem cells, which fall much lower to the pluripotent cells in the cellular hierarchy, have more restricted choice of cell fate. As compared to the concept of mammalian development, transdifferentiation has gained much importance since it does not restrict the fate choice of differentiated cells and, in turn, provides an alternative method to generate desired cellular phenotype. Defined as a stable transition of a differentiated cell type to another, transdifferentiation is considered as a subtype of metaplasia [1]. If one has to produce a differentiated cell from another, it would have to first dedifferentiate back to pluripotent state followed by redifferentiation to the desired phenotype. However, transdifferentiation presents a path that is lineage instructive, bypassing the inducing pluripotent stem cell (iPSC) road.
The concept of reprogramming ruled out the myth that cellular differentiation and identity establishment are unidirectional, whereas transdifferentiation convincingly demonstrated that a stem cell prototype is not necessary for the interconnection between two cellular fates. The never-ending curiosity in the scientific community led to the emergence of a thought process which suggested the possibility of directly converting a terminally differentiated cell to another of an entirely distinct lineage. In spite of being a huge challenge, this task has been successfully accomplished by Wernig and co-workers who provided a proof-of-concept study by deriving functional neurons (ectodermal lineage) from fibroblasts (mesodermal lineage) [2]. Here, we attempt to discuss the different approaches adopted by researchers to accomplish direct cellular fate change using transcription factors.
2 Transdifferentiation
Primarily defined as the interconversion of one differentiated cell to another, transdifferentiation was first reported in 1895 by Wolff. He observed that during the regeneration of crystalline lens, differentiated pigment epithelial cells undergo transdifferentiation to form crystalline lens in newts. In laboratory, transdifferentiation became a possibility when Peter Jones in 1973, while investigating the effect of certain drugs on fibroblast, observed that azacytidine could produce myocytes, adipocytes and chondrocytes [3]. Later in 1987, Davis et al. [4] proved that ‘MyoD’ holds the key in transdifferentiating fibroblast into muscle cells. Though there exist numerous reports on transdifferentiation, the underlying mechanism still stays unclear.
This unconventional differentiation of a cell type to another could be of two variants. One accompanied by cell division and another without, where the former is known as indirect transdifferentiation and the latter being direct. Indirect transdifferentiation might involve a partial dedifferentiation into a common progenitor stage, from where it would further proliferate, generating a completely different phenotypic cells. In contrast, the direct differentiation, devoid of cell division, might have to go through intermediate stages, expressing molecular signature of both cells [5]. The exact mechanism of transdifferentiation is yet to be elucidated. The cells may undergo phenotypic conversion through three possible mechanisms. First, the cells undergoing transdifferentiation will dedifferentiate into an intermediate stage with higher differentiation potential and redifferentiate to a new cell type. The cells of the intermediate stage will display a higher but restricted potency rather than iPSC [6]. This mechanism may or may not be coupled with cell division. For instance, during crystalline lens regeneration of newts, pigment epithelial cells would dedifferentiate to an intermediate state via inactive p53 and retinoblastoma (RB) genes and further differentiate to crystalline lens cells [6]. The second mechanism of transdifferentiation involves the mature cells that will convert directly into another differentiated cell type without dedifferentiating into an intermediate state [7]. This mode of transdifferentiation is mostly manipulated in the laboratory and might have stages where the cells express molecular traits of both the phenotypes. For example, the fate change of cardiac fibroblast into cardiomyocytes is categorized into direct transdifferentiation [7]. The third form is widely observed when a stem cell is transformed into another type, such as the conversion of bone marrow (BM) stem cells into neurons [8] or osteogenic or chondrogenic lineages [9]. In conclusion, transdifferentiation induces the fate conversion of terminally differentiated cells of one lineage to the cells of another lineage.
Different strategies of transdifferentiation that followed include TFs, miRNA or small molecules to achieve clinically relevant cell types as shown in Table 8.1. In the following sections we will discuss on derivation of clinically relevant cell types by transdifferentiation, also use of miRNA and small molecules for direct fate change of cells have been mentioned.
3 Transdifferentiation: A Boon to Everlasting Demand for Pancreatic β-Cells?
Diabetes is one of the most prevalent disorders, wherein β-cells are damaged leading to compromised insulin production. Hence, there is always a huge demand for cell sources from which β-cells can be regenerated. Though pluripotent stem cells are good sources for the derivation of β-cell, they come along with the disadvantage of tumour formation and ethical concerns. Hence a transdifferentiation approach to regenerate β-cells would be an ideal alternative. The mechanisms behind pancreatic development have been studied well from the experiments consisting of generation of mice deficient for a number of pancreatic TFs. Stage-specific expressions of these TFs are the main driving force behind achieving different stages of cells during pancreatic development [10]. Identifying these TFs has facilitated the inter-derm and intra-derm conversion of adult cells towards insulin-secreting β-cells (Table 8.1).
Owing to a common germ layer origin, developmentally related cells can be easily transdifferentiated into β-cells without major epigenomic changes. As the liver and pancreas share a common progenitor during development, hepatic lineage cells have become the first choice of cells to generate β-cells. Intravenous injection of adenoviruses encoding Pdx1 and/or NeuroD, Ngn3, musculoaponeurotic fibrosarcoma oncogene homolog A (MafA) and paired box4 (Pax4) genes successfully transdifferentiated hepatocytes into β-like cells and reversed STZ-induced diabetes in mice [11]. During the course of converting hepatic cells to insulin-producing cells using Pdx1, Pax4 and MafA, Berneman-Zeitouni et al. [12] demonstrated that sequential supplementation of these factors, one day apart from each other, in a hierarchical manner enhanced transdifferentiation potential of cells compared to adding all the factors together. Although, transdifferentiation is not a developmentally organized phenomenon, transcription factor-mediated transdifferentiation from liver to pancreas showed that the event takes place in a progressive and hierarchical manner. Similar to hepatocytes, hepatic oval cells were also redirected to the β-cell lineage by an appropriate combination of high extracellular glucose, specific extracellular matrix proteins (laminin and fibronectin), cytokines (activin A) and ectopic expression of transcription factors (Pdx1, Ngn3, MafA) [13]. Pdx1 is the key transcription factor which marks the appearance of pancreatic anlage during development. Ectopic expression of Pdx1 induced both endocrine and exocrine pancreatic lineages within the liver; however, due to unrestricted expression of Pdx1, complication like fulminant hepatitis was reported in some in vivo studies [14]. Enhanced expression of Ngn3, the gene responsible for endocrine fate determination, converted hepatic progenitors to neoislets which could reverse hyperglycemia in a diabetic mice model [14]. Forced expression of critical transcription factors involved in pancreatic development such as Pdx1, NeuroD or Pdx1/VP16 in cultured intrahepatic biliary epithelial cells induced β-cell phenotype expressing insulin, Glut2 and prohormone convertase 1 and prohormone convertase 2 [15]. Similarly, overexpression of Pdx1, Ngn3 and MafA in primary mouse gallbladder epithelial cells (GBCs) converted them to pancreatic lineage and showed concomitantly reduced expression of GBC-specific genes (Sox17 and Hes1) [16].
Generating β-cells by direct conversion is not restricted only to hepatic lineage cells. They can be even obtained from other endodermal lineages like gastrointestinal cells. As enteroendocrine cells express some of the pancreatic genes like Ngn3 and hormone incretins, they could be an ideal candidate for β-cell reprogramming. Transgenic mice expressing human insulin under the control of the glucose-dependent insulinotropic polypeptide (GIP) promoter produced human insulin specifically in gut K cells, and notably ablation of Foxo1 in enteroendocrine progenitors directly converted them to insulin-expressing cells [17]. Similar to TF cocktail used in deriving β-cells from other endoderm lineages, transient expression of Pdx1, Ngn3 and MafA in the intestine promoted the induction of β-cell phenotype in the intestinal crypt. These derived insulin-secreting cells were found to be glucose responsive and ameliorated STZ-induced hyperglycemia in mice [18].
The other amenable route of transdifferentiation is the interconversion of other pancreatic cells to β-cells. Lineage-tracing studies had shown that the newly formed β-cells during pancreatic ductal ligation can be derived from ductal cells. In-depth understanding of the mechanism showed inactivation of Fbw7 ubiquitin ligase, a tumour suppressor protein, in adult pancreatic ductal cells, stabilized Ngn3 and initiated conversion of ductal cells into predominantly islet β-cells [19]. Transfection of endocrine-specifying TFs Ngn3 or NeuroD in human primary duct cells initiated β-cell programming [14]. Similarly, acinar cell compartment, the largest component of pancreas, is induced to insulin-expressing cells by delivering adenovirus expressing Pdx1, Ngn3 and MafA. In vivo inhibition of the Notch signalling pathway or suppression of Ptf1a (a master regulator of acinar cell fate specification) induced β-cell neogenesis from acinar cells. Among the endocrine cells, forced expression of Pax4 in α-cells led to their conversion into insulin-secreting cells which normalized the STZ-induced hyperglycemia [14]. Using transdifferentiation approach, Herrera’s group showed that α-cells do not undergo senescence and can be converted to insulin-producing cells from puberty through to adulthood, and prior to puberty β-cell reconstitution occurs through reprogramming of somatostatin-producing δ-cells [20].
Transdifferentiation to pancreatic β-cells can also be obtained from cells like skin fibroblasts that are distantly related to the pancreas. Katz et al. induced transdifferentiation of adult human dermal fibroblasts into insulin- and glucagon-expressing cells by using romidepsin histone deacetylase inhibitor and 5-azacytidine (a DNA methyltransferase inhibitor), and the generated β-cells expressed islet factors like NeuroD, Isl1, glucose transporter 1 (Glut1) and Glut2. The above study indicated that fibroblast can be converted to β-cell-like cells bypassing the iPSC route. Later Li et al. [14] derived functional β-cells by a transient exposure of fibroblast to Yamanaka factors (Oct4, Sox2, Klf4 and c-Myc) and then followed by addition of small-molecule epigenetic modulators. Likewise, keratinocytes were also converted into β-cell-like cells by adenoviral introduction of Pdx1, Ngn3 and NeuroD. Even MSCs from dental pulp stem cells were converted to pancreatic lineage by ectopic expression of Pdx1 and Ngn3. All these studies suggest that a landscape of multiple intra-islet and inter-islet cells interconverted to β-cells offer new perspectives for cellular therapy to cure diabetes.
4 Hepatocyte Generation: A Way to Regenerate Liver
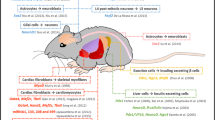
Being the largest organ in the human body and executing multiple metabolic functions, liver regeneration has gained a lot of attention. Due to the shortage of donors for liver transplantation, hepatocytes derived from either differentiation of pluripotent stem cells (PSCs) or transdifferentiation approach have been envisaged as an alternate route for therapy (Fig. 8.1). Several reports are available describing different ways of transdifferentiation to arrive at hepatocyte stage. Zhu et al. [21] generated an intermediate induced multipotent progenitor cells (iMPCs) from human fibroblast, rather than iPSCs. These iMPCs were subsequently differentiated to mature hepatocytes using small molecules. Interestingly, these hepatocytes possess high proliferative potential compared to hepatocytes derived from iPSCs. In vivo transplantation of iMPC-derived hepatocytes into an immune-deficient mouse of human liver failure showed functional improvement without forming tumours [21]. After screening for twelve candidate factors, Sekiya et al. [22] identified three specific combinations of two transcription factors expressed in retroviral vectors, comprising Hnf4α along with either Foxa1, Foxa2 or Foxa3, that could introduce hepatic programme in a non-hepatic cell-like fibroblasts. After transplantation, these transdifferentiated hepatocyte-like cells efficiently rescued the damage hepatic tissues [22]. Later, to overcome the reliance on harmful retroviral vector in transdifferentiation approach, hepatic induction was achieved with mRNAs encoding HNF1α along with any two of the factors FOXA1, FOXA3, and HNF4α in the presence of hepatic growth medium. Induced hepatocytes obtained by overexpressing cocktail of hepatic fate conversion factors HNF1α, HNF4α and HNF-6 along with the maturation factors ATF5, PROX and C/EBPα were found to express phase I and phase II drug-metabolizing enzymes and functional cytochrome p450 members similar to the primary human hepatocytes [23]. In an independent study, inactivation of p19ARF along with overexpression of Gata4, Hnf1α and Foxa3 enabled mouse fibroblasts to become hepatocyte-like cells. These cells upon transplantation in fumarylacetoacetate hydrolase-deficient (Fah−/−) mice rescued about half of the recipients from the lethal effect of the disease [24].
Modes of transdifferentiation. Illustration shows direct transdifferentiation devoid of cell division and indirect transdifferentiation accompanied by dedifferentiation and cell division. Immunostaining images represent human pluripotent stem cell-derived neurons, hepatocytes and cardiomyocytes expressing MAP2, albumin and desmin, respectively
Apart from fibroblast as a starting cell, spermatogonial stem cells (SSCs), subjected to transdifferentiation, led to the expression of hepatic stem cell markers, which later differentiated into mature hepatocytes with distinct morphological, phenotypic and functional characteristics. Mechanistic studies further shed light on the fact that activation of ERK1/2 and Smad2/3 signalling pathways and the inactivation of cyclin A, cyclin B and cyclin E are necessary for such transdifferentiation [25]. Studies also demonstrated the possibility of mesenchymal stem cells (MSCs) to transdifferentiate into hepatocyte-like cells. Though clinically not relevant, transdifferentiation of bone marrow cells to hepatic precursor cell type was obtained by generating bone marrow cells and ES cell hybrids and differentiating them with external cues. Later it was shown that ectopic factors like hepatocyte growth factor (HGF) and oncostatin M (OSM) can induce transdifferentiation of human bone marrow and umbilical cord blood stem cells into functional hepatocyte-like cells in vitro. Similarly, placental derived MSCs were transdifferentiated into hepatic lineage by hepatogenic medium containing HGF, FGF-4 and DMSO. Overexpression of HNF4α improved the hepatic differentiation of human bone marrow MSCs and considered to be an easy way of generating clinically useful hepatocytes. Recent experiments suggested that epigenetic changes involving histone H3 modifications at lysine 9, 14 and 27 are essential to achieve hepatic cells [26]. Transdifferentiation approach for generating hepatocyte-like cells provides an invaluable source of mature hepatocytes for treating liver-related diseases and drug screening.
5 Fate Switching: Can the Neural Circuitry Be Turned On?
Due to ethical issues in obtaining human samples and since certain subpopulations of neurons are species specific, rodent models may not rightly recapitulate human disease progression. For example, amyloid lateral sclerosis (ALS) primarily affects certain motor neurons but spares other subtypes, while in Parkinson’s disease dopaminergic neurons are severely affected. Because the above-mentioned disorders typically appear in adult humans at ages far greater than the short life span in the case of mouse, cellular phenotypes obtained in an animal model may differ significantly from those in case of human diseases. Fortunately, recent advances in cellular reprogramming offer a set of powerful methods to approach the problems of human neurobiology and neurological diseases at the cellular and molecular levels.
More than two decades have been completed after the inception of an idea that nonneural ectodermal cells can be used to reach neural lineage. However, a successful transdifferentiation of neuronal cells took 10 years to become a reality. Functional neurons were derived from cells of astrocytic origin by overexpressing essential neural determinant genes like PAX6, MASH1, NGN2 and DLX2 [27]. Regulation of cell fate decision and modification in epigenetic landscape has been revealed as the underlying phenomenon leading to this intra-germ layer transdifferentiation. Later, it opens up the possibility to derive neurons from other germ layer cells leading to trans-germ layer transdifferentiation.
6 Trans-Germ Layer Fate Conversion: Breaking the Boundaries to Generate Neurons
After achieving remarkable success in intra-germ layer transdifferentiation, the scientific community moved on to break the conventional barriers and push the cells of one germ layer towards the fate of another. The phenomenon essentially comprises of two distinct events: one being the loss of inherent molecular memory and the other acquiring the signatures of an altogether different developmental mark. A milestone in the field of transdifferentiation was created when mouse fibroblasts were converted to neuronal cells by Vierbuchen et al. in the year 2010 [28]. Later, Wapinski et al. [2] mechanistically analysed the conversion of murine fibroblast into neural progenitors by overexpression of Brn2, Ascl1 and Myt1l (BAM factor). It was figured that Ascl1, a basic helix loop helix transcription factor, follows a similar trend of binding in both murine fibroblast and neural progenitors. This study showed that during the initial phases of transdifferentiation, exogenous Ascl1 binds to its neural target genes in the fibroblast genome facilitating the recruitment of other auxiliary factors to their target genes in the late phases of conversion. Apparently, during the process of transdifferentiation, Ascl1 act as an ‘on-target pioneer factor’ binding to its specific target genome regardless of the fact that they are epigenetically silenced [29]. Hence Wapinski et al. [2] hierarchically categorized the BAM factors involved in the neural transdifferentiation. It took only a couple of years to validate similar observation demonstrating the generation of neurons from many different lineages [2]. With time, robust protocols have been formulated that resulted in direct conversion of fibroblasts to stable and functionally active neurons using a set of transcription factors, namely, BRN2, MYT1L, ASCL1, OLIG2 and ZIC1. Nonetheless, attempts for direct conversion of matured neuron from other germ layer cells encountered problems with respect to large-scale expansion [27]. Hence, researchers diverted their efforts generating neural precursor cells by forced expression of lineage-restricted factors. Our group while trying to study the role of transcription factor ZIC3 in reprogramming of human fibroblasts in combination with OCT4, SOX2 and KLF4 colonies resembling neural progenitor-like identity was formed for a very short duration rather than that found in case of iPSC. This study demonstrated that lineage-restricted TF is sufficient to direct the conversion of nonneural cells to neural progenitor state without dedifferentiating them in to ESC state [30]. Attempts were made to derive neural stem-like cells that can profusely proliferate and give rise to both neuronal and glial (oligodendrocytes and astrocytes) cells by overexpressing combinations of Brn2, Sox2 and FoxG1 [31] or Brn4, Sox2, Myc, Klf4 and Tcf3 (Table 8.1) [32]. The above studies emphatically proved that multi-lineage differentiation capacity can be achieved by forcefully expression of TFs in fibroblasts.
As different neurodegenerative disorders demand specific human neuronal subtypes, there is a rapid progress in direct reprogramming to enrich these cells. Pfisterer et al. introduced dopaminergic neural phenotype in fibroblast by overexpressing the basic BAM factors with two more genes LMX1A and FOXA2 that are involved in midbrain and dopamine neuron specification. Later, several studies showed the derivation of multiple neuronal subtypes including motor neurons and peripheral sensory neurons [27]. Schwann cells play an important role in the development and homeostasis of peripheral nervous system and can help in understanding the functions or pathophysiology in vitro and to develop cellular interaction models to study disease mechanisms in coculture with neurons. Using multi-kinase inhibitor, Thoma et al. [33] efficiently converted human fibroblasts into transient neural precursors that were subsequently differentiated into Schwann cells. Altogether, this study shows the proof of principle that conversion of fibroblasts towards a neural cell fate can be achieved solely with small-molecule treatment. Human fibroblasts were also converted to striatal medium spiny neurons (Msns), using [miR-9/9-124] and transcription factors, namely, CTIP2, DLX1, DLX2 and MYT1L [34]. Notch intracellular domain (NICD) overexpression along with treatment with bFGF, forskolin and CNTF was also shown to induce neuronal character in both rat and human MSCs. To date, transdifferentiated neurons have been shown to be derived from fibroblasts, hepatocytes, ESCs/iPSCs and astrocytes of both mouse and human origin [27]. These studies culminate in derivation of either NPCs or neurons directly from the nonneural cells with an advantage of no tumour formation, which will pave the way for future cell therapy in case of numerous neurodegenerative disorders.
7 Haematopoiesis: A New Platform for Transdifferentiation
Haematopoietic stem cells (HSCs) own the property of self-renewal and differentiation into all the haematopoietic lineages. HSCs and their derivatives erythrocytes, platelets and granulocytes can be used for malignant and non-malignant haematological disorders. Due to the difficulties posed by lack of adequate donors and proper matching, the above treatments are being limited. Differentiation of pluripotent stem cells to blood cells and other HSCs is challenged by non-availability of robust protocols. Therefore, transdifferentiation would be an attractive strategy to generate patient-specific transplantable cells. Knockout studies have shown that SCL, RUNX1, ERG and GATA2 are known to be involved in multiple stages of haematopoietic specification, maturation and differentiation [35]. Initial study showed that among various molecular players, two key transcription factors, GATA1 and SPI1, contribute to fate change of haematopoietic progenitor cells to either erythrocytic or myeloid lineage, respectively. This study was followed by many others, which deciphered the interconversion of B and T lymphocytes. For instance, overexpression of just a single leucine zipper transcription factor CEBP-α led to the conversion of B and T lymphocyte progenitors to macrophages, which were functional [1]. However, it is not necessary that a transcription factor will bring about unidirectional transdifferentiation. Depending on the sequence of action of transcription factors, synergism between them may change the cell fate. Also, the starting cells, amenable to fate change, play a role in the success of the process.
By overexpressing HSC-specific transcription factors, Riddell et al. [36] were able to impart HSC characteristics in committed murine blood cells and endothelial cells. A clinically relevant bottleneck to reprogramme blood cells is that the source cells must be from a healthy donor without any diseases in haematopoietic system. Therefore, a developmentally distinct cell-like fibroblast would be an ideal alternative for transdifferentiation into blood cells. Previous studies had shown during reprogramming process OCT4 and human pan-haematopoietic marker CD45 to be predominantly expressed in the human dermal fibroblasts. OCT2 (also called POU2F2) and OCT1 (also called POU2F1) play a role in the development of lymphoid lineage; they are also known to bind similar DNA target motifs to OCT4. Taking into account the above observation, a recent study reported the acquisition of haematopoietic fate in fibroblasts upon overexpression of single factor OCT4. Though these transdifferentiated cells exhibited myeloid and erythroid differentiation potential, lymphoid progenitors could not be established in this system, and long-term engraftment seemed difficult. In addition, use of a pluripotency factor like OCT4 has a high probability of giving rise to partially reprogrammed cells that could in turn lead to tumorigenicity [37]. Later, the expressions of GATA2, GFI1b, ETV6 and c-FOS were shown to induce haemogenic programme in mouse fibroblasts. In addition to this, specific transcription factors like Scl and Lmo2 facilitated the transdifferentiation of mouse embryonic fibroblasts to blood cells, and loss of p53 or p16/p19 enhanced the efficiency of this process. Lack of these factors also led to the development of TER119+ erythroid cells in reprogramming of haematopoietic progenitor cells [38]. The above transdifferentiation approaches that target generation of haematopoietic lineages offer a great potential for the treatment of haematologic and immunologic diseases.
8 Challenges in Cardiac Regeneration: Possible Circumvention by Transdifferentiation?
The restricted therapeutic options for congestive heart failure (CHF) have led scientists to look upon alternative regenerative treatment strategies including stem cell transplantation. Among different approaches, lineage conversion of scar-associated fibroblasts into functional myocardium has been considered the most efficient way. As the adult heart consists majorly of fibroblasts, the cardiac fibroblasts are supposed to be an ideal source of cells for reprogramming. A cocktail of three lineage-specific TFs Gata4, Mef2c and Tbx5, popularly known as GMT, have been reported to induce fibroblasts to cardiomyocytes. However, these minimal TFs were inefficient to produce functional cardiomyocytes, lacking the expression of α-myosin heavy chain (α-MHC). Further to obtain matured functional cardiomyocytes and improve reprogramming efficiency, several additional TFs, such as Mesp1, Hand1, Hand2, Nkx2.5, myocardin (Myocd) and Smarcd3, were used. Not only cardiomyocytes, Nam et al. [39] sought to reprogramme murine fibroblasts into cells of cardiac pacemaker identity by a combination of Gata6, Tbx3, Tbx5 and Rxra. Interestingly, combination of GMT TFs along with miRNAs like miR-133 formed cardiomyocyte phenotype from mouse embryonic fibroblasts as early as day 10 of induction [7]. All these results univocally advocate this alternative approach of generating the functional cardiomyocytes.
During iPSC generation, using Oct4 helps in opening of chromatin from a closed conformation, and when this process is combined with lineage-specific soluble signals, there is a possibility of driving the somatic cells into lineage-specific cells but without entering into pluripotent state. Efe et al. induced myocardium phenotype in MEFs by retroviral overexpression of Oct4, Sox2 and Klf4 (OSK), combined with cytokines BMP4 and small-molecule inhibitor of JAK pathway. The transduced cardiomyocyte expressed Flk1, Nkx2.5 and Gata4 progenitor marker, which later expressed matured marker TropT, α-MHC and α-actinin.
Interestingly, direct reprogramming of non-myocytes to functional cardiomyocytes was also demonstrated successfully. Injection of retroviruses coding for GMT alone or in combination with HAND2 into infarcted myocardium showed newly generated cardiomyocytes derived from resident cardiac fibroblasts. In vivo transdifferentiated cardiomyocytes reduced fibrosis, and the ejection fraction was significantly improved. Recently, Zhouh et al. [40] in an attempt to identify the involvement of kinases in direct reprogramming of fibroblasts in the presence of Gata4, Hand2, Mef2c and Tbx5 (GHMT)], out of 192 protein kinases, Akt/protein kinase B was found to enhance the generation of cardiomyocytes. These findings that decipher the importance of different TFs in direct reprogramming represent an important step towards further application of this technique in cardiac-related disorders.
9 Faulty Transdifferentiation
An intriguing possible explanation for faulty transdifferentiation could be cell fusion and DNA transfer. Ying et al. in 2002, when cocultured neural stem cells (NSC) and embryonic stem cells (ESC), isolated pluripotent stem cell population in which NSC genome has been modified epigenetically. In addition, carrying transgenic marker and chromosome of the pluripotent ESC clearly explains that the NSCs have neither transdifferentiated nor dedifferentiated but simply fused to form a hybrid [41]. Similarly, it was also shown that murine bone marrow stem cells, when cultured in the presence of IL3 with ESCs, spontaneously fuse to generate hybrid cells exhibiting the properties of pluripotent cells. Dolado et al. in 2003 elegantly demonstrated using Cre/lox system that BM-derived cells fused with neural progenitors in vitro. Transplantation of BM-derived cells fuses with hepatocytes in the liver, Purkinje neurons in the brain and cardiac muscles of the heart resulting in the formation of multinucleated cells. Similar incidents would require a through genetic screening to rule out faulty transdifferentiation [42].
During embryonic development, neural crest differentiation witnesses a transformation of ectoderm to mesoderm. Further, a well-established fact is that neural crest cells can differentiate into bone, cartilage, muscle, melanocytes, fibroblasts and even cells comprising the peripheral nervous system which exhibit the possibility of in vivo transdifferentiation. In addition, neural stem cells of embryonic and adult origin can generate neural crest, cartilage and muscles cells [43]. Developmental biology of the kidney, for instance, shows mesodermal to ectodermal transformation generating kidney tubules. It is indeed possible that the mesodermal precursors transform not only mesenchymal and haematopoietic but even ectodermal lineages. However, there is a grey area regarding the classification of this kind of differentiation during early development be it fits into the category of transdifferentiation.
The presence of contaminating population of undesired cells could be one of the major limitations to prove transdifferentiation. One of the possible explanations for the apparent neural transdifferentiation could be possibly due to contaminating neural crest and ventrally emigrating neural tube (VENT) cells. The presence of quiescent crest cells in the peripheral nerves and organs can display faulty transdifferentiation producing neural and nonneural cells. In addition VENT cells have also been shown to contribute to the development of bone, cartilage, cardiac muscle and hepatocytes [44]. Another very likely contamination could be contributed by the circulating haematopoietic stem cells. Perhaps a more stringent selection of purified population of cells could exempt the faulty transdifferentiation.
10 MicroRNAs and Transdifferentiation
Cocktails of transcription factors are quite popular in lineage conversion of cells. However, the non-coding microRNA has shown great promises in recent years. Short segments of non-coding RNA ranging around 20–25 nucleotides, the miRNA inhibits the translation of mRNA by binding to their complementary parts. miRNA binds to either the 3′UTR regions or occasionally the coding region of the mRNA. Traditionally they are more concerned as stabilizers of cell fate or factors that fine-tune differentiation across development. The true potential of miRNA was only unveiled when it was found that they could act as a pioneer factor leading to dedifferentiation or transdifferentiation. Experiments demonstrated by Lim et al. in 2005 opened the doors for the possibility that miRNAs could also act as master regulators of transdifferentiation. Accordingly, miRNA124 was found to induce a partial neuronal transition in HeLa cell line. However, they failed to produce neurons that functionally or morphologically resemble to mature neurons [45].
Enforced expression of miRNA9 and miRNA124 can generate neurons expressing MAP2 from human fibroblast. However, an additional induced expression of transcription factors is required for the functional maturation. In a separate study, miRNA124 enhanced transdifferentiation of fibroblasts into neurons using transcription factors such as BRN2 and MYT1 [46]. Besides neuronal transdifferentiation, cardiac-enriched miRNA, miR1, miR133, miR208 and miR499 alone could drive transdifferentiation towards cardiac lineage [7]. Surprisingly, the inhibition of PTB, a miRNA regulator, alone can produce functionally matured neurons, emphasizing the role of miRNA in transdifferentiation [47].
Owing to the potential of miRNAs in transdifferentiation, it has been interesting to look into the intricate mechanism by which they function. It is quite an irony since miRNA inhibits the mRNA expression, and yet it finds a mechanism to activate a set of genes involved in transdifferentiation. For instance, in the transdifferentiation of fibroblasts into neurons, the miRNA cocktail may downregulate the expression of nonneuronal transcripts enabling a preferential differentiation towards neuronal lineage. When coupled with a transcription factor that drives neuronal differentiation would yield a more efficient transdifferentiation. Possibly the miRNA could be involved in a signalling cascade that directly activates neural genes or suppresses the expression of a gene that is responsible for inhibiting the production of a particular neural subtype.
In the context where a factor is expressed in both the initial and the terminal phases of transdifferentiation, it is difficult to appreciate how elegantly miRNA is involved in the transition of cell fates. For instance, BAF53a (Brg-/Brm-associated factor) is expressed in both neuronal and the fibroblast cells. However, it has been shown previously by the Crabtree’s lab that miR124 and miR9 target BAF53a as neural progenitors differentiate into neurons [46]. Suppression of BAF53a leads to the activation of BAF53b, an activator of neuronal lineage. Though a common set of genes are expressed in the neurons and fibroblasts, it is the post-transcriptional modification by miRNA that makes the difference.
Though the use of miRNA is not the ideal way for transdifferentiation of cells, it enlightens some insight underlying molecular and cellular mechanisms involved in the transition of the cell fate. However, it has its own way to work when compared to transcription factors. miRNAs being smaller in size have the advantage of getting easily transfected into the cells compared to that of transcription factors which are bulky. miRNAs can stably exist in the cytoplasm, while transcription factor or the DNA if transferred could alter the genome. Though not very efficient, it can naturally transfer from one cell to another, and hence it is only adequate to understand the mechanism by which it aided the transition from ESCs to neural crest cells, thereby facilitating drug screening studies [48].
11 Use of Small Molecules: Preventing Genome Intrusion
So far, we have illustrated various study outcomes that reveal the importance of core transcription factors or miRNA for transdifferentiation. However, the discussion would be incomplete without shedding light on a few highly acclaimed small molecules which have contributed to different cell fate conversions. Use of small molecules to achieve cell fate conversion is one of the integration-free approaches which gains an advantage over classical method of transcription factor supplementation because the latter is associated with an unavoidable risk of foreign DNA integration into the host system. In fact, 5-azacytidine is one of the first small molecules that showed to induce muscle, adipocyte and chondroblast conversion of mesenchymal cells. After a long journey of transcription factor-based cellular reprogramming for transdifferentiation, very recently the scientific focus has shifted back to the use of small molecules to achieve the same goals.
It was only in 2013 that the concept of chemical reprogramming was unravelled by Deng and co-workers [49], who demonstrated that a cocktail of valproic acid, CHIR99021, tranylcypromine, forskolin, 3-deazaneplanocin A, 2-methyl-5-hydroxytryptamine hydrochloride and D4476 facilitates reprogramming of mouse somatic cells. The resultant cells were named as chemical-induced pluripotent stem cells (CiPSCs). Later, scientists extrapolated the same strategy and successfully transdifferentiated mouse and human somatic cells to functional neurons. Neural progenitor cells were generated by growing mouse fibroblasts and human urinary cells under physiological hypoxic conditions, along with the administration of small molecules—valproic acid, CHIR99021 and Repsox (VCR). Being inhibitors of histone deacetylase (HDAC), GSK3β and TGFβ, respectively, these molecules alter the pathways and epigenetic landscape of the source cells, pushing them towards an altogether unrelated identity [50]. The same research group went ahead to show that along with VCR, when neuronal differentiation-promoting chemicals like forskolin, JNK inhibitor SP600126, protein kinase C inhibitor G06983 and ROCK inhibitor Y-27632 were added, the mouse fibroblasts acquired a neuronal cell fate, bypassing the progenitor stage [51].
A successful transdifferentiation is a resultant of two independent phenomena—one being the erasure of former epigenetic and molecular memory of the source cell and the other being the establishment of a new identity of a bona fide transdifferentiated cell with desirable functionality and stability. Small molecules have the capability to facilitate both the aforementioned processes and thus enjoy the merit of being the choice of scientific studies. Also, be the transcription factors or small molecules, usage of a minimal number of factors is always preferred owing to reduced intervention. Hence, research group attempted for the goal of transdifferentiation by using a combination of just four small molecules, namely, forskolin, ISX9, CHIR99021 and I-BET151, which robustly converted mouse fibroblasts to active neurons [52].
Fate conversions between glial and neuronal lineages have gained attention more recently. Transcription factors like NeuroD1 have been shown to convert astroglial cells to neuronal identity. The complex cocktail of small molecules consisting of LDN193189, SB431542, TTNPB, Tzv, CHIR99021, VPA, DAPT, SAG and Purmo serves the similar purpose and resulted in the formation of functional neurons within 8–10 days of treatment. These studies also give a deeper insight into the molecular mechanisms governing the cell fate decisions. For instance, the abovesaid cocktail, featuring BMP/TGFβ, GSK3 β and Notch inhibitors, indicated that these pathways play the crucial role in switching human astrocytes to neuronal-like fate [53]. Apart from neurons, functional cardiomyocytes have also been generated using small molecules. miRNA-based transdifferentiation to cardiomyocytes has been complemented with addition of JAK inhibitor I which increased the efficiency of fate switching. OCT4, along with small molecules SB431542 (ALK4/5/7 inhibitor), CHIR99021 (GSK3 inhibitor), parnate (LSD1/KDM1 inhibitor) and forskolin (adenylyl cyclase activator) (SCPF), induced cardiomyocyte phenotype in MEFs [54].
Complementary to the conventional strategies of using TFs, small molecules that target specific signalling pathways or epigenetic stream offer a friendly tool to induce lineage differentiation and facilitate direct reprogramming to derive specific lineage cells of interest. This method could be utilized for basic research, disease modelling, drug screening and for cell-based therapy.
Conclusions
In mammals, different cell types work independently with an assigned specific function. If the whole body is envisioned as an engine, the different cell types are considered as different parts of the engine working synchronously to keep the engine working uninterrupted. As long as different compartments of the engine are working efficiently, the body functions smoothly. However, if any part of the engine is incapacitated, there is always an alternative compartment to take over that particular function, and this process which dictates the overall integrity of the body engine is nothing but ‘transdifferentiation’. Previous notion that adult differentiated cells are sitting passively and stably at the end of a multi-lineage valley has been ruled out by the radical change in our understanding of three important processes: dedifferentiation, transdifferentiation and reprogramming. Although, the goal of replacing lost or damaged cells could be accomplished using either of the processes, the major predicament is to identify the most efficient approach with limited drawbacks. Dedifferentiation and reprogramming events visit a stem cell fate before landing on to the desired cellular phenotype. Derivation of the desired cell type from stem cell fate commands efficient differentiation protocol, without which there is a high probability of developing tumours. Although iPSC reprogramming approach has rejuvenated the alternative means of generating cells of interest, considering few of the limitations fraught with reprogramming to iPSC stage, direct transdifferentiation has become the better choice in few incidences.
Having achieved a remarkable leap in the field of direct cell fate conversion, researchers are now moving towards formulation of transcription factor and miRNA cocktails with minimal number of candidate molecules to arrive at cells of interest. As application to human health and lifestyle betterment is the ultimate goal of all research endeavours, the field of transdifferentiation now needs complete shift in the paradigm towards bringing these complex cell fate switching concepts from bench to bedside.
Abbreviations
- ALS:
-
Amyloid lateral sclerosis
- CHF:
-
Congestive heart failure
- GIP:
-
Glucose-dependent insulinotropic polypeptide
- iMPCs:
-
Induced multipotent progenitor cells
- iPSCs:
-
Inducing pluripotent stem cells
- NPCs:
-
Neuronal progenitor cells
- TF:
-
Transcription factor
References
Slack JM, Tosh D. Transdifferentiation and metaplasia—switching cell types. Curr Opin Genet Dev. 2001;11(5):581–6.
Wapinski OL, Vierbuchen T, Qu K, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155(3):621–35.
Peter J. Out of Africa and into epigenetics: discovering reprogramming drugs. Nat Cell Biol. 2011;13:2.
Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000.
Sisakhtnezhad S, Matin MM. Transdifferentiation: a cell and molecular reprogramming process. Cell Tissue Res. 2012;348(3):379–96. doi:10.1007/s00441-012-1403-y.
Kragl M, Knapp D, Nacu E, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460(7251):60–5.
Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110(11):1465–73.
Lei Z, Yongda L, Jun M, et al. Culture and neural differentiation of rat bone marrow mesenchymal stem cells in vitro. Cell Biol Int. 2007;31(9):916–23.
Ullah M, Stich S, Notter M, et al. Transdifferentiation of mesenchymal stem cells-derived adipogenic-differentiated cells into osteogenic- or chondrogenic-differentiated cells proceeds via dedifferentiation and have a correlation with cell cycle arresting and driving genes. Differentiation. 2013;85(3):78–90.
Gerace D, Martiniello-Wilks R, O’Brien BA, et al. The use of β-cell transcription factors in engineering artificial β cells from non-pancreatic tissue. Gene Ther. 2015;22(1):1–8.
Tang DQ, et al. Genetically reprogrammed, liver-derived insulin-producing cells are glucose-responsive, but susceptible to autoimmune destruction in settings of murine model of type 1 diabetes. Am J Transl Res. 2013;5:184–99.
Berneman-Zeitouni D, Molakandov K, Elgart M, et al. The temporal and hierarchical control of transcription factors-induced liver to pancreas transdifferentiation. PLoS One. 2014;9(2):e87812.
Li Y, Zhao LJ, Xia FZ, et al. Transdifferentiation of hepatic oval cells into pancreatic islet beta-cells. Front Biosci (Landmark Ed). 2012;17:2391–5.
Wei R, Hong T. Lineage reprogramming: a promising road for pancreatic β cell regeneration. Trends Endocrinol Metab. 2016;27(3):163–76.
Nagaya M, Katsuta H, Kaneto H, et al. Adult mouse intrahepatic biliary epithelial cells induced in vitro to become insulin-producing cells. J Endocrinol. 2009;201:37–47.
Hickey RD, Galivo F, Schug J, et al. Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 2013;11:503–15.
Talchai C, Xuan S, Kitamura T, et al. Generation of functional insulin-producing cells in the gut by Foxo1 ablation. Nat Genet. 2012;44:406–12.
Chen YJ, Finkbeiner SR, Weinblatt D, et al. De novo formation of insulin-producing “neo-beta cell islets” from intestinal crypts. Cell Rep. 2014;6:1046–58.
Sancho R, Gruber R, Gu G, et al. Loss of Fbw7 reprograms adult pancreatic ductal cells into alpha, delta, and beta cells. Cell Stem Cell. 2014;15:139–53.
Chera S, Baronnier D, Ghila L, et al. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514(7523):503–7.
Zhu S, Rezvani M, Harbell J, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508(7494):93–7.
Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–3.
Du Y, Wang J, Jia J, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14(3):394–403.
Huang P, He Z, Ji S, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–9.
Zhang Z, Gong Y, Guo Y, et al. Direct transdifferentiation of spermatogonial stem cells to morphological, phenotypic and functional hepatocyte-like cells via the ERK1/2 and Smad2/3 signaling pathways and the inactivation of cyclin A, cyclin B and cyclin E. Cell Commun Signal. 2013;11:67.
Liu WH, Song FQ, Ren LN, et al. The multiple functional roles of mesenchymal stem cells in participating in treating liver diseases. J Cell Mol Med. 2015;19(3):511–20.
Tsunemoto RK, Eade KT, Blanchard JW, et al. Forward engineering neuronal diversity using direct reprogramming. EMBO J. 2015;34(11):1445–55.
Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41.
Zaret KS, Carroll JS. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25(21):2227–41.
Kumar A, Declercq J, Eggermont K, et al. Zic3 induces conversion of human fibroblasts to stable neural progenitor-like cells. J Mol Cell Biol. 2012;4(4):252–5.
Lujan E, Chanda S, Ahlenius H, et al. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109(7):2527–32.
Han DW, Tapia N, Hermann A, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10(4):465–72.
Thoma EC, Merkl C, Heckel T, et al. Chemical conversion of human fibroblasts into functional Schwann cells. Stem Cell Rep. 2014;3(4):539–47.
Victor MB, Richner M, Hermanstyne TA, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84(2):311–23.
Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–44.
Riddell J, Gazit R, Garrison BS, et al. Reprogramming committed murine blood cells to induce hematopoietic stem cells with defined factors. Cell. 2014;157(3):549–64.
Szabo E, Rampalli S, Risuen o RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–6.
Batta K, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9(5):1871–84.
Doppler SA, Deutsch MA, Lange R, et al. Direct reprogramming—the future of cardiac regeneration? Int J Mol Sci. 2015;16(8):17368–93.
Zhou H, Dickson ME, Kim MS, et al. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112(38):11864–9.
Ying QL, Nichols J, Evans EP, et al. Changing potency by spontaneous fusion. Nature. 2002;416(6880):545–8.
Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425(6961):968–73.
Tsai RY, McKay RD. Cell contact regulates fate choice by cortical stem cells. J Neurosci. 2000;20(10):3725–35.
Sohal GS, Ali MM, Ali AA, et al. Ventrally emigrating neural tube cells contribute to the formation of Meckel’s and quadrate cartilage. Dev Dyn. 1999;216(1):37–44.
Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73.
Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–31.
Xue Y, Ouyang K, Huang J, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated MicroRNA circuits. Cell. 2013;152(1–2):82–96.
Banerjee P, Dutta S, Pal R. Dysregulation of Wnt-signaling and a candidate set of miRNAs underlie the effect of metformin on neural crest cell development. Stem Cells. 2016;34(2):334–45.
Hou P, Li Y, Zhang X, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–4.
Cheng L, Hu W, Qiu B, et al. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24:665–79.
Hu W, Qiu B, Guan W, et al. Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell. 2015;17:204–12.
Li X, Zuo X, Jing J, et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell. 2015;17:195–203.
Zhang L, Yin JC, Yeh H, et al. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell. 2015;17:735–47.
Fu Y, Huang C, Xu X, et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015;25:1013–24.
Acknowledgements
The authors thank Manipal University, Manipal, India for supporting this study. Permission/conflict of interest: Author has no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Louis, L.K., Ashwini, A., Kumar, A., Pal, R. (2017). Transdifferentiation: A Lineage Instructive Approach Bypassing Roadways of Induced Pluripotent Stem Cell (iPSC). In: Mukhopadhyay, A. (eds) Regenerative Medicine: Laboratory to Clinic. Springer, Singapore. https://doi.org/10.1007/978-981-10-3701-6_8
Download citation
DOI: https://doi.org/10.1007/978-981-10-3701-6_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-3700-9
Online ISBN: 978-981-10-3701-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)