Abstract
The yeast Blastobotrys adeninivorans (syn. Arxula adeninivorans ) is a dimorphic, asexual hemiascomycete which is phylogenetically very distant from Saccharomyces cerevisiae. It has been shown to be most useful in a wide range of biotechnological applications: as gene donor in enzyme production, as host for heterologous gene expression, and as powerful biological component in biosensors. B. (A.) adeninivorans major advantage is its metabolic flexibility, which enables the utilization of a wide range of different carbon and nitrogen sources. For example, recent analyses of the genome and its transcriptome revealed a new pathway for the assimilation of n-butanol via butyraldehyde and butyric acid as well as new insights into the previously reported purine degradation pathway. Additionally, the synthesis of several secretory enzymes with great biotechnological potential, such as two tannases (Atan1p and Atan2p) and three new cutinases (Acut1p, Acut2p and Acut3p) were identified. Due to these characteristics, B. (A.) adeninivorans can be exploited as a gene donor for the production of enzymes with attractive biotechnological applications. Furthermore, its unusual thermo- and halotolerance as well as differential morphology-dependent glycosylation and the secretion characteristics render B. (A.) adeninivorans attractive as host for heterologous gene expression. Successful expression of bacterial alcohol dehydrogenase (ADH) genes from Rhodococcus ruber and Rhodococcus erythropolis enables B. (A.) adeninivorans to be used as biocatalyst for the synthesis of chiral alcohols as building blocks for the chemical industry. The combination of robustness with its great ability for heterologous gene expression makes B. (A.) adeninivorans a superior choice for the biological component in biosensor applications. Different B. (A.) adeninivorans-based biosensors detecting hormones like estrogens, androgens and glucocorticoides as well as dioxin have been developed and consequently improved in the last decade. This chapter will provide a comprehensive overview on the biology and the biotechnological applications of this yeast.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Blastobotrys adeninivorans
- Arxula adeninivorans
- Heterologous gene expression
- Thermotolerance
- Halotolerance
- n-Butanol
- Tannase
- Cutinase
- Biosensor
1 Introduction
Yeasts are simply organized, unicellular eukaryotes with the ability of rapid adaptation to alternating environmental conditions. Besides the well characterized baker’s yeast Saccharomyces cerevisiae a wide range of non-conventional yeast species exists, that are exhibiting attractive growth properties and biochemical characteristics. For this reason they can be exploited for industrial applications in the field of biotechnology as well as suitable model organisms for plant and animal research. Expression and transformation platforms have been developed to fortify the practical use of yeast species like the traditional baker’s yeast Saccharomyces cerevisiae, Kluyveromyces lactis, Pichia pastoris, Yarrowia lypolytica, Hansenula polymorpha and Arxula adeninivorans for research and biotechnological applications. Using these platforms yeast can be easily deployed as hosts for the production of recombinant proteins or valuable metabolites as building blocks for the chemical industry. Additionally, they are used as gene donors due to their partly wide substrate spectrum (Gellissen 2005; Gellissen et al. 2005; Wolf 1996; Wolf et al. 2003).
In 1984, Middelhoven et al. described the isolation of a yeast species, designated as Trichosporon adeninovorans , from soil by enrichment culturing. In particular, the strain CBS 8244T was found to exhibit unusual biochemical activities being able to assimilate a range of amines, adenine and several other purine compounds as sole energy and carbon source. In parallel, a second strain, LS3 (PAR-4), with similar characteristics (Gienow et al. 1990) was isolated from wood hydrolysates in Siberia (Kapultsevich, Institute of Genetics and Selection of Industrial Microorganisms, Moscow, Russia). Seven additional strains were isolated, three of them from chopped maize herbage ensiled at 25 or 30 °C in the Netherlands and four from humus-rich soil in South Africa (Van der Walt et al. 1990). The new genus Arxula was proposed for these strains, because all representatives are ascomycetous, anamorphic and arthroconidial. Furthermore, they share properties like nitrate assimilation and xerotolerance (Van der Walt et al. 1990). The phylogenetic analysis of the ascosporic yeast genera Sporopachydermia, Stephanoascus, Trichomonascus, Wickerhamiella and Zygoascus and the associated anamorphic genera Arxula, Blastobotrys, Sympodiomyces and Trigonopsis was accomplished by Kurtzmann and Robnett (2007). They compared sequences derived from the large-subunit rDNA genes, the mitochondrial small-subunit rDNA genes, and the genes for cytochrome oxidase II and deduced that Arxula, Blastobotrys and Sympodiomyces are members of the Trichomonascus clade, with the genus Blastobotrys having taxonomic priority for anamorphic states (Table 1).
The genus Blastobotrys includes now both type species of the genus B. terrestris (Van der Walt and Johanssen) Kurtzman and Robnett comb. nov. (Basionym: Arxula terrestris) and B. adeninivorans (Middelhoven, Hoogkamer Te-Niet and Kreger van Rij) Kurtzman and Robnett comb. nov. (Basionym: Arxula adeninivorans ).
So far, approx. 170 articles related to A. adeninivorans have been published. Although this yeast is well characterised and its genome sequence has been known since 2014, it is still hardly known in the public. The purpose of this chapter is to provide novel information on this interesting and useful organism.
2 Physiology and Temperature Dependent Dimorphism
Gienow et al. (1990), Middelhoven (1993) and Middelhoven et al. (1984, 1991, 1992) provided a detailed physiological description of A. adeninivorans. Likewise H. polymorpha this yeast is assimilating nitrate using nitrate reductase and nitrite reductase. A. adeninivorans exhibits a wide spectrum of substrates which can be utilized as sole carbon and energy source including most sugars, polyalcohols, organic acids, adenine, uric acid , butylamine, pentylamine, putrescine, soluble starch, melibiose , melicitose, propylamine or hexylamine. L-rhamnose, inulin, lactose, lactate and methanol on the other hand are compounds which are not assimilated by this yeast. Except of creatine and creatinine all nitrogen compounds are suitable nitrogen sources. Moreover, several nitrogen compounds, like amino acids, purine derivatives and many primary n-alkylamines and terminal diamines, are metabolized as sole source of energy, carbon and nitrogen. Furthermore, metabolic intermediates of alcohols, dialcohols, carboxylic acids, dicarboxylic acids and other nitrogen-free analogous compounds are assimilated by A. adeninivorans. On top of that, this yeast degrades some phenols, hydroxybenzoates, tannic acid and is able to assimilate urotropine as sole nitrogen source (Middelhoven and van Doesburg 2007).
A. adeninivorans synthesizes a vast number of secretory enzymes including RNases, proteases, glucoamylase, lipase, tannase, some acid phosphatases, trehalase , some cellobiases, invertase, β-glucosidase, xylosidase, some cutinases and phytase, which are summarized with some of their biochemical properties in Table 2.
A. adeninivorans and the Siberian wild-type strain LS3 in particular, shows some special features like thermotolerance and temperature-dependent dimorphism, which have an important impact on the biotechnological application of this yeast. Without previous adaptation A. adeninivorans LS3 can grow at temperatures up to 48 °C and survives at 55 °C for some hours (Böttcher et al. 1988; Wartmann et al. 1995a). At temperatures above 42 °C LS3 exhibits a transition from budding cells to mycelia forms, which is reversed to budding cells when the temperature is decreased below 42 °C (Fig. 1). Mutant strain A. adeninivorans 135 with altered dimorphism were selected by Wartmann et al. (2000), which is growing as mycelia already at 30 °C. This mutant was used to examine the morphology-, and not temperature-related effects on gene expression and protein accumulation. It was found that budding cells and mycelia differ in cell dry weight and in their contents of RNA and soluble protein, which are lower in mycelia during the middle and the final phases of the exponential growth. In contrast, the concentration of secreted proteins including glucoamylase and invertase is two-fold higher in mycelia compared to budding cells. The summary in Table 3 indicates that morphology, rather than temperature, is the relevant factor. Moreover, a strong correlation between the morphological status and the iron uptake was found to exist. Two transport systems with different iron affinity are responsible for iron uptake by A. adeninivorans. Budding cells accumulate iron up to seven-fold higher than mycelia at high Fe(II) concentrations (>2 µM), whereas at concentrations <2 µM both cell types accumulate similar amounts of iron. The expression of the AFET3 gene, which encodes a copper-dependent Fe(II) oxidase (Afet3p), was found to strongly depend on iron concentration as well as on the morphological state, but just in a minor way. However, the greater influence of morphology on posttranslational modifications of Afet3p was found. O-glycosylation occurred only in budding cells, but both cell types showed N-glycosylation to some extent. Differential glycosylation of heterologous proteins could be used as a tool to study the influence of O-glycosylation on biological activity or immunological tolerance (Wartmann et al. 2002).
Cell morphology of A. adeninivorans LS3 grown at 30 °C (I), 42 °C (II) and 45 °C (III). The cells were cultured on YEPD medium for 18 h. Influencing and non-influencing factors are indicated in the boxes below. Budding cells (BC) appear at 30 °C, whereas pseudo mycelium (PM) can be observed at 42 °C. At 45 °C mycelium (M) is the predominant growth form
There are also other factors influencing the dimorphism of A. adeninivorans. Compounds like Cd2+ and tocopherol lead to mycelia, whereas NaCl, anaerobic conditions and tunicamycin are enhancing the formation of budding cells. Other factors like Ca2+, pH-value, carbon source and its concentration show no influence on cell morphology (Fig. 1).
Osmotolerance is another interesting property of A. adeninivorans. In presence of ionic (NaCl), osmotic (PEG400) and water stress (ethylene glycol) LS3 is able to grow to osmolarities up to 3.32 osmol kg−1 H2O. At concentrations lower than 3.4 M NaCl showed only limited influence on the growth behaviour. Supplementation with higher concentration (>3.4 M) of NaCl led to a decrease of the specific growth rate, a longer adaptation time as well as a lower cell count during stationary growth phase (Yang et al. 2000). Likewise in other yeast species, the osmotolerance is mediated by compounds of the high osmolarity glycerol (HOG) pathway, which is activated by an elevated osmolarity in the cells´ environment, leading to an increased synthesis of the compatible solutes glycerol, erythritol and mannitol (Böer et al. 2004a).
3 Genetics and Molecular Biology
A. adeninivorans has a DNA content comparable to that of ascomycetous yeasts such as S. cerevisiae (Gienow et al. 1990; Samsonova et al. 1996; Wartmann et al. 2000). Furthermore the fact, that after nitrosoguanidine mutagenesis a high number of auxotrophic mutants is obtained (Samsonova et al. 1989, 1996), along with the quantitative analysis of chromosomal DNA and the determination of genome size (Gienow et al. 1990) show A. adeninivorans to be a haploid organism.
DNA reassociation studies as well as karyotyping were performed in order to assess the complexity of the A. adeninivorans genome (Gienow et al. 1990; Kunze and Kunze 1994a). Genome size measurements, as performed in reassociation kinetics experiments of A. adeninivorans chromosomal DNA, resulted in 16.1 and 16.9 GDa for A. adeninivorans strains LS3 and CBS 8244T, respectively, rendering them the largest genomes reported so far amongst all yeast species, including S. cerevisiae (9.2 GDa). Also the amount of repetitive sequences (33.1% in LS3 and 35.9% in CBS 8244T) exceeds that of other yeasts. Finally, karyotyping demonstrated the existence of four chromosomes with sizes ranging between 1.6 and 4.6 Mb.
As previously mentioned, mutagenesis using UV light or N-methyl-N′-nitro-N-nitrosoguanidine treatment led to a relatively high frequency of auxotrophic mutants and mutants with different catabolite repression (resistant to 2-deoxy-D-glucose) which have subsequently been selected and characterized by Böttcher and Samsonova (1983); Büttner et al. (1990b) and Samsonova et al. (1989, 1996). Heterozygous diploids were generated from auxotrophic mutants of strains LS3 and CBS 8244T using PEG-induced spheroplast fusion as a first step to establish genetic maps (Büttner et al. 1990b; Samsonova et al. 1996). The resulting diploids were then segregated using benomyl treatment, allowing for linkage analysis of a set of markers that were assigned to four linkage groups. Additionally, specific probes for all 32 auxotrophic mutation markers were labelled and hybridized to genomic yeast DNA separated by pulsed field gel electrophoresis, confirming the predicted number of chromosomes (Samsonova et al. 1996).
The whole genome sequence of A. adeninivorans has been analysed and published (Kunze et al. 2014). Both mitochondrial and nuclear genomes were sequenced using the Sanger and 454 pyrosequencing approaches with different shotgun, plasmid and BAC libraries. The mitochondrial genome has a size of 31,662 bp and encodes 24 tRNA and 15 protein coding genes including the seven NADH: ubiquinone dehydrogenase subunits of complex I, the genes encoding the RNA component of RNase P and the two subunits of the mitochondrial ribosomal RNA. As already described and shown by pulsed field gel electrophoresis, the sequencing approach revealed four chromosomes Arad1A, Arad1B, Arad1C and Arad1D with a size of 1,659,397, 2,016,785, 3,827,910, and 4,300,524 Bp, respectively. Regional centromeres could be identified for all of them (Fig. 2). Table 4 shows the comparison of genome data of different representative hemiascomycetes. With 914 introns within 6116 genes, A. adeninivorans is one of the most intron-rich hemiascomycetes sequenced to date. Examples of genes containing at least one intron are ARFC1, AHOG1 and AHSB4. The comparison of 5′-splice site (DS/GUARGU), branch site (HRCUAAC) and 3′-splice site (HAG/R) sequences demonstrates that the consensus sequences are similar to that of S. cerevisiae and filamentous fungi (Böer et al, 2005a).
Schematic representation of the four chromosomes of A. adeninivorans LS3 as they can be seen on the PFGE. The positions of the putative centromere regions are indicated by black lines. The summary of the statistical properties of the four chromosomes given in the table can be found in Kunze et al. (2014)
4 Biochemical Properties and A. adeninivorans as Gene Donor
A. adeninivorans is described as having a wide substrate spectrum that includes the assimilation of many nitrogenous and aromatic compounds such as nitrate and nitrite, purines, tannins and benzoic acid derivatives. The ability to degrade purine compounds is reported in all kingdoms and can occur either aerobically or anaerobically in separate pathways. In the aerobic pathway, the critical step in the degradation of purine bases is the oxidation of hypoxanthine and xanthine to uric acid, catalysed by xanthine oxidase and/or dehydrogenase. The various purine-degradative pathways are unique and differ from other metabolic pathways because they may serve quite different purposes, depending on the organism or tissue. While some organisms degrade the naturally occurring purines to CO2 and ammonia, others perform only some of the steps of the purine degradation pathways, resulting in partial degradation of purines or certain intermediary catabolites. Purine catabolism which is shown in Fig. 3 is a characteristic feature of A. adeninivorans. The purine nucleosides (adenosine, inosine, xanthosine and guanosine) are transported across the membrane and into the cytoplasm by a purine permease. They are then converted to adenine, hypoxanthine, xanthine and guanine, further degraded to uric acid and, after transport into the peroxisomes, to urea. All corresponding genes of this pathway are localized on different chromosomes and are induced by adenine and other pathway intermediates (Jankowska et al. 2013a, b). It has been shown using a strain lacking xanthine oxidoreductase activity that the true inducer of the urate oxidase gene is in fact uric acid (Jankowska et al. 2013b). Interestingly, an adenosine deaminase, needed to transform adenosine to inosine in animals and human, is absent (Fig. 3). This pathway allows A. adeninivorans to use all of these purine derivatives as nitrogen and carbon sources (Jankowska et al. 2013a, b).
Tannin, a plant polyphenol molecule, is widely distributed in the plant kingdom where it protects plants against attack by parasites and herbivores. It inhibits the activity of enzymes by binding and precipitation and is to a greater or lesser extent recalcitrant to biodegradation (Field et al. 1991). While tannins are growth inhibitors for most microorganisms, a few bacteria, fungi and yeasts such as D. hansenii, Mycotorula japonica or Candida sp. are capable of exploiting tannins as a carbon and/or energy source for growth (Aguilar et al. 2007; Bhat Singh and Sharma 1998; Lekha and Lonsane 1997). A. adeninivorans is one of these yeasts that use tannic acid and gallic acid as carbon sources (Sietmann et al. 2010). Genes encoding tannases (ATAN1—ARAD1A06094g, ATAN2—ARAD1A19822g), gallate decarboxylase (AGDC—ARAD1C45804g) and catechol 1,2-dioxygenase (ACDO—ARAD1D18458g) have been identified and His-tagged recombinant enzymes and corresponding gene mutants were used to confirm the activity of these enzymes (data not shown). This demonstrated that the tannic acid catabolism pathway enables this yeast to assimilate tannic acid and other hydroxylated derivatives of benzoic acid by non-oxidative decarboxylation. Interestingly, A. adeninivorans is thus the first eukaryote known to synthesize two tannases , one extracellular (Atan1p) (Böer et al. 2009) and one cell-wall localized (Atan2p—data not shown) which permits effective degradation of extracellular tannic acid as well as the release of gallic acid from both condensed and hydrolysable tannins. Its biochemical parameters (pH optimum at approx. 6.0, temperature optimum 35–40 °C) and the almost complete extracellular localization (≥97%) make Atan1p the superior enzyme for industrial applications. Constructed tannase producing strains are able to accumulate up to 51,900 U L−1 in 42 h with a dry cell weight of 162 g L−1 (Böer et al. 2011).
A genome mining approach (Kunze et al. 2014) was performed to find more interesting features of A. adeninivorans leading to the discovery of the n-butanol degradation pathway which had not been reported to exist in eukaryotes (Fig. 4). The collected data suggest that n-butanol is oxidized to butyraldehyde by an alcohol dehydrogenase (Aadh1p, AADH1—ARAD1B16786g) with a high substrate specificity, and then to butyric acid in a one-way reaction by two aldehyde dehydrogenases (Aald2p, AALD2—ARAD1B17094g; Aald5p, AALD5—ARAD1C17776g). The last steps involve an acyl-CoA ligase, a cytoplasmic acyl-CoA carnitine acyltransferase and a peroxisomal acyl-CoA carnitine acyltransferase for butyryl-carnitine synthesis via a butyryl-CoA intermediate that is transported from the cytoplasm to peroxisomes or mitochondria for ß-oxidation. The one-way reactions of aldehyde dehydrogenase and acyl-CoA ligase are special features of this pathway (Kunze et al. 2014).
n-Butanol degradation pathway of A. adeninivorans. n-Butanol is oxidized to butyric acid via butyraldehyde by an alcohol dehydrogenase (Aadh1p, AADH1—ARAD1B16786g) and one of two aldehyde dehydrogenases (Aald2p, AALD2—ARAD1B17094g; Aald5p, AALD5—ARAD1C17776g). Butyric acid is transported into peroxisome and further degraded in the β-oxidation
Several genes like AEFG1, AFET3, AHOG, AHSB4, AINV, ALIP1, ALYS2, APHO1, ARFC3, ATAL, AXDH and TEF1 as well as the complete rDNA repeat were isolated from gene libraries containing either cDNA or chromosomal DNA from A. adeninivorans strain LS3 via PCR using primers for conserved sequences (Böer et al. 2004a, b, 2005b, c; El Fiki et al. 2007; Kaur et al. 2007; Kunze and Kunze 1996; Rösel and Kunze 1995, 1996; Steinborn et al. 2005; Stoltenburg et al. 1999; Wartmann et al. 2001, 2002, 2003a). Additional genes like GAA encoding glucoamylase were identified from a heterologously expressed cDNA library with S. cerevisiae and K. lactis as hosts using an anti-glucoamylase antibody as probe for product detection. More than 90% of the glucoamylase was found to be secreted, with a 20 times higher level in K. lactis compared to S. cerevisiae using a similar construct for transformation (Bui et al. 1996a, b).
The complementation of respective E. coli and S. cerevisiae mutants was used as an approach for the isolation of additional genes, namely the ALYS2, AILV1, ALEU2 and ATRP1 genes which are suitable selection markers for the A. adeninivorans-based platform (Kunze and Kunze 1996; Steinborn et al. 2007b; Wartmann et al. 1998, 2003b).
In parallel, genes encoding for biotechnologically important secretory enzymes like lipases found in A. adeninivorans were homologously expressed and characterized. This group of temperature-sensitive proteins with a pH optimum at 7.5 hydrolyses ester bounds in triglycerides and other fatty acid esters with highest efficiency for middle-sized chains between C8 and C10 (Böer et al. 2005b).
The AINV gene encoding for an invertase, which is preferentially hydrolyzing β-D-fructofuranosides, has potential to be applied in hydrolysis of sugar cane molasses or sugar beet molasses on an industrial scale. High concentrations of this enzyme have been obtained in recombinant A. adeninivorans strains carbon source independent using the strong constitutive TEF1 promoter (Böer et al. 2004b).
An example for an interesting intracellular protein is the temperature-sensitive xylitol dehydrogenase. This enzyme oxidizes polyols like xylitol and D-sorbitol. Furthermore it can catalyse the reduction of D-xylulose, D-ribulose and L-sorbose. Its biochemical parameters—optimum at low temperatures and weak basic pH values—increase the potential for applications of this enzyme in food manufacturing processes. The respective AXDH gene has already been isolated and successfully overexpressed in A. adeninivorans (Böer et al. 2005c).
An enzyme with acidic pH optimum, transaldolase encoded by the ATAL gene, is another example with potential industrial applications. This temperature-sensitive enzyme, which is using D-erythrose-4-phosphate and D-fructose-6-phosphate as preferred substrates, could be useful in C–C bonding and enantio-selective synthesis of novel sugars (El Fiki et al. 2007).
Recently three genes ACUT1, ACUT2 and ACUT3 encoding for cutinases were isolated from the A. adeninivorans genome (Bischoff et al. 2015). Since cutinases are typically found in plant pathogenic fungi, only a few yeast species like Cryptococcus spp. (Masaki et al. 2005; Suzuki et al. 2013) and Pseudozyma antarctica (Shinozaki et al. 2013; Watanabe et al. 2014) have been found to produce cutinase like enzymes so far. The catalytic triade was identified as S-D-H with a conserved G-Y-S-Q-G domain containing the nucleophilic serine which is interacting with histidine and aspartic acid. Thus the three cutinases belong to the serine hydrolase family with α/β-structure. Recombinant variants outfitted with a 6xhistidine tag were expressed in A. adeninivorans and subsequently purified and characterized. All three cutinases show a pH optimum in the slightly acidic range from 4.0 to 6.5. The temperature optimum is in the range between 20 and 30 °C due to the low temperature stability of the enzymes. The substrate spectrum revealed the highest activity for short chain fatty acid esters of p-nitrophenol and glycerol (C4 and C6). However, the activity for p-nitrophenyl acetate (C2) was low (approx. 10% compared to p-nitrophenyl butyrate), which clearly distinguishes the three cutinases from ordinary carboxyl esterases and lipases. Additionally, the degradation of the natural polymer cutin from apple peels and of the model substrate polycaprolactone was observed (Fig. 5), which is proof of cutinolytic activity. However, the natural function of these cutinases for the yeast and their gene regulation remains unclear.
5 A. adeninivorans as Suitable Host for Heterologous and Homologous Gene Expression
5.1 Transformation and Expression System
An attractive gene expression platform for A. adeninivorans has been established and developed over the last decades starting from a first transformation system based on S. cerevisiae and A. adeninivorans-derived LYS2 genes for selection (Kunze et al. 1990; Kunze and Kunze 1996). Improvements of the transformation system were introduced by Rösel and Kunze (1998) using a vector type (pAL-HPH1) that employed a 25S rDNA fragment from A. adeninivorans targeting sequence for stable integration and the E. coli-derived hph gene (conferring hygromycin B resistance) under the control of the A. adeninivorans-derived TEF1 promoter for dominant selection. Two to ten stable integrations of the hygromycin resistance cassette could be found within the ribosomal DNA.
In the next step auxotrophic strains were used in the combination with their respective gene sequence for complementation, because the usage of dominant marker genes leads to the undesired need for toxic compounds or antibiotics during strain development. Firstly the AILV1 and ALEU2 genes were isolated after selecting the respective auxotrophic strains incapable of synthesizing leucine and isoleucine, which were obtained after treatment with N-methyl-N′-nitro-N-nitrosoguanidine. Strains generated by transformation of these mutants with pAL-AILV1 and pAL-ALEU2m containing the AILV1 and the ALEU2 m gene for complementation were found to harbour 1–3 copies of the heterologous DNA within the rDNA sequences (Steinborn et al. 2005; Wartmann et al. 1998, 2003b). However, at a frequency of 10−6, the ailv1 and aleu2 mutant strains reverted to leucine/isoleucine prototrophy (Samsonova et al. 1989, 1996), which was a great disadvantage. This was eliminated by generating a Δatrp1 gene disruption mutant using a DNA fragment containing the ALEU2m gene flanked by ATRP1 gene sequences of some 750 bp. The resulting auxotrophic host strain A. adeninivorans G1212 [aleu2 atrp1::ALEU2] excels in mitotic stability during cultivation in both rich and minimal medium. The first strain complemented with the ATRP1 gene as selection marker and the 25S rDNA contained just a single copy of the pAL-ATRP1 DNA (Steinborn et al. 2007b). For this reason a novel vector element containing the ATRP1 coding sequence under control of a 53 bp truncated version of the ALEU2 promoter was constructed that provides multicopy integrations (8 or more copies) in A. adeninivorans G1212 [aleu2 atrp1::ALEU2]. In addition the vector design enables the integration of a small vector fragment that consists of yeast DNA only (yeast integration-expression cassette—YIEC) providing high transformation frequencies and a high mitotic stability (Steinborn et al. 2007a).
The construction of expression plasmids followed a two-step cloning strategy. First the heterologous genes are inserted between the respective A. adeninivorans-derived promoter and fungal terminator elements like PHO5 from S. cerevisiae and trpC from Aspergillus nidulans. Subsequently the resulting expression modules (A. adeninivorans promoter—heterologous gene—fungal terminator) are integrated into the respective A. adeninivorans expression plasmid. For this purpose the modules are flanked by unique restriction sites (ApaI–SalI, ApaI–XhoI, SpeI–SacII, SpeI–NotI) allowing a directional integration.
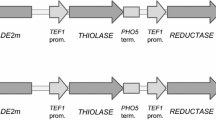
A new set of plasmids was introduced into the latest version of the A. adeninivorans expression platform to circumvent the two-step cloning strategy. Those plasmids already contain the selection marker as well as several combinations of promoter and terminator elements. Figure 6 shows one example plasmid which provides two integration sites for genes with either TEF1 promoter and PHO5 terminator or TIF5a promoter and 60SRPL3 terminator. The genes are integrated after linearization with PacI for TEF1/PHO5 or SpeI for TIF5a/60SRPL3 using isothermal assembly, which avoids complications with restriction sites present in the gene sequence.
5.2 Heterologous Gene Expression
A. adeninivorans is not only a donor of genes encoding for industrially relevant proteins but also a host for heterologous gene expression . An increasing number of heterologous genes have been expressed in A. adeninivorans. First examples were the XylE gene encoding catechol 2,3-dioxygenase from Pseudomonas putida under the control of AILV1 promoter and GFP as well as the HSA gene under the control of the strong constitutive TEF1 promoter (Kunze et al. 1990; Kunze and Kunze 1996). One to two copies of heterologous DNA were found within the 25S rDNA region after transformation of wild-type and mutant strains with pAL-HPH1 and pAL-ALEU2m. Recombinant GFP was localized in the cytoplasm rendering the cells fluorescent, whereas expression of HSA led to secretion of 95% recombinant HSA into the culture medium at levels of 50 mg L−1 after 96 h. In this instance, no difference in secretion levels could be observed comparing budding cells and mycelia, which demonstrates a morphology-independent productivity (Wartmann and Kunze 2003; Wartmann et al. 2003b).
Besides the TEF1 promoter, the strong constitutive AHSB4 promoter was successfully assessed for suitability and was found to exhibit similar expression levels (Wartmann et al. 2003a).
A next example is the expression of a MF1-IL6 fusion under control of the strong TEF1 promoter in A. adeninivorans budding cells and mycelia. Unlike other yeast species (S. cerevisiae, H. polymorpha), A. adeninivorans was correctly processing the MF1-IL6 precursor, which led to the accumulation of recombinant interleukin-6 (IL-6) to more than 95% in the culture medium. A productivity of 210 and 145 mg L−1 was observed in cultivations on shaking flask scale with budding cells and mycelia, respectively (Böer et al. 2007).
A metabolic engineering approach had been done by introducing several genes of the synthesis of polyhydroxyalkanoate (PHA) biosynthetic pathway of Ralstonia eutropha. The genes phbA, phbB and phbC encoding β-ketothiolase, NADPH-linked acetoacetyl-CoA reductase and PHA synthase were established simultaneously enabling A. adeninivorans to synthesize poly-3-hydroxybutyrate (PHB) as well as poly-3-hydroxyvalerate (PHV). The first recombinant yeast strains were able to accumulate up to 2.2% PHV and 0.019% PHB with ethanol as carbon source (Terentiev et al. 2004).
Also promoter assessment studies were performed using lacZ from E. coli, the GFP from Aequorea victoria, the phyK from Klebsiella spec. and the XylE from Ps. putida as reporter genes. In this instance, the GAA, AHOG1, AINV, AXDH as well as the ATAL promoter were analysed on aspects like carbon source, osmolarity of the medium or morphological stage (Böer et al. 2004a, b, 2005c; El Fiki et al. 2007; Hahn et al. 2006; Wartmann and Kunze 2000).
Another successful example is the expression of alcohol dehydrogenase (ADH) from Rhodococcus ruber and Rhodococcus erythropolis as biocatalyst for the synthesis of enantiomerically pure secondary alcohols (Kasprzak et al. 2016). R. erythropolis alcohol dehydrogenase was used for synthesis of 1-(S)-phenylethanol and ethyl (R)-4-chloro-3-hydroxybutanoate either with purified enzyme or with permeabilized cells of genetically modified A. adeninivorans, co-expressing glucose dehydrogenase from Bacillus megaterium for enzyme-coupled co-factor regeneration. One of the major advantages of yeast cells over E. coli is that they are more robust in organic solvents because of their cell wall construction. Ethyl (R)-4-chloro-3-hydroxybutanoate is an important chiral building block in the synthesis of pharmaceuticals such as (−)-macrolactin A (Marino et al. 2002), l-carnitine, (R)-γ-amino-β-hydroxybutyric acid (GABOB) (Song et al. 1995), (+)-negamycin or chiral 2,5-cyclohexadienone synthon, therefore A. adeninivorans expressing R. erythropolis alcohol dehydrogenase as well as B. megaterium glucose dehydrogenase is of great interest for industrial application. 1-(R)-phenylethanol, another chiral building block for the chemical and pharmaceutical industry, was synthetized using A. adeninivorans co-expressing Lactobacillus brevis ADH and GDH from B. megaterium or Bacillus pumilus G6PDH as co-factor regeneration system (Rauter et al. 2014). The combination of ADH and G6PDH showed highest activity. However, together with GDH a higher stability was obtained which in the end leads to higher product formation due to a higher rate of reusability. In this context A. adeninivorans was used for optimization studies in enzymatic synthesis of 1-(S)-phenylethanol with permeabilized cells co-expressing the ADH of R. ruber and the B. megaterium GDH (Rauter et al. 2014). A comparison of the reaction with permeabilized cells, permeabilized immobilized cells and immobilized crude extract of those cells showed that immobilization of permeabilized cells is the best option for high reusability in synthesis reactions which is a key factor for the cost-effective use of enzymes in industrial applications. A summary of the different industrially relevant alcohol dehydrogenases expressed in A. adeninivorans is given in Table 5.
Despite this great potential for industrial applications little research was done on definition of culture and media conditions. Stöckmann et al. (2014) investigated the growth characteristics and culture conditions in a more rational approach. Cultures of A. adeninivorans were inhibited at pH-values below 2.8 and the phosphorus demand has been determined as 1.55 g phosphorus per 100 g dry cell weight. An optimized SYN6 medium was developed which is buffered at pH 6.4 with 140 mmol MES L−1. It provides non-limited cultivation conditions without by-product formation in shake flask cultivations. A maximal specific growth rate of 0.32 h−1 and short fermentations of 15 h were achieved. The rational definition of conditions for a non-limiting oxygen and phosphorus supply as well as the pH stabilization of the medium to non-inhibiting values provide basic conditions for A. adeninivorans cultures characterized by short fermentation times, a complete aerobic metabolism without anaerobic by-products or overflow-metabolites, maximized growth rates, and maximized biomass yields.
6 A. adeninivorans as Bio Compound in Biosensors for the Detection of Hormonal Activities
6.1 A. adeninivorans-Based Biosensors
The utilization of A. adeninivorans for the development of biosensors targeted for the detection of several hormones and pharmaceuticals has been consequently extended in the last years. Since the first evocation of an estrogen-detecting biosensor via a cell-based in vitro assay in 2006 (Hahn et al. 2006), 7 new studies describing the design of biosensors dedicated to estrogens, androgens, progestogens, glucocorticoids and pharmaceuticals using recombinantly produced human hormone receptors have been published (Kaiser et al. 2010; Pham et al. 2012, 2013, 2015, 2016; Gerlach et al. 2014; Chamas et al. 2015). Although most of these published works discuss the development of cell-based in vitro assays followed by enzymatically mediated detection, amperometric detection and fluorometric detection are also described. Additionally, the electrochemical detection of estrogens using an estrogen binding protein from yeast origin recombinantly produced in A. adeninivorans was also reported (Vijayan et al. 2015). Detection of hormones or pharmaceuticals in aqueous samples became of major interest as accumulating studies have shown the negative impact endocrine disruptors can have for the aquatic life (Rempel and Schlenk 2008). All natural and synthetic molecules which can bind to the vertebrate hormone receptors present a potential risk also for human health if released in an uncontrolled way into the environment (Kabir et al. 2015). Among the recently developed whole-cell biosensors, the ones using A. adeninivorans as host present some of the best performance and four of them are already commercially available for routine testing.
6.2 Principle of Action
In the majority of the recently developed A. adeninivorans based biosensors, a similar strategy was used for the detection of hormones. The first step was the construction of a modified A. adeninivorans strain by genomic integration of two expression modules responsible for the constitutive production of the desired human hormone receptor and for the ligand-induced expression of a reporter gene. The human hormone receptor will bind to the ligand and this binding event will be transformed in a measurable signal with the help of a reporter gene. To obtain the inducible mechanism of the reporter gene, a modified version of the A. adeninivorans-based GAA promoter was constructed by inserting two 15 bp sequences called hormone response element (HRE) at the position-107 of the promoter. HRE is a conserved DNA-binding site for the dimerized hormone receptor and only when this dimerized receptor is bound to the HRE the expression of the downstream gene can occur. In the case of the estrogen-related assays, HRE was replaced by the slightly different estrogen response element (ERE) sequence as it shows higher affinity to the estrogen receptor than HRE (Hahn et al. 2006). Hence, the two expression modules used for yeast transformation were TEF1 promoter—hormone receptor gene—PHO5 terminator and GAA[2xHRE or 2xERE]−107 promoter—reporter gene—PHO5 terminator. The mechanism of the biosensor is described in Fig. 7. If present in the cultivation medium, the hormone is transported into the cytoplasm of the modified A. adeninivorans cell by passive diffusion through the membrane. When bound to the constitutively expressed hormone receptor (HR) it induces a conformational change that allows the homodimerization of the receptor in the cytoplasm. The nuclear localization signal present in the receptor will then permit receptor homodimer translocation into the nucleus. Once in the nucleus, the ligand bound receptor dimer can bind to the HRE and activates the transcription of the reporter gene. Three different reporter genes have so far been utilized to produce a measurable signal: the Klebsiella-derived phytase K gene (phyK), the A. adeninivorans-derived tannase gene (ATAN1) and the Discosoma-derived dsRED gene. Both phyK and ATAN1 genes produce an extracellularly located enzyme which can be utilized in an enzymatic reaction. Successful transformation of the substrate into the desired reaction product can then be detected either by spectrometric or amperometric methods. While ATAN1 was successfully utilized for estrogen detection (Kaiser et al. 2010), phyK was solely used in all other biosensors utilizing enzymatically-based detection. Finally, the dsRED gene is responsible for the production of the cytoplasmic fluorescent protein dsRED which can be spectrofluorometrically detected at an emission wavelength of 582 nm if exposed to an excitation wavelength of 542 nm.
Principle of A. adeninivorans-based hormone detection assays. The hormone receptor is constitutively produced in the cytoplasm of the yeast cell. If present, hormones that enter the cytoplasm can bind to the hormone receptor. After ligand-induced homodimerization, the receptor dimer translocates into the nucleus and binds to the HRE present in the GAA promoter, thus inducing the production of the reporter protein
Several improvements have been made to the general mechanism of the assay in order to reliably detect pharmaceuticals. Two genes were integrated in the genome of A. adeninivorans cells to be expressed constitutively: the human arylhydrocarbon receptor gene (hAhR) and the human arylhydrocarbon receptor nuclear translocator gene (hARNT). Only the hAhR protein can bind to pharmaceuticals but it requires heterodimer formation together with hARNT in order to translocate into the nucleus. Additionally, the HRE sequence in the GAA promoter was replaced by the cyp1A1-derived core sequence that moderates the interaction between the promoter and the hAhR-hARNT heterodimer. Reporter gene in this case was phyK both for amperometric and enzymatically-mediated detection.
6.3 Performance of the Assays and Applications
Some of the major characteristics and performances of the developed A. adeninivorans-based biosensors are presented in Table 6. For each of them, the concentration of respective standard ligand giving a half-maximal response (EC50) as well as the limit of detection (LoD) towards this same ligand is indicated. It is also of note that for each class of target compounds, real samples from diverse origins were included in the studies.
One of the major advantages of whole-cell based assays over analytical detection methods is the possibility to determine the total equivalent hormone concentration of a sample. When LC-MS or GC-MS will quantify some standard compounds known to have hormone potency such as 17β-estradiol, the whole cell-based assays add the contribution of all compounds susceptible to bind to the hormone receptor, thereby giving a concentration corresponding to the equivalent concentration of the standard ligand which will have the same biological effect. This is in particular very interesting when considering the wastewater effluent from hospital or agricultural origin where many different compounds can be found and where the endocrine disrupting effect for example on the estrogen receptor may not be due to the 17β-estradiol only. Additionally, these biosensors can help regulation authorities to discover and identify new compounds with endocrine disrupting activity.
The described biosensors have also proven to be working with untreated samples and do not necessitate a time-consuming derivatization or extraction process. This is mainly due to the A. adeninivorans ability to tolerate high salt conditions and allows on-site implementation of biosensors like Estramonitor or Pharma in the wastewater treatment plant for direct monitoring of hormones or pharmaceuticals in effluent water samples.
The family of A. adeninivorans-based biosensors will expand in the next years as compounds like dioxins or bisphenols will be targets of new assays. The construction of such biosensors with modified recombinant human receptors is currently performed. Another perspective will be the combination of already developed biosensors with chromatographic assays for prior separation of compounds, especially when it is of interest to know, which compounds in the sample contribute to the total equivalent hormone concentration. That’s why future studies will aim to first separate a complex sample with the help of thin-layer chromatography before applying one of the developed A. adeninivorans biosensors in order to determine how many compounds possess hormonal activity and for which proportion of the total signal these compounds are responsible. Comparing the obtained retention factors with the retention factors of known chemicals will allow a first identification. Subsequently, transfer of the unknown compounds for analytical identification by mass spectrometry will then lead to complete description of a complex sample.
7 Conclusions
The dimorphic, asexual hemiascomycete A. adeninivorans is an attractive organism for both, basic and applied research and has been shown to have a high potential in interesting biotechnological applications due to the very broad range of substrates, which can be used as carbon and/or nitrogen sources, the growth and secretion characteristics as well as the thermo- and osmotolerance. A. adeninivorans has been intensively exploited as host for heterologous gene expression leading to transformants which, for example, can be used in the chemical industry as biocatalyst for the synthesis of chiral alcohols. Furthermore, the genome of this yeast contains a large pool of genes encoding specialized enzymes with useful biochemical properties making A. adeninivorans an interesting gene donor. In addition, A. adeninivorans emerged to be a powerful tool in bio-based assays for the determination of hormonal activities in environmental samples due to its high robustness combined with high ability in heterologous gene expression.
References
Aguilar, C.N., Rodriguez, R., Gutierrez-Sanchez, G., Augur, C., Favela-Torres, E., Prado-Barragan, L.A., Ramirez-Coronel, A., and Contreras-Esquivel, J.C. 2007. Appl. Microbiol. Biotechnol. 76: 47–59.
Bhat, T. K., Singh, B., and Sharma, O. P. 1998. Biodegradation. 9: 343–357.
Bischoff, F., Litwinska, K., Cordes, A., Baronian, K., Bode, R., Schauer, F., and Kunze, G. 2015. Appl. Environ. Microbiol. 81: 5497–5510.
Böer, E., Wartmann, T., Dlubatz, K., Gellissen, G., and Kunze, G. 2004a. Curr. Genet. 46: 269–276.
Böer, E., Wartmann, T., Luther, B., Manteuffel, R., Bode, R., Gellissen, G., and Kunze, G. 2004b. Antonie van Leeuwenhoek. 86: 121–134.
Böer, E., Gellissen, G., and Kunze, G. 2005a. Arxula adeninivorans In: Gellissen, G. (Ed) Production of Recombinant Proteins, Wiley-VCH Verlag GmbH & Co. KGaA. pp. 89–110.
Böer, E., Mock, H.P., Bode, R., Gellissen, G., and Kunze, G. 2005b. Yeast 22: 523–535.
Böer, E., Wartmann, T., Schmidt, S., Bode, R., Gellissen, G., and Kunze, G. 2005c. Antonie van Leeuwenhoek 87: 233–243.
Böer, E., Steinborn, G., Matros, A., Mock, H.P., Gellissen, G., and Kunze, G. 2007. FEMS Yeast Res. 7: 1181–1187.
Böer, E., Bode, R., Mock, H.P., Piontek, M., and Kunze, G. 2009. Yeast. 26: 323–337.
Böer, E., Breuer, F. S., Weniger, M., Denter, S., Piontek, M., and Kunze, G. 2011. Appl. Microbiol. Biotechnol. 92: 105–114.
Böttcher, F. and Samsanova, I.A. 1983. Systemische Fungizide und antifungale Verbindungen In: Lyr, H. and Polter, C. (Eds.) Systemische Fungizide und antifungale Verbindungen, Akademie Verlag. pp. 255–258.
Böttcher, F., Klinner, U., Köhler, M., Samsonova, I.A., Kapultsevich, J. and Bliznik, X. 1988. Verfahren zur Futterhefeproduktion in zuckerhaltigen Medien. DD 278 354 A1.
Bui, D. M., Kunze, I., Förster, S., Wartmann, T., Horstmann, C., Manteuffel, R., and Kunze, G. Appl. Microbiol. Biotechnol. 44: 610–619.
Bui, D.M., Kunze, I., Horstmann, C., Schmidt, T., Breunig, K. D., and Kunze, G. 1996. Appl. Microbiol. Biotechnol. 45: 102–106.
Büttner, R., and Bode, R. 1992c. J. Basic Microbiol. 32: 159–166.
Büttner, R., Bode, R., and Birnbaum, D. 1987. J. Basic Microbiol. 27: 299–308.
Büttner, R., Bode, R., Scheidt, A., and Birnbaum, D. 1988. Acta Biotechnol. 8: 517–525.
Büttner, R., Scheit, A., Bode, R., and Birnbaum, D. 1989. J. Basic Microbiol. 29: 67–72.
Büttner, R., Bode, R. and Birnbaum, D.. 1990a. Wiss. Z. Ernst-Moritz-Arndt-Univ. Greifswald, Math.-nat.wiss. Reihe 39: 1–21.
Büttner, R., Bode, R., Samsonova, I. A., and Birnbaum, D. 1990b. J. Basic Microbiol. 30: 227–231.
Büttner, R., Bode, R., and Birnbaum, D. 1991a. J. Basic Microbiol. 31: 423–428.
Büttner, R., Bode, R., and Birnbaum, D. 1991b. Zbl. Mikrobiol. 146: 399–406.
Büttner, R., Schubert, U., Bode, R., and Birnbaum, D. 1990c. Acta Biotechnol. 10: 361–370.
Büttner, R., Bode, R., and Birnbaum, D. 1992a. Zbl. Mikrobiol. 147: 225–230.
Büttner, R., Bode, R., and Birnbaum, D. 1992b. Zbl. Mikrobiol. 147: 291–296.
Chamas, A., Nieter, A., Pham, H. T., Giersberg, M., Hettwer, K., Uhlig, S., Simon, K., Baronian, K., and Kunze, G. 2015. Anal. Bioanal. Chem. 407: 8109–8120.
Field, J., Leyendeckers, M.J., Sierra-Alvarez, R., Lettinga, G., and Habets, L. H. 1991. Biotechnol. Bioeng. 37: 247–255.
Fiki, A.E., Metabteb, G. E., Bellebna, C., Wartmann, T., Bode, R., Gellissen, G., and Kunze, G. 2007. Appl. Microbiol. Biotechnol. 74: 1292–1299.
Gellissen, G. 2005. Production of Recombinant Proteins - novel microbial and eukaryotic expression systems, Wiley-VCH Verlag GmbH & Co. KGaA.
Gellissen, G., Kunze, G., Gaillardin, C., Cregg, J. M., Berardi, E., Veenhuis, M., and van der Klei, I. 2005. FEMS Yeast Res. 5: 1079–1096.
Gerlach, T., Knaust, J., Kaiser, C., Körner, M., Hettwer, K., Uhlig, S., Simon, K., Baronian, K., and Kunze, G. 2014. Sci. Total Environ. 490: 1073–1081.
Gienow, U., Kunze, G., Schauer, F., Bode, R., and Hofemeister, J. 1990. Zbl. Mikrobiol. 145: 3–12.
Hahn, T., Tag, K., Riedel, K., Uhlig, S., Baronian, K., Gellissen, G., and Kunze, G. 2006. Biosens. Bioelectron. 21: 2078–2085.
Jankowska, D.A., Faulwasser, K., Trautwein-Schult, A., Cordes, A., Hoferichter, P., Klein, C., Bode, R., Baronian, K., and Kunze, G. 2013a. J. Appl. Microbiol. 115: 1134–1146.
Jankowska, D.A., Trautwein-Schult, A., Cordes, A., Hoferichter, P., Klein, C., Bode, R., Baronian, K., and Kunze, G. 2013b. J. Appl. Microbiol. 115: 796–807.
Kabir, E.R., Rahman, M.S., and Rahman, I. 2015. Environ. Toxicol. Pharmacol. 40: 241–258.
Kaiser, C., Uhlig, S., Gerlach, T., Körner, M., Simon, K., Kunath, K., Florschütz, K., Baronian, K., and Kunze, G. 2010. Sci. Total Environ. 408: 6017–6026.
Kasprzak, J., Bischoff, F., Rauter, M., Becker, K., Baronian, K., Bode, R., Schauer, F., Vorbrodt, H.M., and Kunze, G. 2016. Biochem. Eng. J. 106: 107–117.
Kaur, P., Lingner, A., Singh, B., Böer, E., Polajeva, J., Steinborn, G., Bode, R., Gellissen, G., Satyanarayana, T., and Kunze, G. 2007. Antonie van Leeuwenhoek 91: 45–55.
Kunze, G., and Kunze, I. 1994a. Antonie van Leeuwenhoek 65: 29–34.
Kunze, I., and Kunze, G. 1994b. J. Eur. Microbiol. 212: 24 – 28.
Kunze, I., and Kunze. G. 1996. Arxula adeninivorans In: Wolf, K. (Ed) Nonconventional yeasts biotechnology. Springer Verlag Berlin Heidelberg, pp. 389–409.
Kunze, G., Pich, U., Lietz, K., Barner, A., Büttner, R., Bode, R., Conrad, U., Samsonova, I.A., and Schmidt, H. 1990. Wirts-Vektor-System und Verfahren zu seiner Herstellung. DD 298 821 A5.
Kunze, G., Gaillardin, C., Czernicka, M., Durrens, P., Martin, T., Böer, E., Gabaldon, T., Cruz, J.A., Talla, E., Marck, C., Goffeau, A., Barbe, V., Baret, P., Baronian, K., Beier, S., Bleykasten, C., Bode, R., Casaregola, S., Despons, L., Fairhead, C., Giersberg, M., Gierski, P.P., Hahnel, U., Hartmann, A., Jankowska, D., Jubin, C., Jung, P., Lafontaine, I., Leh-Louis, V., Lemaire, M., Marcet-Houben, M., Mascher, M., Morel, G., Richard, G.F., Riechen, J., Sacerdot, C., Sarkar, A., Savel, G., Schacherer, J., Sherman, D.J., Stein, N., Straub, M.L., Thierry, A., Trautwein-Schult, A., Vacherie, B., Westhof, E., Worch, S., Dujon, B., Souciet, J. L., Wincker, P., Scholz, U., and Neuveglise, C. 2014. Biotechnol. Biofuels. 7: 66.
Kurtzman, C. P., and Robnett, C.J. 2007. FEMS Yeast Res. 7: 141–151.
Lekha, P.K., and Lonsane, B.K. 1997. Adv. Appl. Microbiol. 44: 215–260.
Marino, J.P., McClure, M.S., Holub, D. P., Comasseto, J.V., and Tucci, F.C. 2002. J. Am. Chem. Soc. 124: 1664–1668.
Masaki, K., Kamini, N.R., Ikeda, H. and Iefuji, H. 2005. Appl. Environ. Microbiol. 71: 7548–7550.
Middelhoven, W.J. 1993. Antonie van Leeuwenhoek 63: 125–144.
Middelhoven, W.J., and van Doesburg, W. 2007. Antonie van Leeuwenhoek. 91: 191–196.
Middelhoven, W.J., Hoogkamer-Te Niet, M.C., and Kreger-Van Rij, N.J. 1984. Antonie van Leeuwenhoek. 50: 369–378.
Middelhoven, W.J., de Jong, I.M., and de Winter, M. 1991. Antonie van Leeuwenhoek 59: 129–137.
Middelhoven, W.J., Coenen, A., Kraakman, B., and Sollewijn Gelpke, M.D. 1992. Antonie van Leeuwenhoek 62: 181–187.
Pham, H.T.M., Giersberg, M., Uhlig, S., Hanke, G., Simon, K., Kunath, K., Baronian, K., and Kunze, G. 2012. Sens. Actuators B-Chem. 161: 137–145.
Pham, H.T.M., Kunath, K., Gehrmann, L., Giersberg, M., Tuerk, J., Uhlig, S., Hanke, G., Simon, K., Baronian, K., and Kunze, G. 2013. Sens. Actuat B-Chem. 185: 628–637.
Pham, H.T.M., Giersberg, M., Gehrmann, L., Hettwer, K., Tuerk, J., Uhlig, S., Hanke, G., Weisswange, P., Simon, K., Baronian, K., and Kunze, G. 2015. Sens. Actuat B-Chem. 211: 439–448.
Pham, H.T.M., Chamas, A., Nieter, A., Giersberg, M., Rutten, T., Gehrmann, L., Hettwer, K., Tuerk, J., Uhlig, S., Simon, K., Baronian, K., and Kunze, G. 2016. Sens. Actuat B-Chem. 223: 540–549.
Rauter, M., Prokoph, A., Kasprzak, J., Becker, K., Baronian, K., Bode, R., Kunze, G., and Vorbrodt, H.M. 2014. Appl. Microbiol. Biotechnol. 99: 4723–4733.
Rempel, M.A., and Schlenk, D. 2008. Int. Rev. Cell Mol. Biol. 267: 207–252.
Rösel, H., and Kunze, G. 1995. Curr. Genet. 28: 360–366.
Rösel, H., and Kunze, G. 1996. Yeast. 12: 1201–1208.
Rösel, H., and Kunze, G. 1998. Curr. Genet. 33: 157–163.
Samsonova, I.A., Böttcher, F., Werner, C., and Bode, R. 1989. J. Basic Microbiol. 29: 675–683.
Samsonova, I.A., Kunze, G., Bode, R., and Böttcher, F. 1996. Yeast 12: 1209–1217.
Sano, K., Fukuhara, H., and Nakamura, Y. Biotechnol. Lett. 21: 33–38.
Shinozaki, Y., Morita, T., Cao, X., Yoshida, S., Koitabashi, M., Watanabe, T., Suzuki, K., Sameshima-Yamashita, Y., Nakajima-Kambe, T., Fujii, T. and Kita-moto, H.K. 2013. Appl. Microbiol. Biotechnol. 97: 2951–2959.
Sietmann, R., Uebe, R., Böer, E., Bode, R., Kunze, G., and Schauer, F. 2010. J. Appl. Microbiol. 108: 789–799.
Song, C. E., Lee, J.K., Lee, S.H., and Lee, S.-g. 1995. Tetrahedron: Asymmetry 6: 1063–1066.
Steinborn, G., Gellissen, G., and Kunze, G. 2005. FEMS Yeast Res. 5: 1047–1054.
Steinborn, G., Gellissen, G., and Kunze, G. 2007a. FEMS Yeast Res. 7: 1197–1205.
Steinborn, G., Wartmann, T., Gellissen, G., and Kunze, G. 2007b. J. Biotechnol. 127: 392–401.
Stöckmann, C., Palmen, T.G., Schroer, K., Kunze, G., Gellissen, G., and Büchs, J. 2014. J. Ind. Microbiol. Biotechnol. 41: 965–976.
Stoltenburg, R., Lösche, O., Klappach, G., and Kunze, G. 1999. Curr. Genet. 35: 8–13.
Suzuki, K., Sakamoto, H., Shinozaki, Y., Tabata, J., Watanabe, T., Mochizuki, A., Koitabashi, M., Fujii, T., Tsushima, S. and Kitamoto, H.K. 2013. Appl. Microbiol. Biotechnol. 97: 7679–7688.
Tanaka, A. O., Nobuko; Fukui, Saburo. 1967. J. Ferment. Technol. 617–623.
Terentiev, Y., Breuer, U., Babel, W., and Kunze, G. 2004. Appl. Microbiol. Biotechnol. 64: 376–381.
Van der Walt, J.P., Smith, M.T., and Yamada, Y. 1990. Antonie van Leeuwenhoek 57: 59–61.
Vijayan, V., Giersberg, M., Chamas, A., Mehrotra, M., Chelikani, V., Kunze, G., and Baronian, K. 2015. Biosens. Bioelectron. 66: 379–384.
Wartmann, T., and Kunze, G. 2000b. Appl. Microbiol. Biotechnol. 54: 619–624.
Wartmann, T., and Kunze, G. 2003. Expression of the HSA and GFP gene in budding cells and mycelia of A. adeninivorans In: Wolf, K., Breunig, K. and Barth, G (Eds) Non-Conventional Yeasts in Genetics, Biochemistry and Biotechnology, Springer-Verlag.
Wartmann, T., Kruger, A., Adler, K., Duc, B. M., Kunze, I., and Kunze, G. 1995a. Antonie van Leeuwenhoek 68: 215–223.
Wartmann, T., Kunze, I., Duc, B.M., Manteuffel, R., and Kunze, G. 1995b. Microbiol. Res. 150: 113–120.
Wartmann, T., Rosel, H., Kunze, I., Bode, R., and Kunze, G. 1998. Yeast 14: 1017–1025.
Wartmann, T., Erdmann, J., Kunze, I., and Kunze, G. 2000a. Arch. Microbiol. 173: 253–261.
Wartmann, T., Gellissen, G., and Kunze, G. 2001. Curr. Genet. 40: 172–178.
Wartmann, T., Stephan, U.W., Bube, I., Böer, E., Melzer, M., Manteuffel, R., Stoltenburg, R., Guengerich, L., Gellissen, G., and Kunze, G. 2002. Yeast 19: 849–862.
Wartmann, T., Bellebna, C., Böer, E., Bartelsen, O., Gellissen, G., and Kunze, G. 2003a. Appl. Microbiol. Biotechnol. 62: 528–535.
Wartmann, T., Stoltenburg, R., Böer, E., Sieber, H., Bartelsen, O., Gellissen, G., and Kunze, G. 2003b. FEMS Yeast Res. 3: 223–232.
Watanabe, T., Shinozaki, Y., Yoshida, S., Koitabashi, M., Sameshima-Yamashita, Y., Fujii, T., Fukuoka, T. and Kitamoto, H.K. 2014. J. Biosci. Bioeng. 117: 325–329.
Wolf, K. 1996. Nonconventional yeasts in biotechnology, Springer-Verlag.
Wolf, K., Breunig, K. and Barth, G. 2003. Non-conventional yeasts in genetics, biochemistry and biotechnology, Springer-Verlag.
Yang, X.X., Wartmann, T., Stoltenburg, R., and Kunze, G. 2000. Antonie van Leeuwenhoek 77: 303–311.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media Singapore
About this chapter
Cite this chapter
Bischoff, F., Chamas, A., Litwińska, K., Matthes, F., Böer, E., Kunze, G. (2017). Applications of Blastobotrys (Arxula) adeninivorans in Biotechnology. In: Satyanarayana, T., Kunze, G. (eds) Yeast Diversity in Human Welfare. Springer, Singapore. https://doi.org/10.1007/978-981-10-2621-8_18
Download citation
DOI: https://doi.org/10.1007/978-981-10-2621-8_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-2620-1
Online ISBN: 978-981-10-2621-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)