Abstract
Pili or fimbriae are known virulence factors that facilitate bacterial colonization of specific host tissues and pathogenesis. Corynebacterium diphtheriae, the causative agent of pharyngeal diphtheria, harbors three pilus gene clusters encoding the heterotrimeric SpaA-, SpaD-, and SpaH-type pili. The current model for Gram-positive pilus assembly is based on the studies of the SpaA-type pili, which may serve as a major virulence determinant for C. diphtheriae because they are specifically required for adherence to pharyngeal epithelial cells. The SpaA-type pilus is comprised of the shaft pilin SpaA and minor proteins SpaB and SpaC, which make up the base and tip of the surface structure, respectively. Pilus biogenesis requires tandem sortase enzymes that covalently link SpaABC monomers and subsequently anchor the resulting heterotrimeric polymer to the bacterial peptidoglycan. Here, a decade of research aimed to reveal the assembly, structure, and role of C. diphtheriae pili in pathogenesis is discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
To successfully colonize a host, a bacterium must first adhere to target epithelial tissues. The repulsive force generated by similar charges present on both the bacterium and target cell surfaces, however, presents a challenge (Proft and Baker 2009). To overcome this, prokaryotic organisms assemble proteinaceous structures, known as pili or fimbriae, on the cell surface that recognize cognate host receptors. Pili are polymeric virulence factors required for bacterial colonization and pathogenesis. Their surface expression has been demonstrated to foster progression of infectious diseases that affect periodontal, urinary, gastrointestinal, and respiratory tissue (Sauer et al. 2000). Their importance to pathogenesis makes the surface structures attractive candidates for the development of vaccines and antimicrobial therapies (Maione et al. 2005; Soriani and Telford 2010).
Both Gram-positive and Gram-negative species of bacteria produce pili, but utilize remarkably different mechanisms of assembly (Thanassi et al. 1998; Ton-That and Schneewind 2004). Gram-negative pili are multimeric structures comprised of non-covalently associated subunits that are fastened to the outer-membrane. The chaperone-usher pathway, responsible for the display of uropathogenic Escherichia coli type I and Pap pili on the surface is the best characterized example of Gram-negative surface pilus adhesins among others including type IV pili and curli (Kline et al. 2010). To construct these adhesins, precursors translocated into the periplasmic space first associate with a designated chaperone charged with preventing protein aggregation, and the delivery of substrates to an outer membrane usher which catalyzes both polymerization of subunits and secretion of resulting complexes to the cell surface (Phan et al. 2011; Remaut et al. 2008). Pilin precursors are joined by a process known as ‘donor strand exchange’ in which the N-terminal extension of a preceding subunit fills a gap within the incomplete immunoglobulin fold of an incoming pilin (Sauer et al. 1999, 2002).
The study of pili expressed by Gram-positive bacteria is a relatively recent endeavor. These prokaryotes, unlike their Gram-negative counterparts, lack outer membranes. Their cell envelopes are comprised of a single inner membrane surrounded by a dense layer of peptidoglycan. Sjoquist and coworkers (Sjoquist et al. 1972) first recognized that Gram-positives utilize their cell wall as an organelle to anchor proteins upon observing the Staphylococcus aureus protein A, an MSCRAMM protein known for its ability to bind to the constant region of immunoglobulins, became soluble only after cells were treated with lysostaphin, a peptidoglycan hydrolase. Decades later, Schneewind and colleagues demonstrated that cell wall anchoring of protein A is dependent on a C-terminal cell wall sorting signal (CWSS) comprised of an LPXTG motif, a hydrophobic domain, and a positively charged tail (Schneewind et al. 1992). Soon afterwards, a unique transpeptidase named sortase SrtA in S. aureus was discovered and credited as the catalysis for a transpeptidation reaction that links CWSS-containing substrates like protein A to the cell wall peptidoglycan (Mazmanian et al. 1999, 2001). Finally, the finding that sortase mediates pilus polymerization in Gram-positive bacteria was first demonstrated in Corynebacterium diphtheriae (Ton-That and Schneewind 2003), the causative agent of diphtheria (Klebs 1883) .
This important study has launched subsequent investigations of pili expressed by Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Enterococcus faecalis, Bacillus cereus, and Actinomyces oris (Lauer et al. 2005; Mora et al. 2005; Barocchi et al. 2006; LeMieux et al. 2006; Dramsi et al. 2006; Rosini et al. 2006; Nallapareddy et al. 2006; Budzik et al. 2007; Mishra et al. 2007), establishing C. diphtheriae as a model system for the biogenesis of Gram-positive pili (Mandlik et al. 2008b). This chapter will review the most current knowledge available regarding pilus expression, structure, and sortase-mediated assembly in C. diphtheriae, as well as the role of pili in establishing disease. Pilus assembly is a worthy research endeavor as it will expand our understanding of the infection process potentially leading to the development of new antibiotics or vaccines .
7.2 Pili and Pilus Gene Clusters of Corynebacterium diphtheriae
Gram-positive pili were first observed in 1968 on the surface of Corynebacterium renale using electron microscopy (Yanagawa et al. 1968). Subsequent protein analysis revealed that these structures were remarkably different compared to those expressed by Gram-negative bacteria . The integrity of pili isolated from the cell surface, for example, was highly resistant to heat boiling (Kumazawa and Yanagawa 1972). Pili were also observed in many Gram-positive bacteria, such as Actinomyces viscosus and Actinomyces naeslundii (Girard and Jacius 1974). These pili are required for interbacterial adhesion and bacterial attachment to host surfaces (Yeung 1999) . Despite a flurry of research activities in this area for decades, the mechanism of pilus assembly was not revealed until recently.
In 2003, Ton-That and Schneewind surveyed the available genome of C. diphtheriae NCTC13129 (Cerdeno-Tarraga et al. 2003), a toxin-producing clinical isolate, for open reading frames (ORFs) encoding sortase homologs and CWSS-containing proteins (Ton-That and Schneewind 2003) . They revealed three gene clusters encoding a total of nine surface proteins with the LPXTG motif, termed SpaA-I (Spa for sortase-mediated pilus assembly), and five sortases (SrtA-E) . In addition, a sixth sortase encoding gene named srtF was found elsewhere in the genome (Fig. 7.1). Polyclonal antibodies raised against purified Spa proteins were used to label immobilized cells, followed by staining with IgG-conjugated gold particles, and subsequent detection by electron microscopy. This methodology revealed three distinct pilus structures assembled on the cell surface of C. diphtheriae, designated as SpaA-, SpaD-, and SpaH-type pili based on the major pilin (Ton-That and Schneewind 2003; Gaspar and Ton-That 2006; Swierczynski and Ton-That 2006) . Each pilus is composed of a shaft pilin that displays a tip pilin and is interspersed by a minor pilin also forming the pilus base; for example, the SpaA-type pilus is made of the shaft pilin SpaA, the tip pilin SpaC and the pilus base SpaB (Ton-That and Schneewind 2003; Mandlik et al. 2008a). Pili observed were typically smaller than those seen on the surface of Gram-negative bacteria, ranging from 1–2 μm in length and 1–2 nm in width (Ton-That and Schneewind 2003). Unlike Gram-negative pili, these pili are covalently linked, evident by their resistance to hot SDS and formic acid treatment (Ton-That and Schneewind 2003). Overexpression of shaft pilins resulted in exceedingly long pili (Swierczynski and Ton-That 2006; Ton-That and Schneewind 2003), indicating pilus polymerization is a stoichiometric process. Deletion of a sortase gene within a pilus gene cluster, i.e. srtA within the SpaA-type locus, resulted in the abrogation of pilus polymers, thus designating pilin-specific sortases responsible for assembling specific pilin subunits (Mandlik et al. 2008b) .
Pilus gene clusters of Corynebacterium diphtheriae NCTC13129 and recently sequenced clinical isolates. Homology of pilin and sortase genes is indicated by color; with genes similar to SpaD and SpaH types denoted with primes. Fragmented genes are indicated by asterisks and hatched arrows. (Reprinted from (Trost et al. 2012) with permission)
A recent pangenomic study of 13 C. diphtheriae clinical isolates revealed heterogeneity within pilus gene clusters (Trost et al. 2012). All strains were found to harbor at least two distinct pilus forms with the SpaA-type pili being the most common followed by SpaD-type . Few isolates expressed SpaH-type pili . Interestingly, genes within clusters appeared to have co-evolved. Among all strains tested, the genes encoding SpaABC pilins and their respective sortase were found to be conserved-type specific sortase and pilins (Fig. 7.2), while components of the SpaD-type and SpaH-type were found to be highly divergent in comparison. One strain in particular, the vaccine strain Park-William No. 8 (PW8) contains a spaD cluster with multiple intact and disrupted spaD, spaE, spaF, srtE, and srtB genes, indicating that it was the target of mobile DNA elements . The maintenance of the spa locus throughout evolution underscores its importance for the bacterium’s ability to cause disease. SpaA-type pili specifically facilitate corynebacterial adherence to human pharyngeal epithelial cells, the major site for corynebacterial infection (Mandlik et al. 2007) . This specific adherence is attributed to the two minor pilins of SpaA-type pili, SpaB and SpaC (Mandlik et al. 2007) . Maintenance of these genes is important since mutations affecting SpaA/B/C or SrtA would compromise the fitness of these bacteria. PW8 appears to be an exception as it harbors a frame-shift mutation within spaC preventing the tip pilin from being fully translated (Trost et al. 2012). Interestingly, this correlates with overall decreased virulence compared to other isolates (Iwaki et al. 2010). Low conservation of the spaD and spaH loci suggests that mutations in these regions are better tolerated because they are less important to establishing C. diphtheriae infection. Indeed, adherence assays have demonstrated that while SpaA-type pili target pharyngeal epithelial cells, SpaD and SpaH pili preferentially mediate binding to laryngeal and lung epithelial cells (Mandlik et al. 2007) .
Electron micro graphs of Corynebacterium diphtheriae NCTC13129 and a toxigenic strain. a Corynebacterial cells were immobilized on nickel carbon-coated grids and viewed by an electron microscopy after staining with 1 % uranyl acetate. b–d Cells of C. diphtheriae NCTC13129 b, its isogenic strain over-expressing the pilin shaft SpaA c, and a clinical isolate from Russia (CDC1737; (Popovic et al. 1996)) d were immobilized on nickel carbon-coated grids, stained with antibodies against SpaA (α-SpaA), followed by goat anti-rabbit IgG conjugated to 18 nm gold particles, and stained with 1 % uranyl acetate before electron microscopy. Bars indicate 0.5 μm
7.3 The Archetype SpaA-type Pili: Conserved Pilin Elements and the Mechanism of Sortase-Mediated Pilus Assembly
The SpaA-type pilus, encoded by the gene locus spaA-srtA-spaB-spaC (Ton-That and Schneewind 2003), serves as a model of pilus assembly in Gram-positive bacteria. As mentioned above, immuno-electron microscopic analysis, in combination with genetics and biochemical methods, revealed that SpaA forms the pilus shaft, with SpaC located at the tip. SpaB was observed along the pilus structures (Ton-That and Schneewind 2003) and also at the pilus base (Mandlik et al. 2008a). Consistent with the role of sortase in pilus assembly, a mutant lacking srtA failed to assemble SpaA pili on the cell surface (Ton-That and Schneewind 2003) .
Like all other pilins, SpaA/B/C contain an N-terminal signal peptide and a C-terminal CWSS for sortase recognition and cleavage (Ton-That and Schneewind 2003). Sequence comparison between corynebacterial major pilin proteins, i.e. SpaA, SpaD and SpaH, with the putative shaft pilins of many Gram-positive bacteria revealed several conserved elements, including the pilin motif WxxxVxVYPK and the E-box LXET, which harbor conserved lysine and glutamic acid residues (bold), respectively (Ton-That and Schneewind 2003; Ton-That et al. 2004b). Mutations altering lysine (K190) in the SpaA pilin motif to alanine or arginine completely abrogated pilus assembly (Ton-That and Schneewind 2003), whereas similar mutations of the glutamic acid residue (E446) in the E-box did not dramatically affect pilus polymerization. Instead, these mutations resulted in fragmented SpaA polymers and failed incorporation of SpaB into the SpaA pili (Ton-That et al. 2004b). While it was apparent that the pilin motif is involved in pilus polymerization, it was not clear how the E-box participates in pilus assembly .
Compelling evidence that the pilin motif is essential for pilus polymerization and that both the pilin motif and the LPXTG motif are sufficient and necessary for this transpeptidation, was provided by an experiment in which sortase-mediated polymerization of a hybrid protein was studied (Ton-That et al. 2004b). A region containing the N-terminal signal peptide and the pilin motif of SpaA was fused to the mature domain of staphylococcal enterotoxin B (SEB) at its amino-terminus, whereas its C-terminus was linked to the SpaA-CWSS. This fusion protein was expressed in strains lacking spaA or srtA. Remarkably, SEB polymers were observed by immunoblotting with the ΔspaA mutant but not with the ΔsrtA mutant. A similar phenotype was also observed in a strain harboring a mutation in the lysine residue of the SpaA pilin motif, i.e. K190A. The results have established the specific role of pilin-specific sortase and critical elements for pilus assembly in Gram-positive bacteria (see below) .
7.3.1 A Biphasic Model of Sortase-Mediated Pilus Assembly
Using a classic mutagenesis approach in which S. aureus cells failed to “sort” protein A to the bacterial cell wall were selected, Mazmanian and colleagues identified the first sortase gene termed srtA (Mazmanian et al. 1999), which encodes a transpeptidase enzyme that catalyzes cell wall anchoring of staphylococcal surface proteins (Ton-That et al. 1999) . Further analysis of available bacterial genomes revealed that sortase enzymes were conserved among Gram-positive bacteria (excluding Mycobacterium and Microplasma) (Comfort and Clubb 2004; Dramsi et al. 2005), revealing that the display of cell surface proteins in these organisms relied on a universal mechanism (Ton-That et al. 2004a) .
Homologs of S. aureus SrtA are grouped into class A sortases (often collectively referred to as housekeeping sortases), whereas sortases involved in pilus polymerization, termed pilin-specific sortases like SrtA of C. diphtheriae, belong to class C (Comfort and Clubb 2004; Dramsi et al. 2005). There are additional sortase groups termed B, D, E and F. Class B sortases are involved in iron acquisition, but the others are less well understood (Spirig et al. 2011). It was first determined in C. diphtheriae that the housekeeping sortase SrtF plays an essential role in surface display of pilus polymers generated by a pilin-specific sortase (Ton-That and Schneewind 2003; Swaminathan et al. 2007). By fractionating corynebacterial cells into extracelluar milieu and cell wall compartments, Swaminathan and colleagues showed that deletion of srtF resulted in abundant secretion of SpaA pilus polymers into the culture medium, a phenotype similar to a multiple deletion mutant expressing only SrtA; furthermore, a mutant strain expressing only SrtF failed to produce SpaA polymers (Swaminathan et al. 2007). This work has established a principle for a biphasic model of pilus assembly in Gram-positive bacteria, whereby pilus polymerization catalyzed by a pilin-specific sortase is terminated by cell wall anchoring of the resulting pilus polymers that is catalyzed by non-polymerizing sortase or the housekeeping sortase (Mandlik et al. 2008b) .
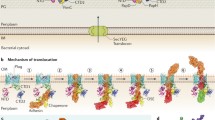
According to this model using C. diphtheriae SpaABC pili as an example (Fig. 7.3), Spa pilin precursors are targeted to the general secretion machinery (Sec) via the N-terminal signal peptide for translocation into the extracellular space. Subunits move across the membrane until they are tethered in place by the C-terminal CWSS. Within the exoplasm, the pilin-specific sortase SrtA recognizes the LPXTG motif of each pilin and cleaves between threonine (T) and glycine (G), resulting in formation of an acyl-enzyme intermediate between the catalytic cysteine residue of the sortase enzyme and the threonine residue of the Spa pilins . Polymerization of SpaA or the cross-linking of subunits SpaC to SpaA occurs by a nucleophilic attack of the pilin motif lysine of a neighboring SpaA-SrtA acyl-enzyme intermediate to the thioester linkage of the next. This process occurs repeatedly to construct the SpaA shaft; pilus assembly, thus, is catalyzed in a bottom-up fashion with SpaC-SpaA linkage as the first transpeptidase reaction. Pilus polymerization is terminated when SpaB tethered the pilus base is transferred to the housekeeping sortase, which catalyzes cell wall anchoring of SpaB, hence the pilus polymers .
Biphasic model of pilus assembly with C. diphtheriae SpaA pili. Shown are the gene locus spaA-srtA-spaB-spaC that encodes the prototype SpaA-type pilus and the housekeeping sortase gene located elsewhere a Pilus assembly on the cell surface occurs by a two-step mechanism b Pilin-specific sortase (black) catalyzes polymerization of Spa pilins with SpaC at the tip and SpaA forming the shaft. SpaB enters the pilus base, which terminates pilus polymerization and triggers cell wall anchoring by the housekeeping sortase (orange) (see text for detail). Heterodimers of SpaB and SpaC are also depicted. Modified from (Chang et al. 2011) with permission
This model has been supported with several lines of evidence. First, as discussed above the mutational analysis of the pilin motif and the SEB fusion protein provided strong evidence for the transpeptidation reaction that crosslinks pilin subunits. Indeed, mass spectrometry analysis of native SpaA pili revealed the predicted isopeptide bond between SpaA subunits, which is formed between the lysine residue K190 of the SpaA pilin motif and the threonine residue within the LPXTG motif of an adjacent subunit (Kang et al. 2009). Similarly, this linkage has been also demonstrated in B. cereus pili using mass spectrometry (Budzik et al. 2008). Presumably, SrtA catalyzes cross-linking to SpaC by a similar mechanism, as this was shown by Budzik and colleagues for the linkage between the tip pilin BcpB and the pilin shaft BcpA (Budzik et al. 2009). Second, the presence of an acyl-enzyme intermediate between pilin and sortase was implicated by mutational analysis of the pilin-specific sortase SrtA of C. diphtheriae (Guttilla et al. 2009) . In this study, several truncations of the membrane anchor domain of SrtA were generated, and the affect of these mutations were analyzed by western blotting with antibodies against SpaA and SrtA. It was found that SrtA mutants loosely bound to the membrane were secreted into the culture medium in complex with SpaA polymers. Third, the two-step pilus assembly, i.e. pilus polymerization by pilin-specific sortase preceding cell wall anchoring by non-polymerizing sortase, is evident by the study of Swaminathan and colleagues with the housekeeping sortase SrtF, as described above. Finally, in agreement with this mode of assembly Mandlik and coworkers elegantly showed that the pilus base SpaB functions as a molecular switch that terminates pilus polymerization, leading to cell wall anchoring of the resulting pilus polymer (Mandlik et al. 2008a). Specifically, deletion of spaB caused a similar phenotype as deletion of srtF, i.e. pilus secretion into the culture medium. Since SpaB is a preferred substrate of SrtF (Mandlik et al. 2008a), formation of a SpaB-SrtF acyl-enzyme intermediate might serve as the signal to end pilus polymerization and begin cell wall anchoring. For this to occur, it was shown that the lysine residue K139 of SpaB functions as a nucleotide for a transpeptidation reaction that links SpaB to the pilus base, similar to the transpeptidation reaction that crosslinks SpaA pilins. SrtF then catalyzes cell wall anchoring of SpaB-linked polymers. SpaB, thus, additionally contributes to control the pilus length because it serves as the rate-limiting step in polymerization termination. Indeed, deletion of spaB is associated with the secretion of abnormally long SpaA polymers into the culture medium. Significantly, studies of pilus assembly in B. cereus, S. agalactiae and S. pyogenes lend further support to this two-step mechanism (Budzik et al. 2007; Nobbs et al. 2008; Smith et al. 2010) .
7.3.2 Sortase Specificity
As aforementioned, unlike the housekeeping sortase gene srtF of C. diphtheriae NCTC 13129 other pilin-specific sortase genes are clustered into three loci. Previous work has shown there is no cross-activity between pilin-specific sortases of different loci, i.e. SrtA is solely required for the assembly of SpaA pili, whereas SrtB and SrtC are specific for the SpaD-type pili (Ton-That et al. 2004b; Ton-That and Schneewind 2003; Swaminathan et al. 2007; Gaspar and Ton-That 2006) . However, specificity of pilin-specific sortases within a pilus type is promiscuous; for example, either SrtB or SrtC is sufficient to catalyze pilus polymerization of SpaDF pilin, but SrtB is specific for the incorporation of the minor pilin SpaE into the SpaD pili (Gaspar and Ton-That 2006). Intriguingly, the housekeeping sortase SrtF is able to catalyze cell wall anchoring of all pilus types to the cell wall peptidoglycan. A long standing question is how sortase specificity is determined? It appears that the LPXTG motif is one determining factor, as this is supported by mutational analysis of the SpaB CWSS (Chang et al. 2011). Like all the base pilins of C. diphtheriae, the SpaB CWSS contains the LAXTG motif, which has been proposed to be the preferred substrate of the housekeeping sortase SrtF (Mandlik et al. 2008a). As SpaB serves as a molecular switch for cell wall anchoring, it would have a greater affinity for SrtF than SrtA. Consistently, deletion of srtF led to increased expression of SpaB that failed to anchor to the cell wall (Mandlik et al. 2008a). However, when the LAFTG motif of SpaB was mutated to the SpaA LPXTG motif, i.e. LPLTG, the mutant SpaB now became a substrate of SrtA, as shown by the ability of SrtA to anchor SpaA pili in the absence of srtF (Chang et al. 2011). This is in agreement with previous studies in S. aureus that show SrtA enzymes recognize the five amino acid peptide LPETG, not the NPQTN peptide, which is the substrate of class B sortase SrtB (Mazmanian et al. 2002) .
The LPXTG motif is not the only determining factor for sortase specificity. Evidently, both SpaA and SpaH pilins contain the LPLTG motif; however, SrtA cannot polymerize SpaH, and neither SrtD nor SrtE catalyzed pilus polymerization of SpaA (Mandlik et al. 2007; Swaminathan et al. 2007). Intriguingly, corynebacterial SrtD is able to polymerize FimA, the major pilin shaft of A. oris type 2 fimbriae (Mishra et al. 2007), when FimA is expressed in C. diphtheriae (Ton-That et al. 2004b). Phylogenetic analysis of FimA and SpaH sequences shows both pilins are closely related (Mishra et al. 2007). While both have the same LPLTG motif, several conserved elements are revealed. It is thus appealing to know if these elements contribute to sortase specificity. Given the proximity of pilin substrates and pilin-specific sortases on the membrane (Guttilla et al. 2009), we speculate that other domains of the CWSS sequence, i.e. hydrophobic domain and positively charged tail, may also be important as they are required for substrate retention within the bacterial membrane (Mazmanian et al. 2001; Mandlik et al. 2008a) .
7.3.3 Pilusosome: A Pilus Assembly Center
Sortases are membrane-bound enzymes working on the outer leaflet of the cytoplasmic membrane (Mazmanian et al. 1999, 2001) . Both sortases and pilin substrates harbor an N-terminal signal peptide; thus, they are subjected to translocation across the membrane by the Sec machinery. Based upon the requirements of protein secretion, the close proximity of sortases and their cognate substrates, specificity of sortase enzymes, and highly organized manner of assembly, it is hypothesized that secretion and pilus assembly machineries are found in close proximity and coupled. A hint for this conjecture came from the study of the pilus base SpaB (Mandlik et al. 2008a). In this study, the CWSS of SpaB was removed, hence preventing it from being inserted into the bacterial membrane. However, SpaB was found to be incorporated into the SpaA pilus structures, although these polymers were secreted into the extracellular milieu. This indicated that SrtA catalyzes the attachment of SpaB to the pilus base before completion of protein translocation suggesting that sortase and the secretion machinery are in close proximity and coupled. In support of this model, immuno-electron microscopic studies have revealed that SrtA, its cognate pilin substrates, and the translocation motor SecA are co-localized (Guttilla et al. 2009). This observation was corroborated by another study that examined co-localization of E. faecalis sortase and SecA (Kline et al. 2009). Interestingly, co-localization was shown to be dependent on a positively charged cytoplasmic domain within the sortase enzyme, suggesting that a retention signal may be responsible for pilusosome maintenance. An outstanding problem is how the orderly pilus assembly is orchestrated within the pilusosome. More experimental work is necessary to elucidate the apparent complex pathway of pilus assembly in Gram-positive bacteria .
7.4 A Structural View of Pilus Assembly
7.4.1 Three-Dimensional Structures of Pilins
The first crystal structure of Gram-positive pilins was solved using the minor pilin GBS52 of Streptococcus agalactiae (Krishnan et al. 2007). This pilin is comprised of two IgG-rev (CnaB) domains termed N1 and N2 that are joined by a short linker region. An in vitro experiment with recombinant GBS52 protein conjugated to fluorescently labeled beads revealed that the IgG-rev (CnaB) regions of the protein were crucial for binding to host target tissues. This was interesting because the S. aureus Cna B repeat region with IgG-rev fold exhibits no adhesive properties (Rich et al. 1998), suggestive of additional functions of the IgG-rev fold.
Shortly after, a crystallization study by the Baker group revealed similar IgG-like domains in the shaft pilin Spy0128 of Group A Streptococcus (GAS) pili (Kang et al. 2007). More important is the discovery of intramolecular isopeptide bonds in Spy0128, a feature not previously reported in any organism (Kang et al. 2007). The crystal structure of Spy0128 is characterized by two IgG-like domains both containing an isopeptide bond formed between lysine (Lys) and aspargine (Asn) residues within hydrophobic regions of the structure. These linkages, confirmed by mass spectrometry, form autocatalytically with assistance from a nearby Asp or glutamate residues. These residues act as proton shuttles permitting Lys to nucleophilicly attack the carbonyl carbon of the Asn R-group resulting in isopeptide bond formation. Significantly, the thermal stability and proteolytic stability of Spy0128 were defected in Spy0128 mutants that are devoid of the isopeptide bonds (Kang and Baker 2009) . As more and more structures of Gram-positive pilins become available, it has become clear that the folding patterns of crystallized pilus proteins are remarkably similar, i.e. IgG-like fold, despite low amino acid identity among known Gram-positive pilins (see (Krishnan and Narayana 2011; Vengadesan and Narayana 2011) for an in-depth analysis) .
A crystal structure of the C. diphtheriae shaft pilin SpaA has been also solved to a resolution of 1.6 A (Kang et al. 2009) (Fig. 7.4). Although the structure exhibits common features, like the IgG-like fold and isopeptide linkages, it has some unique features that are absent from other Gram-positive pilins . The SpaA structure is comprised of three tandem Ig-like domains termed N-terminal (N-domain), middle (M-domain) and C-termimal (C-domain). Both the N and C domains exhibited an IgG-rev fold, while the M-domain is characterized as DEv-IgG fold or CnaA-type fold. Lys-199 and Asn-321 in the M-domain and Lys-363 and Asn-482 in the C-domain form the two intramolecular isopeptide bonds with assistance from acidic residues Asp-241 and Glu-446, respectively. Interestingly, Glu-446 is the conserved residue of the E-Box that involves formation of the Lys-363–Asn-462 intramolecular bond. Furthermore, given that the mutations of Glu-446 severely affect the incorporation of SpaB into the pilus structure (Ton-That et al. 2004b) and the E-Box is in close proximity of the SpaA LPXTG motif, it has been speculated that stability of the C-domain conferred by the intramolecular linkage is required for SpaB incorporation (Kang et al. 2009). Within the crystal, SpaA molecules are arranged in a head-to-toe manner in which the lysine pilin motif abuts the C-terminus of a neighboring protein. SpaA positioning, remarkably, is reminiscent of the proposed ordered assembly proposed to occur in vivo.
Crystal structure of the Corynebacterium diphtheriae shaft pilin SpaA. a The SpaA molecules are stacked end-to-end, in which the C-domain (blue) of one SpaA molecule packs against the N-domain (gold) of the next. The middle domain (M; green) contains a Ca2+ ion (grey sphere). Residues forming isopeptide bonds are shown in red. The lysine pilin motif is labeled K. b Schematic representation of IgG-like folds in each domain with isopeptide bonds showing as a red bar and a disulfide bond as a grey bar. c Shown is the enlargement of end-to-end linkage between two SpaA molecules. The 10 missing residues of the C-terminus is shown with a broken line. Positions of lysine and tryptophan residues in the pilin motif are indicated. (Reprinted from (Kang et al. 2009) with permission)
SpaA exhibits a number of distinguishing features compared to other known pilins. First, the M-domain displays a calcium-binding site that was demonstrated to show high affinity as high levels of chelating agent EDTA could not remove it. The importance of this calcium binding site to the overall SpaA structure has not been explored. In addition, a disulfide bond joining two neighboring β-strands is present within the C-terminus, and is believed to provide additional stability to the protein (Kang et al. 2009). This bond, interestingly, appears to be unique to actinobacterial pili as similar bonds have only been detected in the major shaft pilin FimA of A. oris (Mishra et al. 2011). It remains to be seen if these unique features contribute to the stability and function of SpaA pilins .
7.4.2 Three-Dimensional Structures of Pilin-Specific Sortase Enzymes
Surprisingly, three-dimensional (3D) structures of neither pilin-specific sortase nor the housekeeping sortase of C. diphtheriae are available, considering the well-studied SpaA pilus system in this organism. Nonetheless, structures of different sortase classes solved to date have provided some insights into the mode of sortase catalytic activities. The first 3D structure of sortase was solved with S. aureus SrtA by nuclear magnetic resonance (NMR) (Ilangovan et al. 2001). S. aureus SrtA folds into an eight-stranded anti-parallel β-barrel structure with the active site consisting of His120, Cys184 and Arg197. Based on structural, biochemical and genetic analyses of S. aureus SrtA, sortase activity is dependent on this catalytic triad (Ilangovan et al. 2001; Ton-That et al. 2002; Zong et al. 2004). While the enzymatic activity of S. aureus SrtA is drastically increased by the presence of Ca2+ (Ilangovan et al. 2001), other sortase enzymes tested so far do not require Ca2+ for their activity.
Classes of sortase vary in terms of tertiary structure, but the overall shape of the transpeptidase enzymes is conserved. They all contain the core β barrel and have a similar configuration of the active site (Spirig et al. 2011). However, 3D structures of class C sortases reveal a unique feature that is the presence of a “lid”, first identified in S. pneumoniae pilin-specific sortases SrtC1 and SrtC3 (Manzano et al. 2008). The lid, comprised of a flexible hinge region, a leucine (Leu), and DPW motif hovers over the catalytic triad and surrounding hydrophobic pocket. It has been hypothesized that the lid provides stability to this region as the Asp (D) of the DPW motif has been shown to interact with the reactive residue Arg of the His-Cys-Arg triad. In support of this, deletion of the lid region within the S. pneumoniae SrtC-1 led to protein instability (Manzano et al. 2009). Furthermore, mutations of the Asp and Trp residues abrogate pilus polymerization, presumably due to instability of the sortase enzyme (Manzano et al. 2009). In contrast, studies in A. oris and S. agalactiae showed that mutations of the DWP motif did not affect pilus polymerization (Cozzi et al. 2011; Wu et al. 2012). Intriguingly, Khare and colleagues crystallized the S. agalactiae pilin-specific SrtC1 along with an isogenic mutant in which the lid anchor sequence replaced with IPNTG, the sorting signal of the pilin shaft protein GBS80. (Vengadesan et al. 2011), in place of KDPYS, the lid anchor region of SrtC1. While the mutation did not affect the enzyme integrity or overall structure, the mutant SrtC1 lacked electron density for the introduced ‘IPNTG’ motif exhibited differences around the active site region, speculating the active site and the lid may play a role in sortase specificity (Khare et al. 2011). Thus, it would be revealing to have co-structures of the pilin-specific sortase SrtC1 and its cognate substrate.
7.5 Cellular Adhesion and Tissue Tropism of Corynebacterium diphtheriae Pili
More than three decades ago, C. renale expressing pili were observed to aggultinate trypsinized sheep erythrocytes which could be blocked with anti-pili serum (Honda and Yanagawa 1974). Later, it was also shown that C. renale pili mediated attachment to mammalian cells (Honda and Yanagawa 1975), thus demonstrating that the pili were important for adhesion . Not much was known about adhesive properties of C. diphtheriae pili, except for one study that showed haemagglutination of some C. diphtheriae strains associated with the presence of pili (Ermolayev et al. 1987). Until recently, Mandlik and colleagues employed a battery of sortase and pilin mutants to examine the ability of C. diphtheriae to adhere to different epithelial cells (Mandlik et al. 2007). The SpaA-type pilus was found to bind specifically to pharyngeal epithelial cells, the major site of corynebacterial infection, whereas the SpaD- and SpaH-type pili displayed certain binding specificity to epithelial laryngeal and lung cells , respectively. The specific binding of SpaA pili to pharyngeal epithelial cells is attributed to the minor pilins SpaB and SpaC. This was supported by drastic reduction in binding to pharyngeal epithelial cells by a mutant that lacks spaB and spaC, as compared to strain expressing all three pilins (Mandlik et al. 2007). Secondly, it was demonstrated that latex beads coated with SpaB or SpaC protein adhered to pharyngeal epithelial cells, while those conjugated to SpaA did not. More recently, it was shown that a dimer formed between SpaB and SpaC was observed on the bacterial cell surface (Fig. 7.3), leading to the speculation that long-range adhesion can be mediated by long pilus fibers, while monomeric and heteromeric pilins provide a close surface contact with host cells (Chang et al. 2011). It is also possible that the differential binding mediated by various forms of pili and pilins may contribute to efficient delivery of virulence factors such as diphtheria toxin (Mandlik et al. 2008b).
7.6 Concluding Remarks
Klebs first identified C. diphtheriae as the causative agent of diphtheria in 1883. Since that time, studies focusing on this organism have advanced our understanding of bacterial pathogenesis by contributing to the mechanistic elucidation of diphtheria, as well as the development of diphtheria vaccines. Genomic studies of C. diphtheriae and subsequent investigations of C. diphtheriae covalently-linked pili have expanded our knowledge regarding the arsenal of corynebacterial virulence determinants. We now know the SpaA pilus is the major adhesin that targets corynebacteria to pharyngeal epithelial cells, the main site of infection. Work remains to be seen whether SpaA pili contribute to the establishment of the deadly disease. As pili are a common feature of many Gram-positive bacteria and the mode of pilus assembly is conserved, understanding pilus biogenesis, structural biology, and pilus-mediated pathogenesis will facilitate the development of antimicrobial agents and new vaccines in an era of rampant antimicrobial resistance.
References
Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, Von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B (2006) A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A 103:2857–2862
Budzik JM, Marraffini LA, Schneewind O (2007) Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol 66:495–510
Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O (2008) Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci U S A 105:10215–10220
Budzik JM, Oh SY, Schneewind O (2009) Sortase D Forms the Covalent Bond That Links BcpB to the Tip of Bacillus cereus Pili. J Biol Chem 284:12989–12997
Cerdeno-Tarraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, Chillingworth T, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Quail MA, Rabbinowitsch E, Rutherford KM, Thomson NR, Unwin L, Whitehead S, Barrell BG, Parkhill J (2003) The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res 31:6516–6523
Chang C, Mandlik A, Das A, Ton-That H (2011) Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol Microbiol 79:1236–47
Comfort D, Clubb RT (2004) A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun 72:2710–2722
Cozzi R, Malito E, Nuccitelli A, D’onofrio M, Martinelli M, Ferlenghi I, Grandi G, Telford JL, Maione D, Rinaudo CD (2011) Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J 25:1874–1886
Dramsi S, Trieu-Cuot, P, Bierne H (2005) Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol 156:289–297
Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P (2006) Assembly and role of pili in group B streptococci. Mol Microbiol 60:1401–1413
Ermolayev AV, Fish NG, Birger MO, Sanzhakova IE, Lobanova AN (1987) A study on the adhesive properties of Corynebacterium diphtheriae and Corynebacterium. parvum in the direct haemagglutination reaction. J Hyg Epidemiol Microbiol Immunol 31:313–319
Gaspar AH, Ton-That H (2006) Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol 188:1526–1533
Girard AE, Jacius BH (1974) Ultrastructure of Actinomyces viscosus and Actinomyces naeslundii. Arch Oral Biol 19:71–79
Guttilla IK, Gaspar AH, Swierczynski A, Swaminathan A, Dwivedi P, Das A, Ton-That H (2009) Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in Gram-positive bacteria. J Bacteriol 191:5603–5612
Honda E, Yanagawa R (1974) Agglutination of trypsinized sheep erythrocytes by the pili of Corynebacterium renale. Infect Immun 10:1426–1432
Honda E, Yanagawa R (1975) Attachment of Corynebacterium renale to tissue culture cells by the pili. Am J Vet Res 36:1663–1666
Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT (2001) Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A 98:6056–6061
Iwaki M, Komiya T, Yamamoto A, Ishiwa A, Nagata N, Arakawa Y, Takahashi M (2010) Genome organization and pathogenicity of Corynebacterium diphtheriae C7(−) and PW8 strains. Infect Immun 78:3791–3800
Kang HJ, Baker EN (2009) Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem 284:20729–20737
Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN (2007) Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 318:1625–1628
Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN (2009) The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci U S A 106:16967–16971
Khare B, Fu ZQ, Huang IH, Ton-That H, Narayana SV (2011) The Crystal Structure Analysis of Group B Streptococcus Sortase C1: a Model for the “Lid” Movement upon Substrate Binding. J Mol Biol 414:563–577
Klebs E (1883) Über Diphtherie. Verh Cong Inn Med 2:139–154
Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ (2009) Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 191:3237–3247
Kline KA, Dodson KW, Caparon MG, Hultgren SJ (2010) A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol 18:224–232
Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV (2007) An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure 15:893–903
Krishnan V, Narayana SV (2011) Crystallography of gram-positive bacterial adhesins. Adv Exp Med Biol 715:175–195
Kumazawa N, Yanagawa R (1972) Chemical properties of the pili of Corynebacterium renale. Infect Immun 5:27–30
Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL (2005) Genome analysis reveals pili in Group B Streptococcus. Science 309:105
Lemieux J, Hava DL, Basset A, Camilli A (2006) RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun 74:2453–2456
Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, D’agostino N, Miorin L, Buccato S, Mariani M, Galli G, Nogarotto R, Nardi Dei V, Vegni F, Fraser C, Mancuso G, Teti G, Madoff LC, Paoletti LC, Rappuoli R, Kasper DL, Telford JL, Grandi G (2005) Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science 309:148–150
Mandlik A, Swierczynski A, Das A, Ton-That H (2007) Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol 64:111–124
Mandlik A, Das A, Ton-That H (2008a) The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A 105:14147–14152
Mandlik A, Swierczynski A, Das A, Ton-That H (2008b) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16:33–40
Manzano C, Contreras-Martel C, El Mortaji L, Izore T, Fenel D, Vernet T, Schoehn G, Di Guilmi AM, Dessen A (2008) Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure 16:1838–1848
Manzano C, Izore T, Job V, Di Guilmi AM, Dessen A (2009) Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of gram-positive pathogens. Biochemistry 48:10549–10557
Mazmanian SK, Liu G, Ton-That H, Schneewind O (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763
Mazmanian SK, Ton-That H, Schneewind O (2001) Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol 40:1049–1057
Mazmanian SK, Ton-That H, Su K, Schneewind O (2002) An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A 99:2293–2298
Mishra A, Das A, Cisar JO, Ton-That H (2007) Sortase-Catalyzed Assembly of Distinct Heteromeric Fimbriae in Actinomyces naeslundii. J Bacteriol 189:3156–3165
Mishra A, Devarajan B, Reardon ME, Dwivedi P, Krishnan V, Cisar JO, Das A, Narayana SV, Ton-That H (2011) Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol 81:1205–1220
Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL (2005) Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A 102:15641–15646
Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE (2006) Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807
Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL (2008) Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun 76:3550–3560
Phan G, Remaut H, Wang T, Allen WJ, Pirker KF, Lebedev A, Henderson NS, Geibel S, Volkan E, Yan J, Kunze MB, Pinkner JS, Ford B, Kay CW, Li H, Hultgren SJ, Thanassi DG, Waksman G (2011) Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474:49–53
Popovic T, Kombarova SY, Reeves MW, Nakao H, Mazurova IK, Wharton M, Wachsmuth IK, Wenger JD (1996) Molecular epidemiology of diphtheria in Russia, 1985–1994. J Infect Dis 174:1064–1072
Proft T, Baker EN (2009) Pili in Gram-negative and Gram-positive bacteria—Structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635
Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, Thanassi DG, Waksman G, Li H (2008) Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133:640–652
Rich RL, Demeler B, Ashby K, Deivanayagam CC, Petrich JW, Patti JM, Narayana SV, Hook M (1998) Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry 37:15423–15433
Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, Buccato S, Margarit I, Grandi G, Telford JL (2006) Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol 61:126–141
Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G (1999) Structural basis of chaperone function and pilus biogenesis. Science 285:1058–1061
Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ (2000) Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol 3:65–72
Sauer FG, Pinkner JS, Waksman G, Hultgren SJ (2002) Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543–551
Schneewind O, Model P, Fischetti VA (1992) Sorting of protein A to the staphylococcal cell wall. Cell 70:267–281
Sjoquist J, Meloun B, Hjelm H (1972) Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur J Biochem 29:572–578
Smith WD, Pointon JA, Abbot E, Kang HJ, Baker EN, Hirst BH, Wilson JA, Banfield MJ, Kehoe MA (2010) Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J Bacteriol 192:4651–4659
Soriani M, Telford JL (2010) Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol 5:735–747
Spirig T, Weiner EM, Clubb RT (2011) Sortase enzymes in Gram-positive bacteria. Mol Microbiol 82:1044–1059
Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H (2007) Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol 66:961–974
Swierczynski A, Ton-That H (2006) Type III pilus of Corynebacteria: pilus length is determined by the level of its major pilin subunit. J Bacteriol 188:6318–6325
Thanassi DG, Saulino ET, Hultgren SJ (1998) The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol 1:223–231
Ton-That H, Schneewind O (2003) Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50:1429–1438
Ton-That H, Schneewind O (2004) Assembly of pili in Gram-positive bacteria. Trends Microbiol 12:228–234
Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O (1999) Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96:12424–12429
Ton-That H, Marraffini LA, Schneewind O (2004a) Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta 1694:269–278
Ton-That H, Marraffini LA, Schneewind O (2004b) Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol 53:251–261
Ton-That H, Mazmanian SK, Alksne L, Schneewind O (2002) Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem 277:7447–7452
Trost E, Blom J, De Castro Soares S, Huang IH, Al-Dilaimi A, Schroder J, Jaenicke S, Dorella FA, Rocha FS, Miyoshi A, Azevedo V, Schneider MP, Silva A, Camello TC, Sabbadini PS, Santos CS, Santos LS, Hirata R Jr, Mattos-Guaraldi AL, Efstratiou A, Schmitt MP, Ton-That H, Tauch A (2012) Pangenomic Study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J Bacteriol 194:3199–3215
Vengadesan K, Narayana SV (2011) Structural biology of Gram-positive bacterial adhesins. Protein Sci 20:759–772
Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SV (2011) A Model for Group B Streptococcus Pilus Type 1: the Structure of a 35-kDa C-Terminal fragment of the major pilin GBS80. J Mol Biol 407:731–743
Wu C, Mishra A, Reardon ME, Huang IH, Counts SC, Das A, Ton-That H (2012) Structural determinants of Actinomyces sortase SrtC2 required for membrane localization and assembly of type 2 fimbriae for interbacterial coaggregation and oral biofilm formation. J Bacteriol 194:2531–2539
Yanagawa R, Otsuki K, Tokui T (1968) Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn J Vet Res 16:31–37
Yeung MK (1999) Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med 10:120–138
Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV (2004) Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem 279:31383–31389
Acknowledgments
We thank former and current lab members Anjali Mandlik, Anu Swaminathan, Andrew Gasper, Arlene Swierczynski, Arunima Mishra, I-Hsiu Huang, Elizabeth Rogers, Chenggang Wu, and Chungyu Chang for their invaluable contributions to the pilus work, which was supported by grants AI061381 and DE017382 from the NIH to HTT.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht (outside the USA)
About this chapter
Cite this chapter
Reardon-Robinson, M., Ton-That, H. (2014). Assembly and Function of Corynebacterium diphtheriae Pili. In: Burkovski, A. (eds) Corynebacterium diphtheriae and Related Toxigenic Species. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-7624-1_7
Download citation
DOI: https://doi.org/10.1007/978-94-007-7624-1_7
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-7623-4
Online ISBN: 978-94-007-7624-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)