Abstract
N-acyl homoserine lactone (AHLs) produced by bacteria play a unique role in altering the expression of plant defence genes. AHL signals are constitutively produced by the vast majority of the rhizosphere and other groups of bacteria; and also varied levels of plant response are elicited through different types of AHL signals. Moreover, the defence mechanism of AHL-induced ISR is distinct from other bacterial compound-mediated plant response. It was also evident that the response of plants to bacterial AHLs may depend on plant species and chemical structure of AHLs. However, the question of how plants perceive the AHLs and distinguish between those molecules remains open. To date, no information is available either about plant AHL receptors or how plant cells can incorporate AHL signal molecules. Even though plants produce compounds similar to AHL signals, the precise source, structure and biological significance of these AHL mimics from plants are currently unknown. The specificity of plant mimics to stimulate or inhibit different types of AHL signals needs to be addressed. A thorough understanding of how plants perceive and respond to AHLs needs to be investigated. Copious questions remain to be addressed for the better understanding of quorum sensing of bacteria and trans-kingdom interactions of AHLs with plant cells.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
9.1 Introduction
Rhizosphere microbiome influence plant growth and development; therefore, a collaborative action is essential for establishment of an efficient plant-bacteria interaction. Bacteria employ a variety of chemical molecules as their signals for communication across interspecies, intraspecies and intra-kingdom (Atkinson and Williams 2009). The bacterial communication is mediated by the exchange of small extracellular chemical signals which influence bacterial gene expression and physiological behaviour in a density-dependent signalling mechanism termed quorum sensing (QS). Among them, the best-studied QS mechanisms are from Gram-negative Proteobacteria, which use distinct group of biologically active metabolites, namely, N-acyl homoserine lactone (AHL) autoinducers as signal molecules (Swift et al. 1999; Whitehead et al. 2001). The QS-controlled phenotypes play a vital role for successful bacteria-host interactions, whether symbiotic or pathogenic (Boyer and Wisniewski-Dye 2009). The ecological distribution of AHL producers in natural environments and their potential roles have attracted much attention, and hence the diversity and distribution of AHL producers have been explored in different eco-niches (Cha et al. 1998; Huang et al. 2013; Lv et al. 2013; Viswanath et al. 2015), especially the rhizosphere regions which were reported to harbour high AHL population (DeAngelis et al. 2008; Elasri et al. 2001; Viswanath et al. 2015). The rhizosphere-associated AHL producers play a crucial role in plant health and growth and influence phenotypes such as root colonization and induction of systemic resistance in plants (Hartmann et al. 2004, 2014; Pang et al. 2009). Despite the intense study of AHL signalling in biocontrol bacteria, namely, Pseudomonas spp. (Wood et al. 1997; Chin-A-Woeng et al. 2001; De Maeyer et al. 2011), Rhizobium spp. (Wisniewski-Dye and Downie 2002) and Serratia spp. (Van Houdt et al. 2007), there is only limited information on AHL-dependent regulation in other beneficial plant-associated rhizobacteria. The ability of plant growth-promoting rhizobacteria (PGPRs) such as Pseudomonas, Serratia, Bacillus and non-pathogenic Fusarium oxysporum has been reported to promote plant health mediated through induced systemic resistance (ISR) (Kloepper et al. 2004; De Vleesschauwer and Hofte 2009). Similarly AHL-producing PGPRs triggered induced systemic resistance which had profound effect on the modulation of plant development and defence activity (Hartmann et al. 2014; Schenk and Schikora 2015). The AHL signalling molecules elicit plant response by systemic induction of defence gene expression especially against biotic stress (Schuhegger et al. 2006). To date, reports on diversity of AHL producers among PGPR and AHL-elicited ISR response in multiple plant species are limited. This chapter discusses the current status on the diversity of AHL bacterial communities associated with plant rhizosphere and the mechanism of AHL-elicited ISR-mediated defence response during plant-microbe interaction.
9.2 Induced Systemic Resistance in Plants
Plants develop local defence response due to colonization of beneficial bacteria or infection by pathogenic bacteria, which triggers immunity by the recognition of microbe-associated molecular patterns (MAMP) or effector proteins, resulting in systemic resistance. The two best understood mechanisms of systemic resistance are systemic acquired resistance (SAR) and induced systemic resistance (ISR). The SAR-mediated induced resistance is acquired upon local induction by a pathogen, whereas ISR is triggered by plant-associated beneficial microbes (Berendsen et al. 2012; Pieterse et al. 2014; Schenk et al. 2014). In both the systems, plants activate an elaborate matrix of signal transduction pathways via phytohormones such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET) and abscisic acid (ABA) which act as key signalling molecules.
Induced systemic resistance (ISR) of plants against pathogens has been intensively investigated with respect to the underlying signalling pathways involved in defence response as well as its potential use in plant protection (Choudhary et al. 2007; Heil and Bostock 2002). In plants, ISR defence response is elicited by diverse bacterial determinants including bacterial surface components (flagellin, lipopolysaccharides and exopolysaccharides), volatile organic compounds (acetoin and 2,3-butanediol) and bacterial secondary metabolites (2,4-diacetylphloroglucinol (DAPG) and pyocyanin) (De Vleesschauwer and Hofte 2009; Kloepper et al. 2004; Ryu et al. 2004). The triggered ISR activates defence response through various mechanisms, viz. induction of pathogenesis-related proteins (PRP), phytoalexins, cell wall reinforcement and priming defence responses (Beckers et al. 2009; Ryu et al. 2003; Slaughter et al. 2012; Van Wees et al. 2008).
9.3 N-Acyl Homoserine Lactone (AHL)
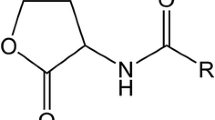
Bacteria use small chemical molecules to synchronize gene regulation within a population in a process called quorum sensing (Bassler 1999). The AHLs are the most common signal molecules used exclusively by Gram-negative bacteria. These molecules are composed of a fatty acyl chain ligated to a lactonized homoserine through an amide band. The length of the acyl side chain ranges from 4 to 18 carbon atoms, and based on the length of the acyl groups, AHLs can be broadly classified as short- or long-chain molecules. Short-chain AHLs have 4–8 carbon atoms in the acyl moiety, while long-chain AHLs have 10–18 carbons. The acyl side group can be substituted with an oxo or hydroxyl group at position C3 which confers signal specificity (Fuqua and Greenberg 2002; Thiel et al. 2009; Waters et al. 2008; Waters and Bassler 2005). Short-chain AHLs are believed to freely diffuse across the cell membrane, whereas AHLs with longer acyl side chains require multidrug efflux pump for transportation (Kaplan and Greenberg 1985; Pearson et al. 1999; Whitehead et al. 2001).
The canonical AHL QS involves two regulatory genes, a luxI family of AHL synthase genes and a luxR family of AHL-responsive transcriptional regulatory genes. Homologous to luxI/luxR QS system have been described in several Gram-negative bacteria, although the AHLs produced by the LuxI homologues as well as the genes regulated by them vary at the species or strain level (Whitehead et al. 2001).
The genes encoding these two proteins are often located adjacent to one another on the chromosome in almost all the AHL-producing proteobacteria (Fuqua et al. 1996; Churchill and Chen 2011; Gelencser et al. 2012). The LuxI proteins synthesize AHL signal molecules using the substrate S-adenosyl methionine for the backbone lactone ring, and acylated carbon chain from fatty acid biosynthesis pathway (Schaefer et al. 1996). LuxR-like proteins are transcriptional regulators which recognize the cognate AHL signals and mediate either activation or repression of QS-dependent gene expression (Fuqua and Winans 1994; Fuqua et al.1996). Also, the activated LuxR proteins up regulate luxI transcription and enhance the rate of AHL synthesis (Fuqua et al. 1996, 2001; Case et al. 2008). The recognition of bacterial AHL receptors to their corresponding AHL signals is highly specific, and hence the AHLs are classified as intraspecies signals among the proteobacteria (Huse and Whiteley 2011; Taga and Bassler 2003). A generalized scheme for an AHL quorum-sensing circuit in a bacterial cell is shown in Fig. 9.1.
9.4 Diversity of AHL-Producing Rhizosphere Bacterial Communities
The rhizosphere habitat provides a favourable environment for QS signalling since it contains significantly higher densities of microorganisms. A wide range of plant-associated PGPR, symbiotic, endophytic, epiphytic and pathogenic bacteria regulate their physiological functions through AHL signals (Ortiz-Castro et al. 2009; Venturi and Fuqua 2013). Recent studies indicate that AHL-based QS is highly prevalent in rhizosphere and endophytic communities of plants (Schaefer et al. 2013). The diversity of AHL-producing bacteria in the rhizosphere-associated bacterial communities of different plant species has been extensively studied and was represented only by the proteobacteria. In general, the proteobacteria group constitutes an estimated two thirds of many temperate plant rhizospheres (Hawkes et al. 2007). AHL-producing proteobacteria have been found to be more common in the rhizosphere than bulk soil (Cha et al. 1998; Elasri et al. 2001; d’Angelo-Picard et al. 2005).
In addition, several endophytic and epiphytic bacteria are also known to produce AHLs; however QS-dependent behaviours are poorly understood in these bacterial groups (Lv et al. 2013; Schaefer et al. 2013). The AHL-producing rhizobacteria were represented by α-, β- and γ-proteobacteria, isolated from the rhizospheres of tobacco (d’Angelo-Picard et al. 2005), potato, strawberry, oilseed rape (Berg et al. 2002), tomato (Steidle et al. 2001), wild oats (De Angelis et al. 2008), wheat (Pierson et al. 1998), cottonwoods (Schaefer et al. 2013), citrus (Trivedi et al. 2011), paddy (Steindler et al. 2008; Vial et al. 2006), cocoyam (De Maeyer et al. 2011), finger millet (Sekar and Prabavathy 2014), mangrove (Viswanath et al. 2015) and wetland plants (Zeng et al. 2014). The majority of the AHL-producing isolates from the plant rhizospheres belonged to the genera Pseudomonas, Rhizobium, Serratia, Burkholderia, Erwinia and Pantoea (Cha et al. 1998; d’Angelo-Picard 2005; Viswanath et al. 2015). The diversity of AHL-producing rhizobacteria was found to be ecological niche specific, e.g. majority of the AHL producers isolated from the mangrove rhizosphere were represented by the genera Vibrio, Halomonas and Photobacterium which were absent in the agriculture plant crops. Similarly, the genera Rahnella, Pantoea, Enterobacter, Erwinia and Burkholderia isolated from agriculture crops were not represented in the mangrove and other wetland rhizospheres (Viswanath 2015; Viswanath et al. 2015; Zeng et al. 2014).
The AHL signalling molecules produced by the rhizosphere bacteria varied from short to long chains and are reported to produce more than one type of AHL molecules, and its AHL profile was not strictly conserved at the genus or species levels. Even though rhizobacterial isolates produced similar group of AHL molecules, the role of AHLs involved in the regulation of phenotypes differed from strain to strain (Fuqua and Greenberg 2002; Gonzalez and Keshavan 2006; Venturi and Subramoni 2009). For example, in Serratia marcescens MG1, C6-HSL regulated swarming motility and biofilm formation, whereas in S. marcescens SS1, the same AHL regulated sliding motility and prodigiosin production (Eberl et al. 1996; Horng et al. 2002). The distribution of AHL molecules among the rhizobacteria was species or strain dependent. This could be due to the acquisition of AHL homologue genes through horizontal gene transfer (Gray and Garey 2001; Lerat and Moran 2004). The production of a similar type of AHL molecules in different genera might help interspecies communication in the natural environment where mixed communities are often represented (Atkinson and Williams 2009). Although the diversity of AHL-producing rhizobacteria has been explored in recent times, still identifying a myriad of AHL signals from bacteria inhabiting diverse plant species is less studied. The predominant occurrence of AHL producers in rhizosphere suggests that AHL QS might be a trait of significant importance in bacterial growth and colonization in the rhizosphere. Therefore, research focus to understand the ecological roles of AHLs in plant-bacteria interaction is needed.
9.5 Interactions of AHL with Plants
In recent years, numerous studies have shown that plants also have evolved means to perceive and respond to AHL signal molecules produced by bacteria. Many of the AHL-regulated phenotypes in bacteria such as biofilm formation, motility and antibiotic and biosurfactant production have profound impact on plant health. Recent reports have revealed that plants have marked response to the AHL signals produced by its associated microbiome. The first indication of plant responses to bacterial AHLs was studied in the legume Phaseolus vulgaris (Joseph and Phillip 2003) and in Medicago truncatula (Mathesius et al. 2003). The exposure of AHLs from symbiotic Sinorhizobium meliloti or pathogenic Pseudomonas aeruginosa at nano to micromolar concentrations induced significant changes in defence and stress management genes and accumulation of over 150 proteins (Mathesius et al. 2003). The influence of AHL molecules in plant defence response was established during interaction of Serratia liquefaciens MG1 and tomato plants (Hartmann et al. 2004; Schuhegger et al. 2006). The rhizobacteria S. liquefaciens MG1 produced short-chain AHLs C4- and C6-HSL when colonizing the tomato root surface, which induced systemic resistance against the leaf-pathogenic fungus Alternaria alternata. The AHLs increased salicylic acid concentration and also induced the ethylene and salicylic acid-dependent defence genes. Similarly, 3-oxo-C6-HSL producing Serratia plymuthica HRO-C48 elicited defence response against damping-off disease caused by Pythium aphanidermatum in cucumber plants and grey mould-causing fungus Botrytis cinerea in tomato and bean plants (Liu et al. 2007; Pang et al. 2009). The production of 3-oxo-C14-HSL by Ensifer meliloti (Sinorhizobium meliloti) associated with Arabidopsis plant roots showed resistance against Pseudomonas syringae (Zarkani et al. 2013). Likewise, Hernandez-Reyes et al. (2014) described the induction of systemic resistance by 3-oxo-C14-HSL-producing S. meliloti in tomato, barley and wheat plants against diverse pathogens. In addition, constitutive expression of AHL genes in transgenic tobacco plants applied with rhizobacterium S. marcescens 90–166 showed increased induced systemic resistance against bacterial pathogens Pectobacterium carotovorum subsp. carotovorum and P. syringae pv. tabaci (Ryu et al. 2013).
Also, the application of synthetic AHLs at a concentration range of 1–10 μM to roots in an axenic system was shown to induce resistance in diverse plants. Tomato plants treated with C4 or C6-HSL directed a systemic induction of genes involved in defence (Schuhegger et al. 2006). Schikora et al. (2011) demonstrated increased systemic resistance against obligate biotrophic fungi Golovinomyces orontii in Arabidopsis thaliana and against Blumeria graminis f. sp. hordei in H. vulgare (barley) plants when treated with synthetic 3-oxo-C14-HSL and 3-oxo-C12-HSL. In addition, the molecule 3-oxo-C14-HSL treated A. thaliana plants showed more resistance towards the hemibiotrophic bacterial pathogen P. syringae pv. tomato DC3000 (Schikora et al. 2011). Likewise, 3-OH-C14-HSL and 3-oxo-C12-HSL showed similar level of defence response against biotic stress in A. thaliana but comparatively weaker than 3-oxo-C14-HSL (Schikora et al. 2011). The degraded product of AHL, namely, homoserine lactone, when added to legume P. vulgaris roots at a concentration of 10 nM increases stomatal conductance and transpiration (Joseph and Phillips 2003). In Trifolium repens (white clover), transcriptional analysis indicated that treatment with 3-oxo-C12-HSL increased transcription of elements associated with auxin-responsive promoters (Mathesius et al. 2003). In Arabidopsis, short-chain (C4- and C6-) AHLs increased the plant’s hormone auxin/cytokinin ratio, which resulted in root elongation (von Rad et al. 2008). Ortiz-Castro et al. (2008) demonstrated that C10-HSL elicited developmental changes in the root system in Arabidopsis plants by altering the expression of cell division and differentiation-related genes, and C12-HSL strongly induced root hair formation. Furthermore, treatment with 3-oxo-C6-HSL and 3-oxo-C8-HSL promoted root elongation in Arabidopsis at concentration range of 1–10 μM (Jin et al. 2012; Liu et al. 2012). In Vigna radiata, 3-oxo-C10-HSL induced the formation of adventitious roots (Bai et al. 2012). H. vulgare (L.) and Pachyrhizus erosus L. (yam bean) plants treated with C6-, C8- and C10-HSL triggered tissue- and compound-specific changes in the activity of important detoxification enzymes (Gotz-Rosch et al. 2015).
The plant response to bacterial AHL signals is dependent on the type of AHL molecules. The length of the AHL side chain is essential for its effect on plants; for example, C4-HSL, C6-HSL, 3-oxo-C6-HSL and 3-oxo-C8-HSL promoted growth in Arabidopsis and barley (Gotz et al. 2007; Liu et al. 2012; Schenk et al. 2012; von Rad et al. 2008), whereas 3-oxo-C10-HSL induced the formation of adventitious roots in mung beans (Bai et al. 2012). On the other hand, C6- and 3-oxo-C6-HSL induced systemic resistance in tomato, cucumber and barley (Pang et al. 2009; Schikora et al. 2011; Schuhegger et al. 2006), while 3-oxo-C12- and 3-oxo-C14-HSL were reported to have resistance-inducing attributes in A. thaliana and M. truncatula (Mathesius et al. 2003; Schikora et al. 2011). The apparent different reactions to long and short chain HSLs may suggest that different receptors or at least different signalling pathways are involved in these responses. Moreover, the reaction of plants to AHLs might also depend on the specific plant-AHL combination. Different acyl length chains of AHL that induce systemic resistance and growth promotion in plants are shown in Fig. 9.2.
Furthermore, plants also control the bacterial QS system by producing compounds that mimic AHL signals. Higher plants including Pisum sativum (pea), Solanum lycopersicum (tomato), Oryza sativa (rice) and M. truncatula secrete compounds that either stimulate or inhibit AHL responses (Bauer and Mathesius 2004; Degrassi et al. 2007; Gao et al. 2003; Perez-Montano et al. 2013; Teplitski et al. 2000).
9.6 Mechanisms of AHL Interaction in Plants
Although the response of plants to AHLs has been more extensively studied, understanding the molecular mechanisms of how plants perceive and respond to AHLs is still unclear. Very recently, possible mechanisms have been proposed to show how AHL signals influence plant defence and reinforce resistance in different plants against bacterial and fungal pathogens.
9.6.1 AHL in Plant Defence
The AHL signals use “priming” mechanisms for the induction of defence response in plants. The following possible mechanisms of plant defence response triggered by AHL molecules have been postulated:
-
1.
Induction of SA-dependent pathway – The AHLs that triggered immune response in plants are activated through SA-mediated systemic resistance. The induction of systemic resistance in tomato against fungal leaf pathogen A. alternata is enhanced due to the increased levels of SA when treated with AHL-producing rhizobacterium S. liquefaciens MG1. Also, enhanced expression of pathogenesis-related 1a (PR1a) and two chitinase genes involved in SA-/ET-dependent pathways were identified in tomato leaves when C6-HSL or C4-HSL was applied to the roots of tomato plants. This strongly emphasizes that the systemic response mediated by short-chain AHL signals in tomato plant functions via an SA-dependent pathway (Schuhegger et al. 2006).
-
2.
Induction via oxylipin-/salicylic acid (SA)-dependent pathway – The oxylipin cis-OPDA, a precursor of JA, and SA involved in plant defence response are elicited by AHL signal molecules (Schenk et al. 2014). The 3-oxo-C14-HSL-treated Arabidopsis plant showed increased accumulation of SA and cis-OPDA on leaves, which resulted in enhanced expression of heat shock proteins, GST6, GSTU19 encoding HSP70 and HSP17 genes and the cytochrome P450-encoding CYP81D11 gene. The lack of enhanced expression of JA-dependent genes such as MYC2 and VSP2 and the ET-dependent genes PR3, ERF5 and ETR1 showed that AHL-treated Arabidopsis plants are independent of JA/ET pathway (Schenk et al. 2014).
-
3.
Induction of stomatal defence response – The induction of SA/cis-OPDA pathway enhanced the stomatal defence response in 3-oxo-C14-HSL-treated Arabidopsis plants when encountered with the bacterial pathogen P. syringae DC3000 pathovar tomato (Pst). Stomatal responses such as an increased rate of stomatal closure and reduced open stomata were observed in AHL-pretreated plants (Schenk et al. 2014). The stomatal defence response in AHL-treated plants was independent of ABA pathway, which was revealed by the lack of RD22, RD29 and RAB18 gene (Montillet et al. 2013; Schenk et al. 2014).
-
4.
Induction via mitogen-activated protein kinase (MAPK) – The 3-oxo-C14-HSL-treated Arabidopsis plant roots induced systemic resistance through altered activation of MAPKs. AHL-treated plants inducted with pathogen-associated molecular pattern (PAMP) flg22 showed altered activation of MAPKs, AtMPK3 and AtMPK6. Further, the altered MAPKs induced high expression of the defence-related WRKY22 and WRKR29 transcription factors, as well as the pathogenesis-related 1 (PR1) gene (Schikora et al. 2011).
-
5.
Induction via cell wall reinforcement – In 3-oxo-C14-HSL-treated Arabidopsis plants, increased level of cell wall components such as callose, phenolic compounds and lignin was observed, which induced resistance through cell wall reinforcement (Schenk et al. 2014). When encountered with fungal plant pathogen B. graminis f. sp. hordei, 3-oxo-C14-HSL-treated barley plants showed induced resistance by the formation of papilla (cell wall apposition) structures, as a result of reactive oxygen species (ROS) accumulation (Schikora et al. 2011). Likewise, inoculation with 3-oxo-C14-HSL-producing S. meliloti, as well as pretreatment with the pure 3-oxo-C14-HSL molecule, primed barley and wheat plants for enhanced reactive oxygen species (ROS) production, which resulted in papilla formation and hence induced the defence response (Hernandez-Reyes et al. 2014).
9.6.2 Role of AHL in Plant Development
The growth promotion activity mediated by AHL signal molecules majorly depended on the induction of phytohormone auxin (von Rad et al. 2008; Bai et al. 2012; Liu et al. 2012). The proteome analysis in AHL-treated M. truncatula plants showed the accumulation of several auxin-induced proteins and enzymes involved in auxin metabolism (Mathesius et al. 2003). Furthermore, the exposure of 3-oxo-C12-HSL to the roots of transgenic T. repens plants with a β-glucuronidase (GUS) reporter gene under the promoters auxin-responsive GH3 promoter and three chalcone synthases substantially increased the expression of auxin-responsive and flavonoid synthesis proteins (Mathesius et al. 2003).
The 3-oxo-C6- and 3-oxo-C8-HSL induced root elongation in Arabidopsis, eventually by the elevated expression of two G-protein receptors, namely, Cand2 and Cand7, which are involved in the activation of signal transduction pathways (Jin et al. 2012; Liu et al. 2012). The addition of 3-oxo-C10-HSL to the roots of mung bean actively accelerated the adventitious root formation by the induction of auxin metabolism via increased accumulation of H2O2, NO and cGMP (Bai et al. 2012).
The response to bacterial AHL QS molecules was very well understood in three different plant species, namely, Arabidopsis, barley and tomato. Two different response patterns, i.e. defence and growth stimulation, are induced by long-side-chain or short-side-chain HSLs, respectively. In accordance with plant response and interruption of AHL signals, it becomes clear that AHL signalling is an important factor in determining the outcome of plant-bacteria interaction. Moreover, production of AHL signal molecule is apparent in total microbiome of plants, including rhizobacteria, epiphytic and endophytic bacteria (Hosni et al. 2011; Kimura 2014; Lv et al. 2013). The AHL signals found in many species of legume-nodulating rhizobia are known to regulate phenotypes, including nodulation, nitrogen fixation, growth rate and polysaccharide production, which are all important for the establishment of a successful bacteria-plant symbiosis (Gonzalez and Marketon 2003). The current understanding of plant interaction with bacterial AHLs was limited to only AHL-producing PGPR strains; it is necessary to explore the role of other AHL-producing bacteria associated with plants. The plant growth is also influenced by AHL derivatives, which are obtained by the hydrolysis of AHL molecules through plant enzyme fatty acid amide hydrolase (FAAH). AHLs presented to the roots are taken up by the plants and hydrolyzed to L-homoserine by the enzyme FAAH. The accumulation of AHL derivative, L-homoserine, positively influenced the plant growth through increased level of nutrient uptake via transpiration and enhanced photosynthetic activity (Palmer et al. 2014). Also, bioengineered tobacco and tomato plants with AHL synthases promoted beneficial plant-bacteria interactions, thereby altering the plant growth and tolerance to biotic and abiotic stress (Barriuso et al. 2008; Mae et al. 2001; Scott et al. 2006).
So, future studies in plant-associated AHL-producing pathogenic, endophytic and epiphytic bacteria will reveal whether these groups have similar effects of modulation in plant development as the rhizobacteria.
9.7 Conclusion
AHL defence response in plants may also have an impact on development of biocontrol or biological agents, which are useful in both integrated agriculture management and organic farming. To ensure food security, agriculture industry has to develop modern plant protection strategies, which provide sufficient yield and quality food and reduce impact of chemical pesticides on the environment. The development of biocontrol agents or biological products from beneficial, soil-borne microorganisms could be a competent approach to support agriculture. Moreover, the knowledge of microbe-plant interactions could contribute the success rate of products in natural environment. The bacterial QS molecules could be of use, since both purified AHL molecules and bacteria with increased production of AHLs have an impact on plant defence mechanisms and portrait the agricultural potential of homoserine lactones. Further, studies are needed to refine our understanding of AHL function in plant interactions under field conditions, where the AHL-producing bacteria or AHLs per se could be in a position to compete with other environmental factors.
References
Atkinson S, Williams P (2009) Quorum sensing and social networking in the microbial world. J R Soc Interface 6:959–978
Bai X, Todd CD, Desikan R, Yang Y, Hu X (2012) N-3-oxodecanoyl-L-homoserine-lactone activates auxin-induced adventitious root formation via hydrogen peroxide- and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol 158:725–736
Barriuso J, Ramos Solano B, Fray RG, Camara M, Hartmann A, Gutierrez Manero FJ (2008) Transgenic tomato plants alter quorum sensing in plant growth-promoting rhizobacteria. Plant Biotechnol J 6:442–452
Bassler BL (1999) How bacteria talk to each other: regulation of gene expression. Curr Opin Microbiol 2:582–587
Bauer WD, Mathesius U (2004) Plant responses to bacterial quorum sensing signals. Curr Opin Plant Biol 7:429–433
Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21:944–953
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Berg G, Roskot N, Steidle A, Eberl L, Zock A, Smalla K (2002) Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68:3328–3338
Boyer M, Wisniewski-Dye F (2009) Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol 70:1–19
Case RJ, Labbate M, Kjelleberg S (2008) AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J 2:345–349
Cha C, Gao P, Chen YC, Shaw PD, Farrand SK (1998) Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol Plant Microbe Interact 11:1119–1129
Chin-A-Woeng TFC, Thomas-Oates JE, Lugtenberg BJJ, Bloemberg GV (2001) Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol Plant Microbe Interact 14:1006–1015
Choudhary DK, Prakash A, Johri BN (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian J Microbiol 47:289–297
Churchill MEA, Chen LL (2011) Structural basis of acyl homoserine lactone dependent signaling. ACS Chem Rev 111:68–85
DAngelo-Picard C, Faure D, Penot I, Dessaux Y (2005) Diversity of N-acyl homoserine lactone-producing and degrading bacteria in soil and tobacco rhizosphere. Environ Microbiol 7:1796–1808
De Maeyer K, D’aes J, Hua GKH, Perneel M, Vanhaecke L, Noppe H, Hofte M (2011) N-acyl homoserine lactone quorum sensing signalling in antagonistic phenazine-producing Pseudomonas isolates from the red cocoyam rhizosphere. Microbiology 157:459–479
De Vleesschauwer D, Hofte M (2009) Rhizobacteria-induced systemic resistance. Adv Bot Res 51:223–281
DeAngelis KM, Lindow SE, Firestone MK (2008) Bacterial quorum sensing and nitrogen cycling in rhizosphere soil. FEMS Microbiol Ecol 6:197–207
Degrassi G, Devescovi G, Solis R, Steindler L, Venturi V (2007) Oryza sativa rice plants contain molecules that activate different quorum-sensing N-acyl homoserine lactone biosensors and are sensitive to the specific AiiA lactonase. FEMS Microbiol Lett 269:213–220
Eberl L, Winson MK, Sternberg C, Stewart GS, Christiansen G, Chhabra SR, Bycroft B, Williams P, Molin S, Givskov M (1996) Involvement of N-acyl-L-hormoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol 20:127–136
Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger PM, Dessaux Y (2001) Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl Environ Microbiol 67:1198–1209
Fuqua C, Greenberg EP (2002) Listening in on bacteria: acyl homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695
Fuqua C, Winans SC (1994) A luxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806
Fuqua C, Winans SC, Greenberg EP (1996) Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751
Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468
Gao M, Teplitski M, Robinson JB, Bauer WD (2003) Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol Plant Microbe Interact 16:827–834
Gelencser Z, Choudhary KS, Coutinho BG, Hudaiberdiev S, Galbats B, Venturi V, Pongor S (2012) Classifying the topology of AHL-driven quorum sensing circuits in Proteobacterial genomes. Sensors 12:5432–5444
Gera C, Srivastava S (2006) Quorum-sensing: the phenomenon of microbial communication. Curr Sci 90:566–677
Gonzalez JE, Keshavan ND (2006) Messing with bacterial quorum-sensing. Microbiol Mol Biol Rev 70:859–875
Gonzalez JE, Marketon MM (2003) Quorum sensing in nitrogen-fixing rhizobia. Microbiol Mol Biol Rev 67:574–592
Gotz C, Fekete A, Gebefuegi I, Forczek ST, Fuksova K, Li X, Englmann M, Gryndler M, Hartmann A, Matucha M, Schmitt-Kopplin P, Schröder P (2007) Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal Bioanal Chem 389:1447–1457
Gotz-Rosch C, Sieper T, Fekete A, Schmitt-Kopplin P, Hartmann A, Schroder P (2015) Influence of bacterial N-acyl-homoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front Plant Sci 6:205
Gray KM, Garey JR (2001) The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387
Hartmann A, Gantner S, Schuhegger R, Steidle A, Durr C, Schmid M, Eberl L, Dazzo FB, Langebartels C (2004) N-acyl-homoserine lactones of rhizosphere bacteria trigger systemic resistance in tomato plants. In: Lugtenberg B, Tikhonovich I, Provorov N (eds) Biology of molecular plant-microbe interactions, vol 4. MPMI, St. Paul, pp 554–556
Hartmann A, Rothballer M, Hense BA, Schroder P (2014) Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front Plant Sci 5:131
Hawkes CV, DeAngelis KM, Firestone MK (2007) Root interactions with soil microbial communities and processes. In: Cardon Z, Whitbeck J (eds) The rhizosphere. Elsevier, New York, pp 1–31
Heil M, Bostock RM (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defenses. Ann Bot 89:503–512
Hernández-Reyes C, Schenk ST, Neumann C, Kogel KH, Schikora A (2014) N-acyl-homoserine lactones-producing bacteria protect plants against plant and human pathogens. Microb Biotechnol 7:580–588
Horng YT, Deng SC, Daykin M, Soo PC, Wei JR, Luh KT, Ho SW, Swift S, Lai HC, Williams P (2002) The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol Microbiol 45:1655–1671
Hosni T, Moretti C, Devescovi G, Suarez-Moreno ZR, Fatmi MB, Guarnaccia C, Pongor S, Onofri A, Buonaurio R, Venturi V (2011) Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J 5:1857–1870
Huang YL, Zeng YH, Yu ZL, Zhang J (2013) Distribution and diversity of acyl homoserine lactone producing bacteria from four different soils. Curr Microbiol 66:10–15
Huse H, Whiteley M (2011) 4-Quinolones: smart phones of the microbial world. Chem Rev 111:152–159
Jin G, Liu F, Ma H, Hao S, Zhao Q, Bian Z, Jia Z, Song S (2012) Two G-protein-coupled-receptor candidates, Cand2 and Cand7, are involved in Arabidopsis root growth mediated b y the bacterial quorum-sensing signals N-acyl-homoserine lactones. Biochem Biophys Res Commun 417:991–995
Joseph CM, Phillips DA (2003) Metabolites from soil bacteria affect plant water relations. Plant Physiol Biochem 41:189–192
Kaplan HB, Greenberg EP (1985) Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163:1210–1214
Kimura N (2014) Metagenomic approaches to understanding phylogenetic diversity in quorum sensing. Virulence 5:433–442
Kloepper JW, Ryu C-M, Zhang SA (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Lerat E, Moran NA (2004) The evolutionary history of quorum-sensing systems in bacteria. Mol Biol Evol 21:903–913
Liu X, Bimerew M, Ma Y, Muller H, Ovadis M, Eberl L, Berg G, Chernin L (2007) Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol Lett 270:299–305
Liu F, Bian Z, Jia Z, Zhao Q, Song S (2012) The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing signals. Mol Plant Microbe Interact 25:677–683
Lv D, Ma A, Tang X, Bai Z, Qi H, Zhuang G (2013) Profile of the culturable microbiome capable of producing acyl-homoserine lactone in the tobacco phyllosphere. J Environ Sci 25:357–366
Mae A, Montesano M, Koiv V, Palva ET (2001) Transgenic plants producing the bacterial pheromone N-acyl-homoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol Plant Microbe Interact 14:1035–1042
Mathesius U, Mulders S, Gao MS, Teplitski M, Caetano-Anolles G, Rolfe BG, Bauer WD (2003) Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci U S A 100:1444–1449
Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Lauriere C, Chevalier A, Castresana C, Hirt H (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11, e1001513
Ortiz-Castro R, Martinez-Trujillo M, Lopez-Bucio J (2008) N-acyl-L-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ 31:1497–1509
Ortiz-Castro R, Contreras-Cornejo HA, Macias-Rodriguez L, Lopez-Bucio J (2009) The role of microbial signals in plant growth and development. Plant Signal Behav 4:701–712
Palmer AG, Senechal AC, Mukherjee A, Ane JM, Blackwell HE (2014) Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem Biol 9:1834–1845
Pang Y, Liu X, Ma Y, Chernin L, Berg G, Gao K (2009) Induction of systemic resistance, root colonisation and biocontrol activities of the rhizospheric strain of Serratia plymuthica are dependent on N-acyl homoserine lactones. Eur J Plant Pathol 124:261–268
Pearson JP, Van Delden C, Iglewski BH (1999) Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210
Perez-Montano F, Jimenez-Guerrero I, Sanchez-Matamoros RC, Lopez-Baena FJ, Ollero FJ, Rodríguez-Carvajal MA, Bellogín RA, Espuny MR (2013) Rice and bean AHL-mimic quorum-sensing signals specifically interfere with the capacity to form biofilms by plant-associated bacteria. Res Microbiol 164:749–760
Pierson EA, Wood DW, Cannon JA, Blachere FM, Pierson LS III (1998) Interpopulation signalling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol Plant Microbe Interact 11:1078–1084
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Ryu C-M, Hu C-H, Reddy MS, Kloepper JW (2003) Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol 160:413–420
Ryu C-M, Farag MA, Hu CH, Reddy MS, Kloepper JW, Pare PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Ryu C-M, Choi HK, Lee C-H, Murphy JF, Lee J-K, Kloepper JW (2013) Modulation of quorum sensing in acyl-homoserine lactone-producing or—degrading tobacco plants leads to alteration of induced systemic resistance elicited by the rhizobacterium Serratia marcescens 90–166. Plant Pathol J 29:182–192
Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Greenberg EP (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A 93:9505–9509
Schaefer AL, Lappala CR, Morlen RP, Pelletier DA, Lu TYS, Lankford PK, Harwood CS, Greenberg EP (2013) LuxR- and LuxI-type quorum-sensing circuits are prevalent in members of the Populus deltoides microbiome. Appl Environ Microbiol 79:5745–5752
Schenk ST, Schikora A (2015) AHL-priming functions via oxylipin and salicylic acid. Front Plant Sci 5:784
Schenk ST, Stein E, Kogel KH, Schikora A (2012) Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal Behav 7:178–181
Schenk ST, Hernandez-Reyes C, Samans B, Stein E, Neumann C, Schikora M, Reichelt M, Mithofer A, Becker A, Kogel KA, Schikora A (2014) N-Acyl-Homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26:2708–2723
Schikora A, Schenk ST, Stein E, Molitor A, Zuccaro A, Kogel KH (2011) N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol 157:1407–1418
Schuhegger R, Ihring A, Gantner S, Bahnweg G, Knappe C, Vogg G, Hutzler P, Schmid M, Van Breusegem F, Eberl L, Hartmann A, Langebartels C (2006) Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ 29:909–918
Scott RA, Weil J, Le PT, Williams P, Fray RG, Von Bodman SB, Savka MA (2006) Long-and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil. Mol Plant Microbe Interact 19:227–239
Sekar J, Prabavathy VR (2014) Novel Phl-producing genotypes of finger millet rhizosphere associated pseudomonads and assessment of their functional and genetic diversity. FEMS Microbiol Ecol 89:32–46
Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B (2012) Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 158:835–843
Steidle A, Sigl K, Schuhegger R, Ihring A, Schmid M, Gantner S, Stoffels M, Riedel K, Givskov M, Hartmann A, Langebartels C, Eberl L (2001) Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl Environ Microbiol 67:5761–5770
Steindler L, Bertani I, De Sordi L, Bigirimana J, Venturi V (2008) The presence, type and role of N-acyl homoserine lactone quorum sensing in fluorescent Pseudomonas originally isolated from rice rhizospheres are unpredictable. FEMS Microbiol Lett 288:102–111
Swift S, Williams P, Stewart GAB (1999) N-acyl homoserine lactones and quorum sensing are widespread in the proteobacteria. In: Dunny GM, Winans SC (eds) Cell–cell signalling in bacteria. ASM Press, Washington, DC, pp 291–314
Taga ME, Bassler BL (2003) Chemical communication among bacteria. Proc Natl Acad Sci U S A 100:14549–14554
Teplitski M, Robinson JB, Bauer WD (2000) Plants secrete substances that mimic bacterial N-acylhomoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact 13:637–648
Thiel V, Kunze B, Verma P, Wagner-Dobler I, Schulz S (2009) New structural variants of homoserine lactones in bacteria. Chembiochem 10:1861–1868
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb Ecol 62:324–336
Van Houdt R, Givskov M, Michiels CW (2007) Quorum sensing in Serratia. FEMS Microbiol Rev 31:407–424
Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448
Venturi V, Fuqua C (2013) Chemical signaling between plants and plant-pathogenic bacteria. Annu Rev Phytopathol 51:17–37
Venturi V, Subramoni S (2009) Future research trends in the major chemical language of bacteria. HFSP J 3:105–116
Vial L, Cuny C, Gluchoff-Fiasson K, Comte G, Oger PM, Faure D, Dessaux Y, Bally R, Wisniewski-Dye F (2006) N-acyl-homoserine lactone-mediated quorum-sensing in Azospirillum: an exception rather than a rule. FEMS Microbiol Ecol 58:155–168
Viswanath G (2015) Diversity of N-acyl homoserine lactone (N-AHL) producing rhizobacteria and transcriptional analysis of AHL regulatory genes, phzI/R and its influence on phenazine biosynthetic gene, phzH in Pseudomonas chlororaphis MSSRF QS59 under biotic and abiotic stress conditions. Ph.D. thesis, University of Madras, Chennai, India
Viswanath G, Jegan S, Baskaran V, Kathiravan R, Prabavathy VR (2015) Diversity and N-acyl-homoserine lactone production by Gammaproteobacteria associated with Avicennia marina rhizosphere of South Indian mangroves. Syst Appl Microbiol 38:340–345
von Rad U, Klein I, Dobrev PI, Kottova J, Zazimalova E, Fekete A, Hartmann A, Schmitt-Kopplin P, Durner J (2008) Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229:73–85
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Waters CM, Lu W, Rabinowitz JD, Bassler BL (2008) Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536
Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP (2001) Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 25:365–404
Wisniewski-Dye F, Downie JA (2002) Quorum-sensing in Rhizobium. Antonie Van Leeuwenhoek 81:397–407
Wood DW, Gong F, Daykin MM, Williams P, Pierson LS III (1997) N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. J Bacteriol 179:7663–7670
Zarkani AA, Stein E, Rohrich CR, Schikora M, Evguenieva-Hackenberg E, Degenkolb T, Vilcinskas A, Klug G, Kogel KH, Schikora A (2013) Homoserine lactones influence the reaction of plants to rhizobia. Int J Mol Sci 14:17122–17146
Zeng Y, Yu Z, Huang Y (2014) Combination of culture-dependent and-independent methods reveals diverse acyl homoserine lactone-producers from rhizosphere of wetland Plants. Curr Microbiol 68:587–593
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this chapter
Cite this chapter
Viswanath, G., Sekar, J., Prabavathy, V.R. (2016). Acyl Homoserine Lactone-Producing Rhizobacteria Elicit Systemic Resistance in Plants. In: Choudhary, D.K., Varma, A. (eds) Microbial-mediated Induced Systemic Resistance in Plants. Springer, Singapore. https://doi.org/10.1007/978-981-10-0388-2_9
Download citation
DOI: https://doi.org/10.1007/978-981-10-0388-2_9
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-0387-5
Online ISBN: 978-981-10-0388-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)