Abstract
In this paper, a novel mathematical morphological approach is proposed, which is combined with an active threshold-based method for the identification of morphological features from images with poor qualities. The algorithm is very fast and needs low computing power. First, a mixed smooth filtering is designed to remove background noises. Second, an active threshold-based method is discussed to create a binary image to achieve rough segmentation. Third, some simple morphological operations, such as opening, closing, filling, and so on, are designed and applied to get the final result of segmentation. After morphological analysis, morphological features, such as contours, areas, numbers, locations, and so on, are obtained. Finally, the comparisons with other conventional methods validate the effectiveness, and an additional experimental result proves the repeatability of the proposed method.
This research is partly supported by the innovative research fund of aerospace, research fund for the program of new century excellent talents in Heilongjiang provincial university No. 1155-ncet-008 and the Natural Science Foundation of Heilongjiang Province under grant No. QC2015084, F201132.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Mathematical morphology
- Automatic image segmentation and analysis
- Active threshold
- Morphological operator
1 Introduction
In the middle of the 19th century, the invention of microscope made people can study on human fundamental biological processes at the cellular or organism level. Nowadays, computer-assisted automatic morphological analysis of cells and tissues from microscopy images, such as cell size, cell morphology, density of staining, and so on, are useful methods to reduce hand data recording and user’s errors for the detection and diagnosis of some diseases [1, 2] or some special vivo cell phenomenon [3]. But for cell microscopy images, problems remain for image segmentation, shape description and analysis, especially the automatic border detection of cell object boundaries as well as surface contours is still a difficult problem [4, 5]. All difficulties are due to the distinguishing qualities of microscopy imaging. It is known that:
-
There is a very low contrast in microscopy image, so that objectives are hard to be separated from background.
-
Cell edge in microscopy images is fuzzy and smooth, and moreover, cells may be contiguous and overlapped.
-
Noises such as salt and pepper noise, Gaussian white noise and remarkable electrical noise are worse than any other digital system.
-
Dust, little bubble, blink point, and blur may be zoomed in by microscopy views, so that cell images may be confused in microscopy images.
For the characters of microscopy images, numerous segmentation methods have, up to now, been proposed; their performances depend largely on the type of images to be processed and on the a priori knowledge relative to the object features. These methods can be roughly classified into several categories, according to the nature of handled features: threshold-based methods, contour-based methods, region-based methods, and methods based on the integration of these techniques.
The threshold-based methods [6, 7] try to separate pixels within or without cell bodies base on the histogram threshold of cell gray-scale. For example, double threshold way, minimum-error way, adaptive threshold way, Otsu way, and so on. Calculations are simple but poor quality images and complex backgrounds may affect the separation result.
The contour-based methods generally deal with the detection and localization of edge points by differential operators and associated regularization processes [8–10], Such as Laplacian operator, Sobel operator, LOG operator, Canny operator, and so on. But they often result in many unclosed frontiers, when they are applied to highly textured objects or weakly contrasted images. Recently, wavelets provide a powerful tool to let microscopy images be studied in nonlinear multiresolution [11, 12]. But, poor quality images and complex algorithms are still problems.
The region-based methods try to detect the areas sharing one or many homogeneity criteria [13, 14], for example, fuzzy C-means, K-means, Mean-Shift, and so on. But they often suffer from their poor localization of boundaries and from over-segmentation. Another interesting region-based technique is based on mathematical morphology [15–17], which is a branch of image analysis based on algebraic, set-theoretic and geometric principles. It is particularly useful in providing basic building blocks to more sophisticated imaging applications. Using mathematical morphology, image is filtered to either preserve or remove features of interest, sizing transformations can be constructed, and information relating to shape, form and size can be easily applied.
Among the mixed methods, one has to mention the integration of two or more methods to conquer the shortage oneself. Integration of threshold and region growing algorithm [18], integration of active contours and mathematical morphology tools [19], integration of wavelet and mathematical morphology algorithm [20, 21], and so on, can perform well.
Our approach in this paper is based on the integration of a region-based method (mathematical morphology) and an active threshold-based method for low quality microscopy images (non-uniform illumination, low contrast, blur or faint image). We based our work on this technique because it is very fast and requires lower computing power. So the final system can run on a very poor computer system, even on an embedded computer-based (such as FPGA, DSP) mini system.
2 Basic Operations in Mathematical Morphology
Morphology processing is a nonlinear filter, its basic concept is to use construction element called as probe to search image information and do morphological transform according to requirement, and so it can be called as morphological filtering. The basic morphological operations are erosion and dilation. Based on them, some important combined operations, such as opening, closing, opening residue and closing residue are defined. They are described as following [15, 16].
A binary image A (i, j) consists of a set of points with the value 1 or 0 at the point (i, j). Usually, let A be an input image, and B a structuring element.
Where the symbols \( \oplus \) and \( \Theta \) are the dilation and erosion operators. The erosion is also expressed by the dilation as:
The superscript C means the complement of the set. The important applications of the basic operations, the opening and the closing of a set A by a set B, are the following:
Where the symbols and are the opening and the closing operators, respectively. It is clear that an opening operation is anti-extensive, which has the ability to remove positive impulses and peaks smaller than the diameter of structuring element, and a closing operation can remove negative impulses and valleys.
3 Relevant Algorithms and Application
The overall procedure of identification of cell morphological features is demonstrated in Fig. 1.
3.1 Pre-processing
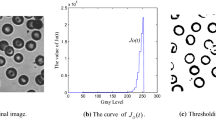
The red, green and blue (RGB) space of the original image is usually got, so images must be transformed to gray-scale first. The gray image (blood cell) is obtained and shown in Fig. 2(a). High noises and very low contrast usually exist, as discussed in Sect. 1. So, it is necessary to pre-process cell images to get rid of the drawbacks exposed above.
A mixed smooth filter is designed to remove background noises. Linear smooth filter can eliminate Gaussian noise and uniformly distributed noises effectively, but may import image blurring. And, median filter can suppress high-frequency noises, but not suit for eliminating Gaussian noise22. So, a combined filter mentioning the integration of a linear filter and a median filter is designed not only for eliminating the aforementioned noises and some kinds of additive noises, but also for reducing calculation volume. For the filter, a 5*5 model is designed, as shown in Eq. (6), and the combined filter can be expressed in Eq. (7). In equations, ai,j is the gray value at (i, j) in image; g(x, y) is the medium value of 5 elements; and MED is a sampling medium value operator.
In the pre-processing, smooth filtering is along the cross directions as shown in Eq. (6). This filter model can keep the details in horizontal and vertical directions and eliminate the details in diagonal line, which corresponds with man’s visual sense. After smooth filtering, g(x, y) is set as the new gray value at (i, j). Repeatedly, filtering is carried out in whole images. Base on this method, both effective noise suppression and simplifying calculation are satisfying. After smooth filtering, operate histogram equalization to increases the contrast of the image. The pre-processing result is as shown in Fig. 2(b).
3.2 Segmentation
The segmentation is in two steps, in the context of morphological segmentation, the initialization is achieved by means of active threshold-based method first to get the binary image by a primary segmenting, and the second step relates to the localization of boundaries by applying the morphological operators.
After applying pre-processing, the threshold value is calculated by a iteration process, as following:
① Getting initial threshold \( T_{0} \): \( T_{0} = \frac{1}{2}(Z_{\hbox{max} } + Z_{\hbox{min} } ) \).· \( Z_{\hbox{max} } \) and \( Z_{\hbox{min} } \) are maximum gray value and minimum gray value of images individually.
② Dividing image into background and foreground based on \( T_{k} \), and calculating the mean gray values of background and foreground, \( E_{a} \) and \( E_{b} \), individually.
③ Getting next new threshold \( T_{k + 1} \): \( T_{k + 1} = (E_{a} + E_{b} )/2 \).
④ If \( T_{k + 1} = T_{k} \), the best threshold is obtained, else turn to ② to continue the iterative loop. End the iterative operation, best threshold \( T_{n} \) is obtained:
In Eq. (8), i (i = 0, 1…, 255) is the gray value of a certain pixel; is the amount of the pixels, where the gray value is i; and is the n time and n-1 time iterative results individually.
Base on the best threshold, rough cells segmentation from background is done, and a binary image is shown in Fig. 2(c). The process can be expressed in Eq. (9):
\( T(i,j) \) is the gray value at \( (i,j) \) in the image; \( BW(i,j) \) is the result of binary operation at this point.
The second step of segmentation is mathematical morphological operation. In Fig. 2(c), it is clearly to see that some discontinuous concave convenes exist at the edge of cells, conglutination or overlap phenomenon appears among the cells, and fracture frontiers are still at cell edges. Moreover, holes in cells and specks in background are in wide distribution. So obeying Eqs. (4) and (5), an opening operation of basic morphological operations is applied (Structuring element is “disk, 4*4”), and a closing operation is executed subsequently (“disk, 2*2”). The result is shown in Fig. 3(a). But still, it is found that bigger holes in cells and bigger specks still exist. So, Morphological reconstructions are operated to solve these problems. Border objects are removed firstly. Next, 8-connected domain is selected to search the boundary of cells and the background, then set background as “0” and the other as “1” to finish the segmentation, as shown in Fig. 3(b).
3.3 Morphological Analysis
Obeying the 4-neighbourhood rule to explore the whole binary image to identify the separate connected regions (set cell regions to “1”, set background to “0”), numbered 1~N, N is the total amount of cell regions. Counting the pixels amount of “1” (named n) to calculate the average area of cells m, m = n/N. If the area of a separate connected region is smaller than 1/3 m (Depend upon the given cell species), this region should be regarded as a false cell and set to “0” to be removed. After removing false cells, new real amount and average area of all cell regions are computed, marked N1, \( \overline{m} \). The following step is to get morphological features of cells. Gravity model approach is adopted to get the center point of each cell, which is significant for position finding and tracing of cells (Fig. 4(a)). For example, if one cell region numbered n, center point \( (x_{n} ,y_{n} ) \) is got from Eq. (10).
Next, counting the areas of all close regions, if one is bigger than \( 5/3\overline{m} \) (Depend upon the given cell species), which is regarded as a remarkable cell. After that, cell information such as serial number, location of the center point, and area is recorded. This information may indicate abnormal cells or cell proliferation. Finally, 8-connected domain is selected to extract the cell contour, as shown in Fig. 4.
4 Results and Comparisons
Cell morphological features are obtained after the proposed mathematical morphological operation in Sect. 3. For the experimental cell image, information is got as following: Pixels: 512*512; Amount: 47 cells; Average area of cells: 1554; Biggest area: 3469, No. 20, (207, 384). Abnormal cells: No. 20, 24, 38 and 46. Moreover, base on the algorithm, lots of other cell morphological features can be learned easily, such as cell tracing, cell growth analysis and so on.
In this paper, it is confirmed that identification of cell f morphological features from microscopy images applying proposed mathematical morphological algorithm is better than of the conventional methods. The comparison is show in Fig. 5.
In Fig. 6, all results are processed from Fig. 2. Hereinto, (a) is got by applying the Sobel operation, (b) is got by applying the Laplacian operation, (c) is got by applying the Canny operation, and (d) is got by applying the proposed method. It is easy to see that (a) and (b) do not got the cell edges for the potential reasons of low contrast and smooth edges in microscopy images. And, (c) represents too many edges and unclosed frontiers for the potential reasons of terrible noises and interferences such as dust, little bubble, blink point, and blur in microscopy images. In (d), by applying the proposed method, clear cell edges are detected and cell morphological features are identified. It is indicated that the proposed method can perform much better for the identification of cell morphological features from microscopy images than the others do obviously.
To prove the universal property of method, another experiment is done for the identification of one endothelial cell image. The result is shown in Fig. 6. Figure 6(a) is the original image of endothelial cells, and (b), (c) are the results by applying the same proposed method. It is clear to see that the proposed method performs well too.
5 Conclusions and Expectations
For the identification cell morphological features from microscopy images with poor qualities, we proposed an attractive mathematical morphological approach combining with an active threshold-based method. This method can be taken into both account some shortages of microscopy image and morphological characters of cells. By applying some simple morphological operations, such as opening, closing and filling, the cell morphological features are identified. The results not only demonstrate the validity of this approach for the identification of cell morphological features from microscopy images, but also highlight the need for techniques such as these by illustrating the ambiguity associated with the conventional and manual measurement of such properties. Especially, the algorithm is very fast and requires lower computing power. So that, a mini automatic imaging and analysis system (FPGA&DSP-based) based on the algorithm is under design.
References
Leland, D.S., Ginocchio, C.C.: Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 20, 49–78 (2007)
Hall, K.K., Lyman, J.A.: Updated review of blood culture contamination. Clin. Microbiol. Rev. 19, 788–802 (2006)
Wingate, R., Kwint, M.: Imagining the brain cell: the neuron in visual culture. Nat. Rev. Neurosci. 7, 745–752 (2006)
Stephens, D.J., Allan, V.J.: Light microscopy techniques for live cell imaging. Science 300, 82–87 (2003)
Germain, R.N., Miller, M.J., Dustin, M.L., et al.: Dynamic imaging of the immune system: progress, pitfalls and promise. Nat. Rev. Immunol. 6, 497–507 (2006)
Sahoo, P.K., Soltam, S., Wong, A.K., et al.: A survey of thresholding techniques. Comput. Vis. Graph. Image Process. 41, 233–260 (1988)
Otsu, N.: A threshold selection method from gray-level histogram. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979)
Canny, J.: A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698 (1986)
Witkin, A.P.: Scaling-space filtering. In: International Joint Conferences on Artificial Intelligence, vol. 2, pp. 1019–1022 (1983)
Marr, D., Hildreth, E.: Theory of edge detection. Proc. R. Soc. Lond. B207, 187–217 (1980)
Mallat, S.: multifrequency channel decompositions of images and wavelet models. IEEE Trans. Acoust. Speech Signal Process. 37, 2091–2110 (1989)
Mattlat, S.: A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans. Pattern Anal. Mach. Intell. 7, 674–693 (1989)
Cheng, Y.: Mean shift, mode seeking, and clustering. IEEE Trans. Pattern Anal. Mach. Intell. 17, 790–799 (1995)
Comanieiu, D., Meer, P.: Mean Shift analysis and applications. In: Proceedings of the Seventh IEEE International Conference on Computer Vision, vol. 2, pp. 1197–1203 (1999)
Serra, J.: Introduction to Mathematical Morphology. Comput. Vis. Graph. Image Process. 35, 283–305 (1986)
Breen, E.J., Jones, R., Talbot, H.: Mathematical morphology: a useful set of tools for image analysis. Stat. Comput. 10, 105–120 (2000)
Lee, J.R., Smith, M.L., Smith, L.N., et al.: A mathematical morphology approach to image based 3D particle shape analysis. Mach. Vis. Appl. 16, 282–288 (2005)
Pavlidis, T., Liow, Y.T.: Integrating region growing and edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 12, 225–233 (1990)
Elmoataz, A., Schupp, S., Clouard, R., et al.: Using active contours and mathematical morphology tools for quantification of immunohistochemical images. Signal Process. 71, 215–226 (1998)
Goutsias, J., Heijmans, H.: Nonlinear multiresolution signal decomposition scheme-part I: morphological pyramids. IEEE Trans. Image Process. 11, 1862–1876 (2000)
Heijmans, H., Goutsias, J.: Nonlinear multiresolution signal decomposition scheme-part II: morphological wavelets. IEEE Trans. Image Process. 11, 1897–1913 (2000)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this paper
Cite this paper
Xia, W., Hu, Z., Zhai, H., Kang, J., Song, J., Sun, G. (2016). A Novel Approach for the Identification of Morphological Features from Low Quality Images. In: Che, W., et al. Social Computing. ICYCSEE 2016. Communications in Computer and Information Science, vol 623. Springer, Singapore. https://doi.org/10.1007/978-981-10-2053-7_4
Download citation
DOI: https://doi.org/10.1007/978-981-10-2053-7_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-2052-0
Online ISBN: 978-981-10-2053-7
eBook Packages: Computer ScienceComputer Science (R0)