Abstract
In contrast to the situation in land plants and animals, very little information is available concerning the molecular mechanisms underlying multicellular development in the brown algae. Historically, one of the reasons for this has been the lack of an effective model organism for the latter group that would permit the application of powerful genomic and genetic approaches to explore these processes. This situation has changed in recent years with the emergence of the filamentous brown alga Ectocarpus as a model organism. This chapter describes the genetic and genomic resources that are currently available for this organism and describes some of the additional tools that are under development. Potential additional models that would provide access to the biological diversity within the brown algae are also discussed, with a particular focus on the evolution of multicellular complexity within this group.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Brown Algal Model Systems
Whilst brown algae as a group clearly exhibit complex multicellularity (see Chap. 16), the level of developmental complexity is variable across the group, ranging from species with relatively simple filamentous thalli to species with complex organs consisting of multiple tissue types. Recently evolved groups such as the Fucales and Laminariales (fucoid algae and kelps) exhibit the highest degree of developmental complexity, but the members of these groups are also large organisms with long life cycles that are difficult to cultivate under laboratory conditions. Experimental work on brown algae from the Fucales and Laminariales has therefore mostly involved either manipulation of ex situ material for short periods in the laboratory or experimentation under field conditions. Fucoid species have been used as models to study zygote polarisation and early embryogenesis. The large size of fucoid egg cells and the fact that fertilisation is external, and therefore easily observable under the microscope, have permitted very detailed characterisation of early developmental processes at the cellular level. These studies have led to several important advances, including elucidation of the physical determinants of zygote polarity, the relationship between initial cell polarisation and the first cell cycle, and the discovery of a role for the cell wall in determining cell identity (Berger et al. 1994; Bouget et al. 1998; Coelho et al. 2002; Corellou et al. 2001a, 2005; Kropf et al. 1988; Shaw and Quatrano 1996). However, our understanding of the molecular circuitry that regulates these cellular events is much less complete (Fowler et al. 2004; Corellou et al. 2001b), making it difficult to make meaningful comparisons with systems from other multicellular lineages such as green plants or animals. Until recently, progress on understanding brown algal developmental processes at the molecular level has been held back by several factors, including a lack of gene sequence information (genome data) and the absence of tools for the manipulation of gene function. The long life cycles of brown algae from the Fucales and Laminariales have also limited the scope for genetic analysis.
These limitations do not apply to all the brown algae. The filamentous brown alga Ectocarpus has been studied for many years and is emerging as the model of choice for the application of genomic and genetic approaches to diverse questions concerning the biology of this group of organisms (Charrier et al. 2008; Cock et al. 2011; Coelho et al. 2007, 2012a; Cock et al. 2010a). From a developmental point of view, Ectocarpus exhibits significantly less complexity than members of the Fucales and Laminariales, and this is true of the group Ectocarpales in general. However, recent phylogenies indicate that the Ectocarpales is a sister group to the Laminariales, the two lineages only having diverged about 100 million years ago (Silberfeld et al. 2010). It is therefore likely that the regulatory circuits that control development in members of the Ectocarpales and the Laminariales share many common features, despite the simpler bodyplans of the former.
Multicellular Development During the Ectocarpus Life Cycle
Like many brown algae , Ectocarpus has a haploid-diploid life cycle that involves alternation between two multicellular organisms, the sporophyte and the gametophyte (Fig. 1; Kornmann 1956; Müller 1964, 1967; Peters et al. 2008). Both sporophyte and gametophyte thalli consist of uniseriate, branched filaments. Gametophyte germlings are made up of a rhizoid and an upright filament, the latter consisting of cylindrical cells. The upright filament grows and branches to produce the mature thallus, which carries plurilocular gametangia in which the gametes are produced. The developmental program of the sporophyte is slightly more complex in that it produces a basal structure consisting of round and elongated cells before producing upright filaments. The upright filaments of the sporophyte resemble those of the gametophyte but are less profusely branched. The sporophyte upright filaments bear two types of reproductive structures, plurilocular sporangia containing mito-spores (which will germinate to produce clones of the parent sporophyte) and unilocular sporangia where a single meiotic event produces the meio-spores that are the initial cells of the gametophyte generation. Overall, during its life cycle Ectocarpus produces about eight different cell types (not including zoids; Fig. 1), significantly less than the 14 reported for kelps (reviewed in Bell and Mooers 1997).
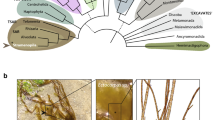
Morphologically distinct cell types found in the sporophyte and gametophyte generations of the model brown alga Ectocarpus. The left and right panels show sporophyte and gametophyte cell types, respectively. a Upright filament cell (sporophyte), delineated with a dotted red line. b Rhizoid cell (sporophyte), delineated with a dotted red line. c Prostrate base cells, round (asterisk) and elongated (double asterisk). d Unilocular sporangium initial cell (asterisk). e Plurilocular sporangium initial cell (asterisk). f Upright filament cell (gametophyte), delineated with a dotted red line. g Rhizoid cell (gametophyte), delineated with a dotted red line. h Mature plurilocular gametangium. i Plurilocular gametangium initial cell (asterisk). The central drawings indicate where each cell type occurs in the multicellular sporophyte (brown) and gametophyte (green) bodyplans
Ectocarpus sporophytes can be derived from zygotes (i.e. formed by the fusion of two gametes) or can develop parthenogenetically from a gamete that has failed to find a partner of the opposite sex (in which case they are called partheno-sporophytes). Being derived from a single gamete, most partheno-sporophytes are haploid (Fig. 2, but see Bothwell et al. 2010). With Ectocarpus, therefore, it is possible to obtain both the gametophyte and sporophyte generations as haploid individuals, a feature that greatly facilitates genetic analysis of life-cycle-related developmental processes.
Representation of the Ectocarpus life cycle showing the alternation between the diploid sporophyte and the haploid, dioicous gametophytes during the sexual cycle and the production of haploid partheno-sporophytes by gamete parthenogenesis (parthenogenetic cycle). Note that both the sporophyte and gametophyte generations exhibit multicellular development
Genetic Analysis Using Ectocarpus
Ectocarpus has been used as a genetic model for several decades, earlier studies demonstrating Mendelian inheritance of a number of natural biological characters including sexuality, viral sequences that insert into the genome and membrane lipid composition (Müller 1967, 1991; Bräutigam et al. 1995; Müller and Eichenberger 1997). All stages of the life cycle can be grown in the laboratory and it takes about 3 months to complete the sexual life cycle under these conditions. Individuals are raised in Petri dishes containing either natural or artificial seawater. Thalli usually become fertile when they are less than a centimetre in size. The small size of the thallus not only allows multiple individuals to be maintained in a small space, but also facilitates screens for genetic mutants, particularly when the screens are carried out during early development. Protocols have been developed for both ultraviolet (Coelho et al. 2011) and chemical (EMS and ENU) mutagenesis along with screening methodologies for developmental mutants. Classical genetic analysis of mutants is possible, with protocols having been developed both for carrying out genetic crosses and for the isolation of meiotic progeny (Coelho et al. 2012b, c). Using these approaches, large segregating populations can be generated for mapping experiments and the first mutant allele has recently been identified by positional cloning using these tools (unpublished results, see Chapter “Independent Emergence of Complex Multicellularity in the Brown and Red Algae”). Inbred lines can also be created by repeated crosses between siblings, although this is not required for many analyses because it is possible to work with haploid individuals.
There are also considerable genetic resources associated with Ectocarpus, including a collection of more than 2000 strains held at the Station Biologique de Roscoff (France). These strains represent worldwide diversity within the genus and include several collections of populations from single sites, providing access to information about local population structures and diversity. Strains can be stored under low light and low temperature conditions for at least 1 year before the culture needs to be refreshed. In addition, an alternative stock maintenance method based on cryopreservation has recently been developed (Heesch et al. 2012).
Ectocarpus as a Genomic Model
The small size of the Ectocarpus genome (214 Mbp) compared to those of most other brown algal species also represents an important advantage. The complete sequence of the genome is available and the transcribed regions have been characterised using data generated by several different approaches including Sanger expressed sequence tags (91,000), deep Illumina-base RNA-seq experiments, whole genome tiling arrays and deep Illumina sequencing of small RNAs (Cock et al. 2010b and unpublished data). An EST-based microarray has also been developed to allow near-genome-wide assessment of changes in transcript abundances (Dittami et al. 2009). A high quality genome reference sequence has been established based on these diverse transcriptomic data and on manual annotation of a large proportion of the genes in the genome. Genome data and genetic data have been combined to generate a sequence-anchored genetic map, which both provides a chromosome-scale assembly of the genome and represents an important resource for ongoing genetic analyses (Heesch et al. 2010).
Tools for the Analysis of Ectocarpus Gene Function
The major current bottleneck for Ectocarpus as a model system is the lack of tools to investigate gene function. Despite considerable investment, it has proved to be very difficult to develop a reliable genetic transformation protocol for this model organism. The recent demonstration that injection of double stranded tubulin RNA into Fucus zygotes led to disruption of the microtubule cytoskeleton (Farnham et al. 2013) suggests a possible alternative approach. The results of the Fucus experiments indicate that brown algae possess a functional RNA interference (RNAi) system and this hypothesis is supported by the presence of putative dicer and argonaute genes in the Ectocarpus genome (Cock et al. 2010b). Current work is aimed at adapting the Fucus protocol for use in Ectocarpus.
Another promising approach is the reverse genetic technique referred to as Targeting Induced Local Lesions in Genomes (TILLING). In this approach, a large population of individuals, each carrying hundreds of different mutations is screened for individuals carrying a mutation in a specific gene of interest by analysis of amplified polymerase chain reaction fragments (Kurowska et al. 2011). A recently completed pilot study in which a mutant population produced by UV and chemical mutagenesis was screened using a three-dimensional pooling and next-generation-sequencing-based approach indicates that this approach is feasible in Ectocarpus (unpublished results).
The availability of a tool that will allow gene function to be investigated experimentally, whether it be based on transformation, RNAi or TILLING, will represent an important step forward for Ectocarpus as a model organism. The combination of such a tool with existing resources and technologies, such as genome information, mutant screens and positional cloning, is expected to lead to significant progress in our understanding of diverse aspects of brown algal biology, including the developmental processes that underlie multicellular development in this lineage. It is also clear, however, that it will be important to complement experiment work using Ectocarpus with data from more developmentally complex brown algae. This will be required both to test the generality of information obtained using Ectocarpus and to investigate how developmental processes operate in more morphologically complex brown algae.
Additional Brown Algal Models
The development of an RNAi protocol for Fucus (Farnham et al. 2013) is likely to stimulate renewed interest in fucoids for brown algal research. The lack of a complete genome sequence for this genus remains a limiting factor, but an increasing amount of transcriptomic data is being generated (Coyer et al. 2009; Pearson et al. 2010) and this will be an important resource for future work. It is likely that Fucus-based research will continue to focus on early developmental events because of the problems associated with extended culture in the laboratory, but this should nonetheless provide access to many interesting questions relevant to the evolution of multicellularity.
As the most developmentally complex group among the brown algae , kelps are of particular interest for questions related to the emergence of complex multicellularity. Potential model species from within this group include Undaria, Laminaria, Saccharina and Macrocystis (Cock et al. 2012). Saccharina and Macrocystis are of particular interest because they are accessible to genetic analysis, whilst at the same time having a high potential for biotechnological and aquaculture applications (Li et al. 2007; Westermeier et al. 2010, 2011; Gutierrez et al. 2006). The Saccharina genome is estimated to be around 600 Mbp, whereas that of Macrocystis is larger (about 1 Gbp), comparable in size to those of fucoid species (Kapraun 2005; Phillips et al. 2011; Peters et al. 2004).
If an effective kelp model can be developed, it will be particularly interesting to compare processes related to multicellularity in this group with information obtained for Ectocarpus, both at the level of genome evolution and, where possible, from a mechanistic point of view. Analysis of the genome sequence of Ectocarpus has identified a number of features that may be related to the transition to multicellularity (see Chapter “Independent Emergence of Complex Multicellularity in the Brown and Red Algae”). It will be interesting to determine whether these features are also observed in a kelp genome, with perhaps more marked trends such as expansions of key gene families. In the longer term, it will also be of great interest to investigate how the developmental genes that are currently being identified in Ectocarpus function in the more morphologically complex kelps.
Summary
-
1.
Ectocarpus has emerged as a model system for the brown algae, allowing the application of genetic and genomic approaches to this group of organisms.
-
2.
The life cycle of Ectocarpus involves two multicellular stages, the sporophyte and gametophyte generations, with a total of about six different cell types.
-
3.
Genomic and genetic resources that have been developed for Ectocarpus include a complete genome sequence, extensive transcriptomic data, protocols to produce and screen mutant populations, a genetic map, and a large collection of strains.
-
4.
Methods being developed to analyse gene function in Ectocarpus include RNA interference and Targeting Induced Local Lesions in Genomes (TILLING).
-
5.
In the future, extension of work carried out in Ectocarpus to more developmentally complex brown algal models, such as kelp species, should provide further insights into the emergence of complex multicellularity in this group.
References
Bell G, Mooers AO (1997) Size and complexity among multicellular organisms. Biol J Linn Soc 60:345–363
Berger F, Taylor A, Brownlee C (1994) Cell fate determination by the cell wall in early Fucus development. Science 263(5152):1421–1423
Bothwell JH, Marie D, Peters AF, Cock JM, Coelho SM (2010) Role of endoreduplication and apomeiosis during parthenogenetic reproduction in the model brown alga Ectocarpus. New Phytol 188(1):111–121
Bouget FY, Berger F, Brownlee C (1998) Position dependent control of cell fate in the Fucus embryo: role of intercellular communication. Development 125(11):1999–2008
Bräutigam M, Klein M, Knippers R, Müller DG (1995) Inheritance and meiotic elimination of a virus genome in the host Ectocarpus siliculosus (phaeophyceae). J Phycol 31(5):823–827
Charrier B, Coelho S, Le Bail A, Tonon T, Michel G, Potin P, Kloareg B, Boyen C, Peters A, Cock J (2008) Development and physiology of the brown alga Ectocarpus siliculosus: two centuries of research. New Phytol 177(2):319–332
Cock JM, Coelho SM, Brownlee C, Taylor AR (2010a) The Ectocarpus genome sequence: insights into brown algal biology and the evolutionary diversity of the eukaryotes. New Phytol 188(1):1–4
Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury J, Badger J, Beszteri B, Billiau K, Bonnet E, Bothwell J, Bowler C, Boyen C, Brownlee C, Carrano C, Charrier B, Cho G, Coelho S, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami S, Doulbeau S, Elias M, Farnham G, Gachon C, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper F, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez P, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli C, Nelson D, Nyvall-Collén P, Peters A, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing S, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder D, Ségurens B, Strittmatter M, Tonon T, Tregear J, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P (2010b) The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465(7298):617–621
Cock JM, Peters AF, Coelho SM (2011) Brown algae. Curr Biol 21(15):R573–575
Cock JM, Arun A, Godfroy O, Macaisne N, Strittmatter M, Peters AF, Coelho SM (2012) Genomics of brown algae: current advances and future prospects. Genes Genomics 34:1–5
Coelho S, Taylor A, Ryan K, Sousa-Pinto I, Brown M, Brownlee C (2002) Spatiotemporal patterning of reactive oxygen production and Ca(2+) wave propagation in Fucus rhizoid cells. Plant Cell 14(10):2369–2381
Coelho S, Peters A, Charrier B, Roze D, Destombe C, Valero M, Cock J (2007) Complex life cycles of multicellular eukaryotes: new approaches based on the use of model organisms. Gene 406(1–2):152–170
Coelho SM, Godfroy O, Arun A, Le Corguillé G, Peters AF, Cock JM (2011) OUROBOROS is a master regulator of the gametophyte to sporophyte life cycle transition in the brown alga Ectocarpus. Proc Natl Acad Sci U S A 108:11518–11523
Coelho SM, Scornet D, Rousvoal S, Peters N, Dartevelle L, Peters AF, Cock JM (2012a) Ectocarpus: a model organism for the brown algae. Cold Spring Harb Protoc 2012:193–198
Coelho SM, Scornet D, Rousvoal S, Peters N, Dartevelle L, Peters AF, Cock JM (2012b) Genetic crosses between Ectocarpus strains. Cold Spring Harb Protoc 2012(2):262–265
Coelho SM, Scornet D, Rousvoal S, Peters NT, Dartevelle L, Peters AF, Cock JM (2012c) How to cultivate Ectocarpus. Cold Spring Harb Protoc 2012(2):258–261
Corellou F, Brownlee C, Detivaud L, Kloareg B, Bouget FY (2001a) Cell cycle in the Fucus zygote parallels a somatic cell cycle but displays a unique translational regulation of cyclin-dependent kinases. Plant Cell 13(3):585–598
Corellou F, Brownlee C, Kloareg B, Bouget FY (2001b) Cell cycle-dependent control of polarised development by a cyclin-dependent kinase-like protein in the Fucus zygote. Development 128(21):4383–4392
Corellou F, Coelho SM, Bouget FY, Brownlee C (2005) Spatial re-organisation of cortical microtubules in vivo during polarisation and asymmetric division of Fucus zygotes. J Cell Sci 118(Pt 12):2723–2734
Coyer JA, Hoarau G, Beszteri B, Pearson G, Olsen JL (2009) Expressed sequence tag-derived polymorphic SSR markers for Fucus serratus and amplification in other species of Fucus. Mol Ecol Resour 9(1):168–170
Dittami S, Scornet D, Petit J, Ségurens B, Da Silva C, Corre E, Dondrup M, Glatting K, König R, Sterck L, Rouzé P, Van de Peer Y, Cock J, Boyen C, Tonon T (2009) Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol 10(6):R66
Farnham G, Strittmatter M, Coelho SM, Cock JM, Brownlee C (2013) Gene silencing in Fucus embryos: developmental consequences of RNAi-mediated cytoskeletal disruption. J Phycol 49:819–829
Fowler JE, Vejlupkova Z, Goodner BW, Lu G, Quatrano RS (2004) Localization to the rhizoid tip implicates a Fucus distichus Rho family GTPase in a conserved cell polarity pathway. Planta 219(5):856–866
Gutierrez A, Correa T, Muñoz V, Santibañez A, Marcos R, Caceres C, Buschmann AH (2006) Farming of the giant kelp Macrocystis pyrifera in southern Chile for development of novel food products. J Appl Phycol 18:259–267
Heesch S, Cho GY, Peters AF, Le Corguillé G, Falentin C, Boutet G, Coëdel S, Jubin C, Samson G, Corre E, Coelho SM, Cock JM (2010) A sequence-tagged genetic map for the brown alga Ectocarpus siliculosus provides large-scale assembly of the genome sequence. New Phytol 188(1):42–51
Heesch H, Day JG, Yamagishi T, Kawai H, Müller DG, Küpper FC (2012) Cryopreservation of the model alga Ectocarpus (Phaeophyceae). Cryo Lett 33(5):327–336
Kapraun DF (2005) Nuclear DNA content estimates in multicellular green, red and brown algae: phylogenetic considerations. Ann Bot 95:7–44
Kornmann P (1956) Über die Entwicklung einer Ectocarpus confervoides-Form. Pubbl Della Stn Zoologica di Napoli 28:32–43
Kropf DL, Kloareg B, Quatrano RS (1988) Cell wall is required for fixation of the embryonic axis in Fucus zygotes. Science 239(4836):187–190
Kurowska M, Daszkowska-Golec A, Gruszka D, Marzec M, Szurman M, Szarejko I, Maluszynski M (2011) TILLING: a shortcut in functional genomics. J Appl Genet 52(4):371–390
Li X, Cong Y, Yang G, Shi Y, Qu S, Li Z, Wang G, Zhang Z, Luo S, Dai H, Xie J, Jiang G, Liu J, Wang T (2007) Trait evaluation and trial cultivation of Dongfang No. 2, the hybrid of a male gametophyte clone of Laminaria longissima (Laminariales, Phaeophyta) and a female one of L. japonica. J Appl Phycol 19:139–151
Müller DG (1964) Life-cycle of Ectocarpus siliculosus from Naples, Italy. Nature 26:1402
Müller DG (1967) Generationswechsel, Kernphasenwechsel und Sexualität der Braunalge Ectocarpus siliculosus im Kulturversuch. Planta 75:39–54
Müller DG (1991) Mendelian segregation of a virus genome during host meiosis in the marine brown alga Ectocarpus siliculosus. J Plant Physiol 137:739–743
Müller DG, Eichenberger W (1997) Mendelian genetics in brown algae: inheritance of a lipid defect mutation and sex alleles in Ectocarpus siliculosus (Ectocarpales, Phaeophyceae). Phycologia 36(1):79–81
Pearson GA, Hoarau G, Lago-Leston A, Coyer JA, Kube M, Reinhardt R, Henckel K, Serrão ET, Corre E, Olsen JL (2010) An expressed sequence tag analysis of the intertidal brown seaweeds Fucus serratus (L.) and F. vesiculosus (L.) (Heterokontophyta, Phaeophyceae) in response to abiotic stressors. Mar Biotechnol (NY) 12 (2):195–213
Peters AF, Marie D, Scornet D, Kloareg B, Cock JM (2004) Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J Phycol 40(6):1079–1088
Peters AF, Scornet D, Ratin M, Charrier B, Monnier A, Merrien Y, Corre E, Coelho SM, Cock JM (2008) Life-cycle-generation-specific developmental processes are modified in the immediate upright mutant of the brown alga Ectocarpus siliculosus. Development 135(8):1503–1512
Phillips N, Kapraun DF, Gómez Garreta A, Ribera Siguan MA, Rull JL, Salvador Soler N, Lewis R, Kawai H (2011) Estimates of nuclear DNA content in 98 species of brown algae (Phaeophyta). AoB Plants 2011:plr001
Shaw SL, Quatrano RS (1996) The role of targeted secretion in the establishment of cell polarity and the orientation of the division plane in Fucus zygotes. Development 122(9):2623–2630
Silberfeld T, Leigh JW, Verbruggen H, Cruaud C, de Reviers B, Rousseau F (2010) A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Mol Phylogenet Evol 56(2):659–674
Westermeier R, Patiño DJ, Müller H, Müller DG (2010) Towards domestication of giant kelp (Macrocystis pyrifera) in Chile: selection of haploid parent genotypes, outbreeding, and heterosis. J Appl Phycol 22:357–361
Westermeier R, Patiño DJ, Murúa P, Müller DG (2011) Macrocystis mariculture in Chile: growth performance of heterosis genotype constructs under field conditions. J Appl Phycol 23:819–825
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Cock, J. et al. (2015). Emergence of Ectocarpus as a Model System to Study the Evolution of Complex Multicellularity in the Brown Algae. In: Ruiz-Trillo, I., Nedelcu, A. (eds) Evolutionary Transitions to Multicellular Life. Advances in Marine Genomics, vol 2. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9642-2_8
Download citation
DOI: https://doi.org/10.1007/978-94-017-9642-2_8
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9641-5
Online ISBN: 978-94-017-9642-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)