Abstract
E.B. Ford’s 1964 book Ecological Genetics was a call for biologists to engage in multidisciplinary work in order to elucidate the link between genotype, phenotype, and fitness for ecologically relevant traits. In this review, we argue that the integration of an ecological genomics framework in studies of phenotypic plasticity is a promising approach to elucidate the causal links between genes and the environment, particularly during colonization of novel environments, environmental change, and speciation. This review highlights some of the questions and hypotheses generated from a mechanistic, evolutionary, and ecological perspective, in order to direct the continued and future use of genomic tools in the study of phenotypic plasticity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
E.B. Ford’s 1964 book Ecological Genetics was a call for biologists to engage in multidisciplinary work in order to elucidate the link between genotype, phenotype, and fitness for ecologically relevant traits. It became rapidly clear that methodologies were the main limiting factor in meeting this goal, but recent next generation sequencing (NGS) technologies have reinvigorated interest in this field (Feder and Mitchell-Olds 2003; Orsini et al. 2013). Ecological genetics has given way to ecological genomics (Fig. 5.1), or the investigation of the entire set of genes that interact to produce the phenotype and shape the evolution of species and communities (Ungerer et al. 2008). Ecological genomics is limited less by technology than by the complexity of statistical tools required to quantify the voluminous data, the interdisciplinary knowledge required to fully understand the production of even a single phenotype, experimental constraints (the space required to perform carefully controlled experiments), and the nature of the organism (challenges to raising and breeding the organism, structures that inhibit DNA extraction, etc.), but initial work has been promising (Tollrian and Leese 2010; Whitehead et al. 2012; Andrew et al. 2013).
Ecological genomics is the telling of a complex story about the mechanisms governing the production of a phenotype, but moves beyond functional genomics (e.g., Dalziel et al. 2009) to ask questions concerning the evolution of the phenotype and its role in the greater community (Table 5.1). To date the main focus of ecological genomics has been on the genetic and molecular basis of ecologically relevant traits and their evolutionary consequences. Such an approach cannot capture the full story. Ecological genomics, to be successful, must recognize that genes can only go so far in producing a phenotype – the environment proposes the phenotype in a manner that cannot be separated from the genome (Moczek 2012). Ecological genomic approaches must therefore consider the role of phenotypic plasticity.
Phenotypic plasticity, the environmentally sensitive production of alternative phenotypes by a single genotype (DeWitt and Scheiner 2003) (Fig. 5.2), reminds us that individuals can exhibit phenotypic differentiation not only among genotypes, but also across environments. The conceptual framework of phenotypic plasticity broadens the scope of ecological genomics (Table 5.1) by asking questions and generating novel hypotheses that more fully integrate the environment into the production and evolution of the phenotype. Although plasticity has been the topic of numerous reviews (e.g., Bradshaw 1965; Alpert and Simms 2002; West-Eberhard 2003; Ghalambor et al. 2007; Fusco and Minelli 2010; Pfennig et al. 2010; Moczek et al. 2011; Fitzpatrick 2012; Moczek 2012) and theoretical work (e.g., Lande 2009; Thibert-Plante and Hendry 2011; Espinosa-Soto et al. 2011), only recently have researchers been able to focus on the integration of phenotypic plasticity with ecological genomic approaches (examples of initial forays into ecological genomics include Evans and Wheeler 2000; Renn et al. 2008; McCairns and Bernatchez 2010; De Boer et al. 2011; Richards et al. 2012; Schwartz and Bronikowski 2013).
Some examples of phenotypic plasticity. (a) Genetically-identical clones of Daphnia produce morphological defenses (right) in the presence of predator kairomones. (b) Sea urchin larvae raised under different pH display morphological plasticity (top) at a pH of 7.0, but resist such change at higher pH. This robustness to acidity is due to underlying transcriptional plasticity (bottom) that upregulates biomineralization genes at low pH. At a pH of 7, however, this upregulation disappears. (c) Slijper’s (1942a, b) two-legged goat learned to walk upright on its hindlimbs, resulting in numerous plastic changes to other phenotypes. (a) Shows the morphology of a regular goat, (b) shows the morphology of the two-legged goat, for (a) hindlimb skeletal structure; (b) pelvic musculature, showing the elongated gluteal tongue (gt) and tendon reinforcements (t); (c) thoracic skeleton, showing a transverse, horizontal, and ventral view (left to right); (d) pelvic bones, showing the kangaroo-like ischium (i) of the two-legged goat. (d) Transcriptional plasticity in killifish for the HMGB1 gene (black) with fluctuating temperature (grey) (Figure (a) Reproduced from Laforsch and Tollrian (2010), image kindly provided by C. Laforsch. Published with kind permission of © Elsevier Inc. 2010. All Rights Reserved) (Figure (b) Reproduced from Martin et al. (2011; doi:10.1242/jeb.051169). Published with kind permission of © The Company of Biologists Ltd. 2011. All Rights Reserved) (Figure (c) Reproduced from West-Eberhard (2003; Fig. 3.13, p. 53) with kind permission of Oxford University Press after Slijper (1942a, b). Published with kind permission of © Koninklijke Nederlandse Akademie Wetenschappen 1942. All Rights Reserved) (Figure (d) Reproduced from Podrabsky and Somero (2004; rightmost box of Fig. 4E). Published with kind permission of © The Company of Biologists Ltd. 2004. All Rights Reserved)
Although plasticity involves a single genotype, genomic tools are essential for understanding the molecular basis for how alternative phenotypes may be produced. Given the goal of linking patterns of phenotypic and genotypic variation to patterns of environmental variation, testing the predicted evolutionary consequences and patterns of plasticity will be necessary to understand community-level processes driven by plasticity, and to provide evidence for the fitness consequences of plastic variation in evolving populations. In this review we identify questions and hypotheses about the role of phenotypic plasticity in adaptive evolution and speciation that can be tested with an ecological genomics framework, illustrating how such integrated approaches will enable a depth of insight into ecological and evolutionary questions we would not have considered asking in the twentieth century.
5.2 Genomics and Plasticity
5.2.1 Why Genomics?
Given that genotypically identical individuals can produce different phenotypes under different environmental conditions, it might seem strange to approach plasticity from a genomic perspective. After all, a phenotype cannot be explained solely through a genetic “blueprint”. Along with genes, offspring also inherit epigenetic modifications (Hackett et al. 2013), the internal cellular environment of the gamete/embryo (including lipids, polysaccharides, free nucleotides, transcripts, mitochondria, symbionts, and minerals) (West-Eberhard 2003), the external environment of the developing embryo (Refsnider and Janzen 2012), and/or the environment in which juveniles are reared (Dawkins 1976; Rossitter 1996). Each of these components is important in shaping the phenotype. Each of these components also has the capacity to influence an individual’s fitness. And each of these may be passed on in a relatively stable form for several generations (Crews et al. 2012). Furthermore, individuals can shape their own internal and external environments, which can have phenotypic effects (Dawkins 1982). Therefore, how one differentiates between genetic and environmental effects will depend on one’s starting point; the relationship between genotype and the environment is more integrated than the term “genomics” implies (West-Eberhard 2003; Moczek 2012). This integration has led to concepts like phenotypic accommodation, which questions our ability to discover genes that are “for” certain phenotypes (West-Eberhard 2005).
Of course, this does not imply that the gene is irrelevant, or even equivalent to the actions of the environment, when it comes to the production of phenotypic diversity and its association with fitness. It is the gene that evolves. Selection operates at the level of the phenotype but acts on genetic variation (Lande and Arnold 1983). The environment can produce the effects that it does because gene products are built by selection in such a way as to be so affected. Indeed, genomic tools have established a functional link between gene expression and physiological, morphological, and behavioral plasticity (Aubin-Horth and Renn 2009). A genomics and developmental perspective of plasticity, therefore, enquires into the mechanistic basis, hierarchical interactions, genetic architecture, and robustness of plasticity, while remembering that phenomena other than plasticity exist (Table 5.2). In the context of ecological genomics, the integration of these facets under the predictive framework of the ecological theory of adaptive divergence provides a means to move beyond the notion that plasticity is common in nature and towards actually understanding (and predicting) its role in adaptive evolution (Schluter 2000).
5.2.2 The Mechanisms of Plasticity
There are at least two distinct forms of environmental induction. First, the environment can force the phenotype by virtue of chemical and physical laws (passive induction). For instance, temperature can cause phenotypic changes through enzyme kinetics and diffusion rates, while low nutrient availability can impact growth and morphology. Second, phenotypic change can be wrought through a complex interaction between environmental cues, sensors for the cues, signaling molecules that transfer information about the environment, and all of the machinery involved in phenotypic modification (active induction) (Windig et al. 2003). Understanding the actual form of induction will therefore be key to understanding the causal link between genotype and phenotype.
The active pathway from cue → receptor → signal → translation of signal → phenotype is being elucidated in a few species (Beldade et al. 2011). For example, predator-secreted cues (kairomones) are known to induce morphological defenses in Daphnia species (Fig. 5.2), but little is known about the structure of the receptors associated with these chemical compounds (Peñalva-Arana et al. 2009; Akkas et al. 2010; Miyakawa et al. 2010). Activated kairomone receptors stimulate neural pathways to release hormones into the hemolymph (Barry 2002; Weiss et al. 2012). These hormones, including juvenile and insulin signaling hormones (Miyakawa et al. 2010), target polynucleated cells that control production of the inducible structures (Beaton and Hebert 1997; Barry 2002; Simon et al. 2011), resulting in increased transcriptional activity and post-translational modifications of structural proteins (Schwarzenberger et al. 2009; reviewed in Tollrian and Leese 2010). Plasticity, in turn, comes with a cost to the immune system (Yin et al. 2011). This summary represents decades of research in an easily-reared model organism with a sequenced genome, whose plasticity has been known since the early 1900s (Woltereck 1909), and yet the number of genes involved, their function, and their fitness consequences are only beginning to be determined. Even less is known of plasticity in ecologically important non-model species, reinforcing the significance of ecological genomics as an approach to understanding the consequences of plasticity.
Overall, phenotypic plasticity is possible because of the environmental sensitivity of gene expression or protein, lipid, and RNA activity, and/or variation in the levels of environmental components that are required for the production of a “normal” phenotype. This environmental sensitivity, in turn, may be driven by epigenetics (Richards et al. 2010), exploratory behavior coupled with intra-individual selection (Frankenhuis and Panchanathan 2011; Snell-Rood 2012), and/or the evolved coordinated response to the stimulation of environmental sensors (Tollrian and Leese 2010). Ecological genomic studies are revealing that the development of alternative phenotypes by a single genotype may be common but is amazingly complex, involving the interplay of numerous plastic and non-plastic reaction norms moving through developmental trajectories (Beldade et al. 2011; Sommer and Ogawa 2011; Valena and Moczek 2012; Zhou et al. 2012).
The hierarchy of plasticities (Bradshaw 1965), including transcriptional and proteomic plasticity, protein activity plasticity, physiological and morphological plasticity, and behavioral plasticity. Note that in this hierarchy higher-level reaction norms can affect lower-level reaction norms, and vice versa, as indicated by the two-directional arrows. Behavioral plasticity especially can alter the rest of the hierarchy, as behavioral plasticity can bring organisms into new environments
5.2.3 Interactions of Reaction Norms
A major challenge for the ecological genomics of plasticity will be elucidating the relationship between reaction norms at all levels of the phenotype, from molecular plasticity to physiological and morphological plasticity, to behavioral plasticity. This interaction is known as the hierarchy of plasticities (Bradshaw 1965) (Fig. 5.3).
5.2.3.1 Molecular Plasticity
Gene expression can be measured as a molecular phenotype (Ranz and Machado 2006) that responds to the environment (Gracey et al. 2004; Greenberg et al. 2012; Yampolsky et al. 2012). Measuring transcriptional plasticity has its advantages: thousands of phenotypes can be measured simultaneously from a small sample, revealing plastic phenotypes that a priori predictions may not have anticipated. Since transcripts are gene copies, candidate genes involved in plasticity can be identified (Pavey et al. 2010). For instance, Podrabsky and Somero (2005) subjected killifish to different temperatures and found a tight negative correlation with high mobility group box one protein (HMGB1) transcript abundance, identifying HMGB1 as a putative global temperature sensor on top of its previously described roles as regulatory protein and cytokine (Müller et al. 2001; Vezzoli et al. 2011) (Fig. 5.2). This hypothesis could not have been generated without the integration of transcriptional plasticity.
Proteomic plasticity measures protein abundance for the entire proteome under different environmental conditions. Proteomic plasticity has been well-documented in several organisms, although the integration of proteomics with ecological genomics is currently limited (Diz et al. 2012). As with transcriptional plasticity, proteomic plasticity can identify potential candidate genes for plasticity (including some not found in the transcriptome nor annotated from the genome; Findlay et al. 2009; Schrimpf et al. 2009), can measure thousands of phenotypes simultaneously, is closely associated with the genome, and can uncover unanticipated plastic phenotypes. Unlike transcript abundance, protein abundance is one step closer to the expression of the macrophenotype (Diz et al. 2012).
The importance of transcriptional plasticity for ecological genomics has been questioned in light of advances in proteomics, on biological rather than methodological grounds. For instance, it has been suggested that the control of protein production is more essential than the control of transcript abundance, as it imposes heavier costs (Malakar and Venkatesh 2012). However,estimates of the costs of protein production suggest they are minimal (Stoebel et al. 2008; Shachrai et al. 2010; Eames and Kortemme 2012), but may increase with stress (Vilaprinyo et al. 2010). Furthermore, evidence for selection against long introns in highly expressed genes indicates that transcription is also costly (Castillo-Davis et al. 2002) and can influence energy reserves and fitness (Wagner 2007; Lang et al. 2009). Altogether, studies that aim to understand the fitness consequences of molecular plasticity may shed more light on the adaptive link between transcript and protein abundance.
Developmental noise has also been used to defend a proteomic rather than a transcriptomic perspective. Genes involved in plasticity tend to be transcriptionally noisy; a decoupling between transcript and protein abundance is predicted to evolve as a strategy to reduce the impact of transcriptional noise on the phenotype (Raser and O’Shea 2005; Maier et al. 2011). Indeed, correlations between transcript and protein abundance tend to be low (Diz et al. 2012), although this varies with the type of gene and the type of regulation investigated (Lee et al. 2011; Maier et al. 2011). However, experiments on yeast have demonstrated that plasticity is not as noisy as once thought. There is a negative relationship between how vital the gene is for cellular functions and the amount of noise it generates. This has been achieved through the selection of certain genetic architectures, with greater noise being associated with particular chromatin dynamics and promoter types (Lehner 2010), epigenetic modifications (Viñuelas et al. 2012), and translational efficiencies (Bajić and Poyatos 2012). Overall, noise provides a biologically relevant reason why protein abundance should not be ignored, but this should not preclude efforts to understand the ecological genomics of transcriptional plasticity.
Despite noise, transcript abundance tends to drive protein abundance, linking these two phenotypes together in the hierarchy of plasticities. However, this relationship is in practice difficult to determine. Plasticity in the expression of one gene can have pleiotropic effects on other genes (Zhou et al. 2012), making it difficult to determine which plastic phenotypes are adaptively responding to environmental change, and which are responding via pleiotropy. Since pleiotropic genes may be less vital and therefore more prone to noise, pleiotropy could mask a positive relationship between transcript and protein abundance for adaptively plastic genes. Furthermore, the causal link between transcript and protein abundance may take several forms, further diminishing our ability to measure their relationship. For instance, increased transcript abundance may maintain protein levels if protein degradation increases, while a lack of transcriptional plasticity may allow proteomic plasticity (Beldade et al. 2011). Protein abundance, in turn, may affect transcript expression in a similar manner (Tomanek and Somero 2002; Tomanek 2008). Even if transcript and protein abundance are not correlated, the fitness consequences of unnecessary plasticity should be of ecological interest (Lang et al. 2009). In short, to understand the hierarchy of plasticities, both transcriptional and proteomic plasticity must be measured for a single gene, and the relationship between these reaction norms ascertained through techniques such as RNA interference, morpholinos, and methylation (Juliano et al. 2005; Zhou et al. 2007; Wang et al. 2012). If, for instance, protein abundance changes across environments, what happens to protein abundance when transcript production is suppressed across environments in adult organisms?
Along these lines, other sources of molecular plasticity such as metabolomics and epigenomics will become increasingly incorporated into ecological genomics studies (Bossdorf et al. 2008; Sardans et al. 2011). The epigenome is of special interest as techniques for sequencing methylated regions of DNA have only recently been established (reviewed in Bock 2012). Recent studies on plasticity and the epigenome have shown that epigenetic modifications can produce alternative phenotypes within a single individual (Herrera and Bazaga 2012), can plastically prepare offspring for uncertain future conditions (Angers et al. 2010), and can transfer plastic changes induced in one generation to future generations. The latter is particularly interesting, as plastic modifications to the phenotype in one generation can arise in later generations, even if the later generations never experience the inducing environment (Stern et al. 2012). The effects of the epigenome on plasticity are context-specific, and in some systems have been known to limit the development of alternative phenotypes (Roberts and Gavery 2012). However, even in such cases methylation and histone modifications are induced by the environment, and can be measured as a form of intergenerational plasticity. There are still many questions to answer regarding the relationship between plasticity and the epigenome, but epigenetics does seem to be an important mechanism in at least some forms of plasticity (Richards et al. 2010; Valena and Moczek 2012).
Proteins may have their own reaction norms apart from protein abundance. Protein movement, half-life, and enzyme efficiency are all influenced by the environment. Their degree of plasticity, however, is dependent on their amino acid sequences. Changes to amino acid sequences can alter reaction norms, increasing or decreasing plasticity in protein behavior (Powers and Schulte 1998). Finally, interactions between proteins, genes, non-coding RNA, lipids, etc., can be influenced by the environment and may affect the macrophenotype (Hayward et al. 2007; Tomanek 2008; Deredge et al. 2010).
The integration of molecular plasticity in ecological genomics is driven by several questions: what is the relationship between transcriptional and proteomic plasticity for particular ecologically relevant genes? How does this relationship affect ecologically important traits? Where in the pathway from gene to protein do mutations that alter plasticity lie? Comparing gene and protein sequences for populations with different reaction norms can begin to address these questions. For instance, killifish adapted to cooler waters had an amino acid substitution at site 311 of their lactate dehydrogenase B enzyme that altered the kinetic properties of the enzyme relative to warm-adapted fish (Powers and Schulte 1998). The complex nature of molecular plasticity will be sure to challenge researchers attempting to answer these questions for years to come, but the tools to investigate them are now available.
5.2.3.2 Hierarchy of Plasticities
Transcriptional studies can discover functional relationships between molecular plasticity and physiological, morphological, or behavioral plasticity (e.g. Schwarzenberger et al. 2009; Martin et al. 2011) (Fig. 5.3). What is less appreciated is the relationship between phenotypes that resist environmental change and molecular plasticity.
Non-plastic phenotypes may resist change despite environmental perturbations, and this resistance to the environment may be evolutionarily important. The production of non-plastic traits has been analyzed in some organisms across different environments. For instance, in sea urchin larvae Paracentrotus lividus, morphology was relatively insensitive to decreasing pH. This non-plasticity, however, was maintained by transcriptional plasticity for genes involved in biomineralization. At a pH of 7 morphology was disrupted by pH, and this was associated with a breakdown of gene expression regulation (Martin et al. 2011) (Fig. 5.2). This type of study shows the breadth of reaction norm interactions, and reminds us that the environment may influence the phenotype even if plasticity cannot be readily observed (see plastic compensation).
The highest rung on the plasticity hierarchy is behavioral plasticity. Although behavioral plasticity is difficult to define (see Glossary), it has long been expected that behavioral plasticity can drive plastic changes in other phenotypes, and may be an important first step in the generation of phenotypic variation (Price 2003; West-Eberhard 2003). For instance, a goat born with congenital limb defects learned to walk on two legs, which sparked numerous plastic changes to its musculature and skeleton (West-Eberhard 2003) (Fig. 5.2), while stickleback ecotypes may have evolved morphological differences via behavioral plasticity in diet acquisition (Wund et al. 2008). One intriguing recent hypothesis suggests that exploratory behavior, for cells and for organisms, is likely an important generator of individual differences in plasticity. Individuals that stochastically sample the environment before developing an appropriate phenotype may plastically respond early in development if the environments they sample are homogeneous, leading to reduced environmental sensitivity during later stages of development. Individuals that stochastically sample an unpredictably heterogeneous environment, however, may maintain a propensity for plasticity in later stages of development (Frankenhuis and Panchanathan 2011). Thus the hierarchy of plasticities cannot be conceived as an inflexible chain, but rather every level of the hierarchy can induce plastic changes at every other level.
5.2.4 Genetic Architecture
Genetic architecture of plasticity is concerned with the number, placement, and effect size of genes involved in the development of alternative phenotypes. Most studies that discuss the genetic architecture of plasticity have yet to address any of these subjects. Experimental work has shown that plasticity can be influenced by single genes of large effect. For instance, Caenorhabditis briggsae normally develop into hermaphrodites across all temperatures, but mutations in the she-1 gene (such as v49, which produces an early stop codon, or vDf2, which is a 5′ deletion) can lead to the development of XX females at 25°C and XX hermaphrodites at lower temperatures (Guo et al. 2009). Plants ordinarily exhibit density-dependent plasticity in stem length, but the transgenic addition of an oat phytochrome A gene to tobacco induces long stems even under low densities, while Brassica rapa mutants for phytochrome B exhibit small stems even under high densities (Schmitt et al. 1995). Other examples could be given (Beldade et al. 2011). However, the genetic architecture of plasticity involves more than comparing phenotypic differences between mutant lines; it involves determining the entire complement of genes involved in plasticity, and these are likely more numerous than mutational studies could ever determine.
Given the complex nature of plastic responses, genes involved in plasticity can include any of the components of a plastic response, from cue reception to signal transduction to phenotype production. This can include protein-coding and RNA-coding genes, such as regulatory genes and genes involved in epigenetic modifications. Comparative approaches are ill-prepared to identify this diversity of genes. Quantitative Trait Loci (QTL) and expression QTL studies can identify genomic regions associated with divergent plastic phenotypes between genotypes, but cannot capture loci involved in plasticity that lack genetic or phenotypic variation. Gene expression studies have identified thousands of genes induced by a single environmental variable, but it can be difficult to differentiate between transcripts that produce the induced macrophenotype and transcripts that pleiotropically respond to environmental change or plastic changes in other phenotypes (Aubin-Horth and Renn 2009; Fraser 2011). This task is further limited by the lack of ecological annotation for genes that exhibit molecular plasticity (Pavey et al. 2012). In short, standard approaches for quickly ascertaining the number of genes involved in plasticity (e.g., QTL analysis, microarrays, RNA-sequencing) cannot provide basic information regarding the genetic architecture of plasticity, but can identify those loci that lead to divergent plastic responses or those genes whose expression is environmentally sensitive. To provide a complete picture of the genetic architecture of plasticity, gene expression studies need to be extended across multiple tissues and developmental stages under contrasting environments (Beldade et al. 2011). The transcriptome, proteome, metabolome, epigenome, etc., and their interactions, must all be considered, and the relevance of individual genes or gene networks for the plastic response must be ascertained through gene silencing methods (ex. Zhou et al. 2007) or other functional approaches. The focus of genes under selection, or genes producing divergent plastic responses, although important, cannot preclude research on functionally important genes that lack variation, or non-genetic aspects of the organism that are involved in the production of alternative phenotypes. As seen in the example of Daphnia given above, this will take a coordinated effort by a multitude of researchers with different areas of specialization, all focused on a single species. Such research is already under way, and the results are promising (some recent examples: Bossdorf et al. 2010; Meister et al. 2011; Greenberg et al. 2012; Srinivasan and Brisson 2012).
5.2.5 Robustness of Plasticity
Robustness (often called canalization) tends to be used to describe phenotypes that resist environmental change and are thus non-plastic, but plasticity itself can be robust to stochastic, environmental, and genetic perturbations (Waddington 1953a, b; Gibson and Wagner 2000; Debat and David 2001), as adaptations to maintain a consistent plastic response. Stochastic robustness occurs whenever the reaction norm is resistant to developmental noise. Such resistance can occur via alterations to the surrounding genomic structure, or by loose causal links between molecular plasticity and higher levels of plasticity (Raser and O’Shea 2005; Lehner 2010). Environmental robustness includes a lack of discontinuous change in reaction norm shape under unnatural extreme environments, or the maintenance of reaction norm shape under one environmental variable when a second environmental variable is introduced. For instance, if temperature-induced plasticity is maintained despite changes in salinity, that reaction norm is robust to salinity. How environmental robustness for plasticity occurs, and how species can evolve such robustness, has never, to our knowledge, been explored. It is important to remember that robust reaction norms at one level of the hierarchy may be driven by non-robust reaction norms at other levels of the hierarchy. Finally, reaction norms are tested against diverse genetic backgrounds. Studies of natural populations have revealed that individual genotypes often have distinct reaction norms, indicating a relative lack of genetic robustness (Landry et al. 2006; Bentz et al. 2011). This lack of robustness permits evolution. The alleles generating these changes, however, have rarely been examined, and the relative degree of genetic or environmental robustness for plasticity has not been measured. Going forward, reaction norms induced by a single environmental variable need to be measured when held against other environmental variables or genotypes, and the role of molecular plasticity in maintaining a plastic reaction norm against environmental, genetic, and stochastic perturbations needs to be measured (e.g., Lehner 2010).
Some important concepts in the evolution of plasticity are shown for simple linear reaction norms. Stars and circles represent phenotypic optima within each environment. Ancestral denotes a stable environment that the population was initially adapted to. Derived denotes a novel stable environment that the population has colonized. Solid lines indicate the reaction norm of the population prior to colonization, and the dashed line indicates the reaction norm of the population after evolving in the derived environment. (a) Plasticity-Mediated Population Persistence occurs when plasticity pre-exists and moves the colonizing population towards its new fitness optimum. It may then evolve under directional selection to maximize fitness in both environments (adaptive plasticity). (b) If the derived environment is stable, the population may evolve the loss of plasticity (genetic assimilation, dashed line), such that a return to the ancestral environment would induce no plastic change. (c) Some environments may induce phenotypic changes that move the population away from their phenotypic optimum (maladaptive plasticity, solid line). Selection may then work to bring the population back to its optimum (genetic compensation), potentially causing a loss of plasticity (dashed line). (d) Maladaptive plasticity may not be detected (plastic compensation, solid line), if adaptive plasticity in an underlying trait (dash-dotted line) counteracts maladaptive plasticity on the affected phenotype (arrow). If the underlying trait was not plastic, maladaptive plasticity would be evident in the solid line
5.3 Evolution and Plasticity
The ecological genomics of plasticity is concerned not only with the production of ecologically-relevant traits, but also the consequences of plasticity for population differentiation and evolutionary novelty. Figure 5.4 and the glossary define some important terms (adaptive plasticity, maladaptive plasticity, neutral plasticity). We favor fitness-based rather than historically based definitions of adaptation, as they avoid unnecessary and often untestable assumptions. Some recent findings of the evolutionary significance of plasticity will be discussed below.
Plasticity-Mediated Population Persistence (PMPP) occurs when plasticity enables colonization of a novel environment. In this diagram, the likelihood of persistence across an environmental range is shown for genotype A and genotype B. This likelihood is directly related to the abundance of transcript induced by the environment. Genotype A (dashed line) produces transcript under a narrow set of environments, and so can persist in a narrow set of environments. Genotype B (solid line) can produce transcript under a greater range of environments. Both genotypes can persist in the environmental range at which they evolved (black box). At the edge of Genotype A’s tolerance range (grey box), both genotypes could colonize, but Genotype B has an advantage due to its greater level of transcript abundance. Under extreme environments (open box) Genotype A cannot successfully colonize, while Genotype B has a small likelihood of survival (PMPP) (Modified from Pavey et al. (2010, Fig. 2) with kind permission of © New York Academy of Sciences 2010. All Rights Reserved)
5.3.1 Plasticity and Population Persistence
Baldwin (1896, 1902) hypothesized that adaptive phenotypic plasticity could enable individuals to colonize novel environments (Plasticity-Mediated Population Persistence – PMPP, Pavey et al. 2010) (Fig. 5.5). This has recently been supported by theoretical (Ghalambor et al. 2007; Thibert-Plante and Hendry 2011) and empirical (Yeh and Price 2004; Hahn et al. 2012) research. For example, Daphnia lumholtzi plastically produce head spines in the presence of predators. Its invasive success in North America appears to be mediated by this plasticity: in the presence of native non-plastic Daphnia pulicaria, D. lumholtzi is an inferior competitor, but when predators are introduced D. lumholtzi has a competitive edge (Engel and Tollrian 2009). Numerous other studies have implicated plasticity in invasive success, although it is not always clear if plasticity pre-existed or evolved after colonization (Bachmann et al. 2012; Hanshew and Garica 2012; Molina-Montenegro et al. 2012; Mozdzer and Megonigal 2012; Purchase and Moreau 2012; but see Matzek 2012), an important distinction to make when assessing the role of plasticity in population persistence.
There are predictions regarding the likelihood of PMPP. For instance, organisms that adjust their phenotype post-dispersal are more likely to colonize new environments than individuals that adjust their phenotype irreversibly pre-dispersal (Thibert-Plante and Hendry 2011). Post-dispersal plasticity may also facilitate PMPP by reducing the genetic swamping of migrants (migration load), as migrants and their offspring can plastically adjust to their new surroundings, taking on the phenotypes of residents and limiting selection against interbreeding (Thibert-Plante and Hendry 2011).
One underexplored area of PMPP involves the role of cryptic genetic variation (CGV), a form of standing genetic variation (SGV). Under normal environmental conditions, individuals may exhibit similar phenotypic traits despite genotypic differences, due to the suppression of genetic variation via phenotypic capacitors (Levy and Siegal 2008) or the accumulation of neutral mutations in unexposed regions of the reaction norm (Ghalambor et al. 2007). In either case, novel environments may expose CGV in plasticity, increasing heritability for the phenotype and thereby permitting rapid evolution. PMPP will occur for those individuals whose CGV exhibits plasticity in the adaptive direction. This has likely occurred in the colonization of freshwater environments by marine threespine sticklebacks: freshwater salinities exposed CGV in body size, resulting in the rapid parallel evolution of smaller body sizes in freshwater populations (McGuigan et al. 2011). New genomic tools have allowed the mechanisms governing CGV production to be elucidated (Iwasaki et al. 2013), and its evolutionary significance to be tested. For instance, ribozymes selected for their ability to bind to a particular substrate were replicated via mutagenic Polymerase Chain Reaction (PCR) to introduce genetic variation into the ribozyme population. Following ten generations of replication, ribozymes were again selected for their ability to bind to the same substrate, thereby favoring mutations that had no phenotypic effect (CGV). The wild-type and CGV populations were then introduced to a new substrate. The evolution of enzymatic efficiency in the presence of this new substrate was measured over several generations of moderately-mutagenic PCR, and ribozymes were genotyped each generation. CGV enabled more rapid evolution on this new substrate by “pre-adapting” certain ribozyme genotypes to this new environment (Hayden et al. 2011). Future work is clearly moving away from (albeit important) heritability studies, towards tracking the cryptic alleles responsible for rapid evolution in novel environments.
There are at least six potential consequences of CGV for PMPP under post-dispersal plasticity. First, since selection only favors adaptive plasticity, the colonizing population will have reduced genetic diversity at those loci compared to the ancestral population. This could potentially decrease future evolutionary potential for that phenotype. Second, CGV could increase the likelihood of PMPP relative to small or recently bottlenecked populations that exhibit little CGV (and therefore exhibit plasticity in the same, possibly maladaptive, direction). Third, CGV may increase the likelihood that colonists experience stabilizing rather than directional selection, as the random nature of CGV may produce some individuals with a perfect environment-phenotype match (Ghalambor et al. 2007). Fourth, founder effects and drift could play an important role during PMPP – different colonizing populations from the same ancestral population could have different likelihoods of persistence and be subject to different selection strengths or forms of selection (stabilizing or directional), depending on the subset of CGV present among dispersers. Fifth, CGV could increase the heritability of a trait under new environments, resulting in rapid evolution (Neyfakh and Hartl 1993; Chown et al. 2009; McGuigan et al. 2011). Finally, individuals with different genotypes could produce similar adaptive phenotypes in the novel environment, and be selected together. This could increase their likelihood of reproduction, producing reaction norms comprised of the cryptic alleles from several individuals. This in turn could produce new reaction norms, potentially causing genetic assimilation or increased niche breadth. CGV in reaction norms are clearly important for adaptive evolution and must be included in theories of PMPP and adaptive divergence.
Individual-level differences in plasticity that permit the PMPP of certain individuals may exist in the absence of genetic variation. Identical genotypes that experience different levels of environmental heterogeneity early in life may have altered abilities to respond plastically to novel environments later in life (Frankenhuis and Panchanathan 2011). Exploratory behavior is therefore the non-genetic equivalent of CGV. Experiments that actively uncover the alleles generating CGV, or experimentally account for the prior history of the organism, are needed to differentiate between these genetic and non-genetic processes.
The role of maladaptive plasticity in population persistence has also been relatively ignored (Morris and Rogers 2013). Presumably maladaptive plasticity would decrease the possibility of persistence and increase the likelihood of extinction (plasticity-mediated population extinction, PMPE). PMPE could occur if the phenotype is forced away from its optimum (Ghalambor et al. 2007), or if the environmental context that favored adaptive plasticity were to change. For instance, freshwater snails Physella virgata have evolved adaptive plasticity in shell morphology, such that in the presence of fish predators they can produce crush-resistant rotund shells. These changes come at the cost of reduced fecundity and increased leech predation, and can be induced by non-predatory sunfish. Snails introduced to ponds containing non-predatory sunfish may therefore be less likely to persist because of plasticity (Langerhans and DeWitt 2002). If populations are able to persist despite maladaptive plasticity, this could have some interesting consequences for adaptive divergence (see below).
5.3.2 PMPP and Adaptive Divergence
Ecological speciation results from a combination of colonization of distinct environments and adaptive divergence due to divergent selection. Ironically, theoretical work has shown that post-dispersal plasticity facilitates colonization but inhibits adaptive divergence (Thibert-Plante and Hendry 2011), as plasticity enables migrants to successfully interbreed with residents. Migrant and resident populations therefore remain phenotypically distinct but genetically homogeneous. Pre-dispersal plasticity, however, is unique in that divergent selection predates genetic divergence, as migrants are selected against when competing with residents. Pre-dispersal plasticity can therefore facilitate adaptive divergence, but it reduces the likelihood of colonization (Thibert-Plante and Hendry 2011).
Divergent selection can occur within a single environment in the absence of migration, if the population experiences a fitness minimum. The theory of adaptive speciation states that population size can alter the fitness landscape. Colonizing populations may first evolve under directional selection, allowing population size to increase over time. As population size increases, the adaptations that occurred under directional selection become less favorable. Directional selection therefore moves the population towards a fitness minimum, at which point individuals on either side of the minimum experience divergent selection and follow different evolutionary trajectories (Dieckmann et al. 2004). Given that plasticity can facilitate colonization to new environments, and that plasticity can occur in response to demographic changes (Svanbäck et al. 2009), the role of PMPP in adaptive speciation needs to be addressed. These sorts of models have opened the door to many exciting theoretical and empirical opportunities for researchers testing predictions about plasticity in cases of ecological or adaptive divergence.
5.3.3 PMPP and Evolutionary Rescue
Evolutionary rescue occurs when populations adapt to stressful environments after a period of population decline, such that population size increases. Theoretical work has shown that plasticity can promote evolutionary rescue by slowing the rate of population decline, permitting time for adaptive changes to occur (Chevin et al. 2013).
Reaction norms can evolve in height (a → c) or slope (a → b–e). a (solid line) represents the initially expressed reaction norm upon colonizing a new environment. It does not attain the phenotypic optimum (star), and so is subject to directional selection. The reaction norm can then evolve to meet the optima in both environments (d) or overshoot the optima in the ancestral environment (e). If plasticity is selected against in the derived environment, plasticity could be lost (d → c → b). d could also represent a reaction norm under stabilizing selection
5.3.4 Adaptive Plasticity and Adaptive Divergence
Baldwin (1896, 1902) hypothesized that reaction norms could evolve post-colonization. The likelihood and form of such evolution depends on whether plasticity results in stabilizing (Fig. 5.6d) or directional (Fig. 5.6a) selection (Ghalambor et al. 2007). Stabilizing selection occurs when plasticity brings the phenotype to its fitness maximum. It can reduce the likelihood of reaction norm evolution or lead to the loss of plasticity, depending on whether plasticity is expressed or not expressed in the new environment. Directional selection, which occurs when plasticity does not bring the phenotype to its fitness maximum, could result in the evolution of reaction norm height or slope (Fig. 5.6). One would expect an increased slope (Fig. 5.6d, e) if the population routinely migrated between its ancestral and newly-colonized habitat, if the colonized environment fluctuated beyond the conditions experienced in the ancestral environment, or if gene flow between environments was high (Berrigan and Scheiner 2003; Crispo 2008), and a decreased slope (Fig. 5.6b, c) if the maintenance of plasticity was costly and the environment was stable (see below). Furthermore, polymorphisms in reaction norms may be maintained by fluctuating environments, if different reaction norms produce phenotypes that are optimal in contrasting environments. In short, the nature of environmental fluctuations in the colonized environment, the costs to plasticity, the mutations available to selection, and the strength of selection, will, among others, determine the form that the evolved reaction norm takes. Despite this, there is good evidence that plasticity does evolve, leading to adaptive differences between populations (Crispo 2007; McCairns and Bernatchez 2010; Pfennig et al. 2010; Schwander and Leimar 2011; Svanbäck and Schluter 2012).
5.3.5 Genetic Assimilation and Adaptive Divergence
Plasticity can generate dramatic phenotypic divergence, as seen in the case of a two-legged goat whose musculature changed rapidly upon assuming a bipedal form of locomotion (Slijper 1942a, b). West-Eberhard (2003, 2005) proposed that such phenotypic divergence can come under genetic control. That is, genetic changes could occur that result in the loss of adaptive plasticity, canalizing one possible phenotype across environments. This genetic assimilation (Waddington 1953a, b) (Fig. 5.4) is expected to evolve if the environment remains stable, and: (1) plasticity is costly to maintain when it is not required; (2) neutral mutations accumulate in the unexpressed portion of the reaction norm, such that plasticity is lost in other environments; (3) hybridization is permitted due to plasticity, but hybrids incur some fitness cost (genetic assimilation via reinforcement); or (4) selection reduces the environmental threshold required to induce the phenotypic change (Waddington 1956; West-Eberhard 2003). Differentiating between the causes of genetic assimilation has proven difficult, and some possibilities may not even be plausible. For example, could costs to plasticity be reduced rather than plasticity itself (DeWitt et al. 1998)?
Genetic assimilation has important consequences for ecological genomics. The flexible stem model of evolution (West-Eberhard 2003; Pfennig et al. 2010), in which a plastic ancestral population births phenotypically divergent non-plastic populations, predicts that population phenotypes may not always be built from the “ground up”, but may reflect canalized ends of the same reaction norm. Day et al. (1994) tested plasticity for trophic morphology on benthic and limnetic sticklebacks from Paxton Lake, British Columbia, fed on a “benthic” diet of worms or a “limnetic” diet of plankton. Limnetics, which have a more diverse diet, exhibited significantly greater plasticity in gill raker length than did benthics. Intriguingly, when fish were fed the diet of their contrasting ecotype, plasticity partially moved them in the direction of that ecotype, suggesting that benthic and limnetic individuals were derived from a plastic ancestor. Wund et al. (2008, 2012) complemented this work by comparing diet-induced plasticity in marine, solitary limnetic, and solitary benthic sticklebacks. The marine ancestor was highly plastic, producing a benthic or limnetic morphology depending on the food source (but see Svanbäck and Schluter 2012), but the solitary populations were also plastic. This leads to the intriguing possibility that plasticity was not costly in derived populations, but was reduced in benthic-limnetic species pairs via reinforcement. At the moment this is simply speculation, but the flexible stem model allows such hypotheses to be generated and tested. Phylogenetic studies lend further support to the reality of genetic assimilation (Schwander and Leimar 2011).
5.3.6 Maladaptive Plasticity and Adaptive Divergence
Non-plasticity may evolve for reasons other than genetic assimilation. For instance, adaptive plasticity could become costly if other environmental variables were to change. Populations of Daphnia melanica plastically adjust their melanin production with depth as an adaptation to ultraviolet radiation. This plasticity makes Daphnia visible to predators at shallow depths. Populations recently exposed to predators have rapidly evolved the loss of melanin production plasticity (Scoville and Pfrender 2010). Non-plasticity could also evolve if novel environments were to move the phenotype away from its optimum (maladaptive plasticity). Selection can work via mutation to overcome maladaptive plasticity, such that populations that originally exhibited maladaptive plasticity can produce the same phenotype as their ancestors through a novel developmental pathway (Fig. 5.4). This has been called genetic compensation (or cryptic evolution) and has been demonstrated in Kokanee salmon (Oncorhynchus nerka) (Grether 2005; Fitzpatrick 2012). Genetic compensation may produce a non-plastic reaction norm but it does not need to (Grether 2005), and may explain phenomena like countergradient variation (Conover and Schultz 1995). Genetic compensation can be distinguished from genetic assimilation, in that the pathway of genetic assimilation is phenotypic divergence between populations via adaptive plasticity → genetic divergence → phenotypic divergence via loss of plasticity, while the pathway of genetic compensation is phenotypic divergence between populations via maladaptive plasticity → genetic divergence → phenotypic similarity in divergent environments.
Maladaptive plasticity may be overcome in the absence of novel genetic input. Plastic compensation (Morris and Rogers 2013) (Fig. 5.4), defined as adaptive plasticity overcoming maladaptive plasticity, is likely a common phenomenon that has been underrepresented in discussions of maladaptive plasticity. In plastic compensation, a phenotype that should express maladaptive plasticity does not, or does so transiently, due to an adaptive plastic response in some other phenotype. Plastic compensation may therefore prevent maladaptive plasticity from being identified. Plastic compensation likely comes with a cost. For instance, in the brittlestar Amphiura filiformis, the ability to regenerate limbs (the otherwise maladaptively plastic phenotype) was maintained despite decreasing pH. However, this could only be maintained at low pH by digesting muscle tissue for energy (the cost) to presumably fuel increased rates of biomineralization (the adaptively plastic phenotype) (Wood et al. 2008). The pathway of plastic compensation can be described as phenotypic divergence between populations via maladaptive plasticity → phenotypic similarity between populations for the otherwise maladaptively plastic phenotype via adaptive plasticity in some other phenotype → possible genetic divergence to reduce costs. If plastic compensation occurs immediately, the initial step (phenotypic divergence via maladaptive plasticity) may never be observed. A key test of plastic compensation involves the inhibition or deletion of the adaptively plastic phenotype, which should lead to the expression of maladaptive plasticity.
Intriguingly, plastic compensation may be maintained across generations via heritable epigenetic modifications that keep the compensating phenotype induced even in the absence of the environmental inducer (Stern et al. 2012). This could be considered the epigenetic form of genetic assimilation, in which a once-plastic phenotype becomes constitutively produced across environments for several generations, a phenomenon known as epigenetic assimilation (Sollars et al. 2003; Ruden et al. 2005).
5.3.7 Plasticity and Reproductive Isolation
The final component of ecological speciation is reproductive isolation. Fitzpatrick (2012) helpfully reminds us that plasticity may result in reproductive isolation prior to adaptive divergence, if plasticity occurs pre-dispersal. If individuals follow a basic rule such as “breed only with individuals that are phenotypically similar to conspecifics,” there should be little reproduction between genetically identical but phenotypically distinct populations. The opposite, however, that plasticity may confound measures of reproductive isolation, has been rarely noted (but see Crispo et al. 2011). For instance, reproductively isolated populations may colonize the same environment, inducing similar plastic changes that reduce the phenotypic differences between them. If reproductive barriers are pre-zygotic, these plastic changes could alter reproductive behavior and increase hybridization. Environmental disturbance can also affect reproductive isolation. In Lake Victorian haplochromine cichlids, increased turbidity has been shown to reduce reproductive isolation between species, largely because the cues females use to find preferred mates can only be detected under broad spectrum light (Seehausen et al. 1997, 2008). Mate choice behavior is therefore plastic; measures of reproductive isolation in common garden experiments may not reflect actual levels of reproductive isolation. For cichlids, any measure of reproductive isolation performed in clear laboratory waters would overestimate the degree of reproductive isolation experienced under murkier natural conditions.
5.3.8 Summary
Ecological genomics approaches are vital for understanding the multitudinous consequences of phenotypic plasticity for evolutionary biology. Understanding how populations differ in their molecular reaction norms, and how this generates macrophenotypic divergence across environments; ascertaining the direction of plasticity evolution through phylogenetic studies; identifying the alleles that generate reaction norm differences and the alleles that contribute to CGV; testing different molecular strategies for persisting in novel environments; mapping alternative developmental pathways that produce similar phenotypes in different environments; and above all determining how alternative plastic phenotypes are generated, and how these pathways are altered in derived populations and species; all require an ecological genomics framework if we wish to move from conjecture and modelling to testing these ideas in nature.
5.4 Ecology of Plasticity
Phenotypic plasticity involves the environmentally-sensitive production of a phenotype, and therefore the ecological conditions experienced by the organism, both abiotic and biotic (including conspecifics), must be measured and incorporated into the ecological genomics of plasticity. Just as plasticity will not evolve without genetic variation, it also cannot evolve without certain environmental conditions. Demography, which can be influenced by the environment, also has consequences for the evolution of plasticity. Furthermore, plasticity in one organism has ecological and evolutionary consequences for other members within the community, thereby giving a role for plasticity in community and ecosystem processes. Finally, unnatural environments, often eschewed by ecologists, can shed important light on the mechanisms and evolutionary consequences of plasticity. Altogether, current research reveals the importance of integrating ecology into the ecological genomics of plasticity.
5.4.1 Plasticity and the Environment
Plasticity is expected to evolve under predictably varying environments (Berrigan and Scheiner 2003). If the environment does not vary, plasticity is not expected to evolve. If the environment varies in an unpredictable manner, bet-hedging strategies may evolve (Starrfelt and Kokko 2012). Predictable variation alone is not sufficient: for active induction, environments must also produce reliable cues for the environmental change, cues that can be detected by the organism and translated into a phenotypic response (Berrigan and Scheiner 2003). Reliable cues must then permit adequate time between the reception of the cue and the production of the plastic phenotype. If environmental change outpaces phenotypic change, or if the time-lag between cue reception and phenotype production is too long, non-plastic strategies may be favored (Padilla and Adolph 1996). These models show the importance of generating precise measurements of the environments experienced by organisms. This is easier said than done, as motile organisms may reduce the temporal environmental variation that they experience by moving throughout a spatially heterogeneous landscape. Thus environmental metrics taken at a single site may not measure the environment as experienced by an organism. Basic ecological measures are sorely needed for many species to test the relationship between environmental heterogeneity and plasticity evolution.
5.4.2 Plasticity and Demography
Environmental change can influence plasticity directly, as shown above, but it can also influence plasticity by affecting population migration and abundance. Environments which favor gene flow between subpopulations, for instance, are predicted to favor the evolution of plasticity (Crispo 2008). Population abundance can influence plasticity in at least four ways. (1) Large populations, which have greater opportunities for mutations, are more likely to harbor SGV, which can favor PMPP. (2) Large populations are also more likely to evolve beneficial mutations that positively affect reaction norms and reduce the costs of plasticity (Stern 2010). (3) Small populations produce fewer mutations and therefore are more likely to evolve pleiotropic and costly reaction norms (Stern 2010). (4) Population size can alter fitness landscapes, with individuals experiencing reduced fitness as the population increases (Dieckmann et al. 2004). If populations routinely experience fluctuations in population size, plasticity may evolve to reduce the effect of population size on fitness. For instance, models have shown that population fluctuations due to predator–prey dynamics facilitates the evolution of plasticity rather than adaptive speciation (Svanbäck et al. 2009). The challenge for researchers in the next few years will be to incorporate increasingly more complex population dynamics into their study of the ecological genomics of plasticity.
5.4.3 Plasticity and Community Genetics
Community genetics is an emerging subdiscipline within ecological genomics (Fig. 5.1). It involves studying how certain genotypes affect the distribution, abundance, and evolution of other genotypes within a community, and what genes underlie heritable community traits (Whitham et al. 2008; Hersch-Green et al. 2011). Community genetics of plasticity has received increased interest over the last several years (Rowntree et al. 2011; Tétard-Jones et al. 2011), particularly as plasticity’s role in community ecology has been documented (Agrawal 2001; Fordyce 2006) (Fig. 5.7). Plasticity can, among other things, drive selection in one species to overcome plastic changes in another species (ex. induced plant defenses and herbivore tolerance – Mithöfer and Boland 2012); induce plastic changes in another species (ex. parasite modifications of host phenotypes, or behavioral plasticity during competition – Dawkins 1982; Grangier and Lester 2012); cause coevolution of reciprocal plasticity between two or more species (antagonistically or mutualistically – Reimer and Tedengren 1996; Agrawal 2001; Freeman et al. 2009); alter the composition or abundance of other species within the community (ex. irreversible barnacle plasticity affects mussel and algal abundance – Raimondi et al. 2000, Fig. 5.7); determine community composition via dominant plasticity (Ashton et al. 2010) or limits to plasticity (ex. homeostatic mechanisms limit species distributions latitudinally – France 1992; Molina-Montenegro and Naya 2012); affect community interactions under changing climates, by altering species in such a way that their interactions are maintained (Cresswell and McCleery 2003; Charmantier et al. 2008) or disrupted (Post and Forchhammer 2008); can place novel selection pressures on species that exist in communities invaded by plastic species (Strauss et al. 2006; Lankau 2012); and can alter fitness landscapes, such that plastic resident organisms reduce the likelihood of successful colonization by plastic invasive species (Peacor et al. 2006).
Plasticity can affect the community in numerous ways. This example shows the direct (solid lines) and indirect (dashed lines) positive and negative interactions between four species of an intertidal community, that are influenced by plasticity in one species. (a) Whelks pass over young barnacles that are too small to consume. The barnacles, in response, plastically develop a bent morphology that inhibits whelk predation. Since mussels utilize empty barnacle shells for space, their abundance is negatively affected. Encrusting algae, which compete with mussels for space, increase in density. (b) Whelks pass over adult barnacles that developed in the absence of whelks. They consume the barnacle, leaving behind an empty shell that can be colonized by mussels. The mussels outcompete encrusting algae, such that algal density declines while mussel density increases (Reproduced from Raimondi et al. (2000, Fig. 2). Published with kind permission of © John Wiley and Sons 2003. All Rights Reserved)
These multitudinous interactions between plastic genotypes and other members of the community have led to recent studies on plasticity from a community genetics perspective (Schweitzer et al. 2008; Utsumi 2011). Experimental work is limited, but a recent study likely foreshadows things to come: the interactions between barley, aphids, and rhizobacteria were measured, along with barley and aphid plasticity across rhizobacterial environments. QTL for barley and aphid plasticity were mapped on to the barley genome, thus identifying gene regions in one species that influenced plasticity in another species, as mediated by a third species (Tétard-Jones et al. 2011). Predictions regarding the role of plasticity in community genetics are few, but recent modeling work suggests that plasticity may facilitate community stability in tritrophic systems to a greater extent than genetic variation (Kovach-Orr and Fussmann 2012). Community genetics is ripe for studies on phenotypic plasticity, but requires nuanced predictions and experimental data before patterns can be ascertained.
5.4.4 Plasticity Under Unnatural Ecological Conditions
Although the ecological genomics of adaptive plasticity emphasizes the need to study ecologically relevant traits under natural conditions, there are several reasons why one would want to study plasticity under non-natural conditions. (1) Subjecting multiple populations to an extreme environment that only a subset of populations has colonized could elucidate the mechanisms that have enabled survival in the extreme environment. For instance, one could find that populations not adapted to these extreme environments may nevertheless plastically adjust to survive in these new environments, or that stress-induced plasticity is only present in non-native populations, motivating research into how that stress was overcome. (2) Subjecting organisms to unrealistically extreme environments can allow the study of symmorphosis. (3) Unnatural environments may allow researchers to predict how organisms will respond to future environmental change (Reekie et al. 1994; Martin et al. 2011). For instance, exposing congeneric marine species to elevated temperatures revealed that cold-adapted marine organisms live well below their maximum thermal tolerance, while warm adapted marine organisms are negatively affected by very small increases in temperature (Somero 2005, 2010). (4) Studying plasticity using a single manipulated environmental variable is an important first step in elucidating how a particular environmental variable influences the phenotype. Once the production of that phenotype by that variable is understood, multienvironmental variables can be used to determine the influence of environmental interactions on plasticity. In other words, decomposing the environment into its different variables is an important, albeit unnatural, means of learning about plasticity. (5) Subjecting organisms to unnatural environments may allow researchers to uncover how different reaction norms interact to produce the phenotype. Sea urchins subjected to natural pH exhibited plastic compensation in morphology via transcript abundance. Under extremely unnatural acidities, transcriptional activity broke down and morphological plasticity was induced (Martin et al. 2011) (Fig. 5.2). Without the unnatural environment, however, the link between transcript abundance and morphological plasticity would not have been made.
5.4.5 Summary
The environment both shapes plasticity, and is shaped by plasticity, particularly when that environment consists of other genes. The next several years will likely see an increase in the use of genomic tools to test key predictions about the types of mutations that occur in large versus small populations, or the sorts of plastic genes that shape community structure. However, the use of genomics will be limited without precise measures of the ecological conditions experienced by natural populations.
5.5 Fitness of Plasticity
The adaptive or maladaptive nature of plasticity implies fitness consequences for plasticity. Fitness consequences of plasticity can be assessed both indirectly and directly under an ecological genomics framework.
5.5.1 Indirect
Indirect methods for assessing the fitness consequences of plasticity involve everything from modelling the possibility of plasticity evolution to discovering patterns for plasticity among taxa. Plasticity models generally compare plastic and non-plastic phenotypes under distinct stable or fluctuating environments. There are three main types of models (Scheiner 1993): optimality models, which provide cost-benefit analyses of plasticity (Stearns and Koella 1986); quantitative genetic models, which assess the evolution of plasticity given certain selection regimes and genetic variances/covariances for plastic traits (Via and Lande 1985); and gametic models, which assess the consequences of pleiotropy, epistasis, or linkage on the evolution of plasticity (De Jong 1990). Recent theoretical work has involved the evolution of plasticity given dispersal rates (Scheiner and Holt 2012; Scheiner et al. 2012), colonization events (Thibert-Plante and Hendry 2011), spatial heterogeneity (Chevin and Lande 2011), and an environment that contains genes (Wolf et al. 2003), and the consequences of plasticity for demography (Chevin et al. 2013). Although models are only as good as their assumptions, they have led to several testable predictions about the requirements for plasticity evolution, including the presence of genetic variation, high gene flow, low costs, and a predictable and reliable environmental cue (Crispo 2008).
Studies on natural populations can also provide indirect evidence for the fitness consequences of plasticity. For instance, phylogenetic analyses have shown that plastic traits can arise or become assimilated within a clade (Schwander and Leimar 2011). If this occurs in parallel within a clade, it suggests positive fitness for plasticity or its loss. Finding the ecological relevance of an induced phenotype can also indirectly test the adaptive nature of plasticity, particularly if the induced phenotype is difficult to produce. For instance, the induced defense morphology of Daphnia is clearly relevant to the environment that induces it, requiring receptors for predator abundance and the coordinated action of numerous underlying phenotypes. The fact that such a system evolved in association with this environment implies its positive fitness consequences. Finally, comparisons of plasticity that involve multiple populations adapted to different environmental regimes can provide indirect support for the adaptive nature of plasticity or non-plasticity. Three such patterns include: (1) Positive correlations between the degree of plasticity exhibited by a population and the extent of environmental heterogeneity experienced by that population. For instance, the climatic variability hypothesis suggests that plants and ectotherms at high latitudes should exhibit greater temperature-induced plasticity for tolerance and acclimation phenotypes, than populations that reside at lower latitudes. This is because temperature fluctuations are greater at higher latitudes, requiring an increased capacity to maintain homeostasis. Evidence for this hypothesis has been found (Compton et al. 2007; Sunday et al. 2011; Molina-Montenegro and Naya 2012). (2) The parallel evolved loss or gain of plasticity from a known ancestral population. For instance, tiger snakes (Notechis scutatus) from mainland Australia feed on relatively small prey, while island colonists consume larger prey. This dietary switch is facilitated by ancestral plasticity in head shape. Head shape plasticity has subsequently been lost in older colonized populations, due to costs associated with the production of smaller heads (Aubret et al. 2004; Aubret and Shine 2009, 2010). (3) Finally, patterns at a genomic level can be assessed. For instance, plasticity was hypothesized to buffer against the effects of selection, resulting in higher rates of evolution for genes whose expression was environmentally sensitive. Leichty et al. (2012) used microarrays to assess genes involved in the production of environmentally-induced morphs of tadpoles. Using 454 sequencing, they then sequenced “biased” (plastic) and “unbiased” (non-plastic) genes, and compared rates of evolution between these genes for multiple plastic and non-plastic amphibian species. Contrary to expectations, they found that plastic genes had higher substitution rates even in non-plastic species, leading to the intriguing hypothesis that non-essential genes in non-plastic species may rapidly accumulate mutations. This rapid evolution then becomes a precondition for the evolution of plasticity, permitting the co-option of these non-essential genes for novel plastic functions under heterogeneous environments (Leichty et al. 2012). Although selection on these plastic genes was not assessed, the repeatability of these patterns in other taxa suggests that plasticity can have positive fitness effects (Hunt et al. 2011).
5.5.2 Direct
Direct methods for assessing the fitness consequences of plasticity involve comparisons of fitness between plastic and non-plastic genotypes. This can be assessed in several ways.
5.5.2.1 Estimates of Fitness in Natural Populations
Long-standing field studies can provide measures of plasticity for individuals, and can measure heritability, fitness, and selection for those plastic phenotypes. Seasonal plasticity in bighorn sheep mass was measured over a 25 year span in both parents and offspring, and was found to have a genetic basis. Selection was measured for summer and winter mass changes, and revealed that plastic individuals had a higher fitness coming out of the winter than less plastic individuals (Pelletier et al. 2007; see also Nussey et al. 2005). Although field studies provide compelling examples of selection under ecologically relevant conditions, true differences in plasticity are difficult to measure due to limited environmental control.
5.5.2.2 Common-Garden/Mesocosm Experiments
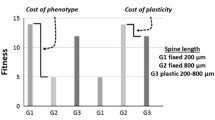
Genotypes that differ in their degree of plasticity can be raised in several (often reciprocal) common gardens or mesocosms that manipulate some environmental variable, such that plastic changes are induced between common gardens. The fitness of each plastic and non-plastic genotype can be assessed for each environment (Griffith and Sultan 2012; Matesanz et al. 2012). These genotypes can occur as polymorphisms within a population or between populations, or can be the result of genetic manipulation. For instance, plant genotypes that exhibited plasticity in stem length in response to conspecific density had consistently high fitness at low and high densities, whereas plant genotypes that could produce only long or only short stems had high fitness only at specific densities (Schmitt et al. 1995). Morphologically plastic and non-plastic species of Daphnia were raised together and apart in the presence and absence of predators. The plastic species had higher fitness in the presence of predators, but lower fitness in the absence of predators when competing with the non-plastic species. This reduced fitness in the plastic species was not measured when species were raised apart (Engel and Tollrian 2009).
5.5.2.3 Experimental Evolution
Artificial selection experiments, in which plasticity is selected by researchers (Scheiner and Lyman 1991; Scheiner 2002; Kelly et al. 2006), or compared between domestic and wild lineages (Morris et al. 2011; Debes et al. 2012; Solberg et al. 2013), increases our confidence that plasticity is heritable and can evolve. Experimental evolution studies, in which genotypes freely evolve under controlled conditions, have shown that plasticity can evolve when it benefits the organism rather than the researcher (Garland and Kelly 2006). For instance, viruses raised in a combination of single-infection and coinfection conditions experimentally evolved greater plasticity than viruses raised in single-infection conditions alone, and this increased plasticity conferred greater fitness under both environments (Leggett et al. 2013). Plasticity has also been shown to evolve in silico for digital organisms (Clune et al. 2007). A genomics approach to the experimental evolution of plasticity would ideally track both the phenotypic and genetic changes that occur under various forms of environmental change to address questions regarding the rules of plasticity evolution, including the types of genes or chromatin structures involved, the importance of pleiotropy, the nature of parallel plasticity evolution, etc. (Bell 2010; Dettman et al. 2012).
5.5.2.4 F2 Selection Experiments
It can be difficult in practice to determine if fitness differences between plastic and non-plastic genotypes are due to plasticity or to other phenotypic differences. For instance, in comparisons between the fitness of invasive plants and their ancestral counterparts, plastic tetraploids had greater fitness than less-plastic diploids, but non-plastic phenotypes affected by tetraploidy could have conferred this fitness benefit (Hahn et al. 2012 – but see their plausible explanation for why this was not the case). If distinct populations can be hybridized, this difficulty can be circumvented. Recombination in the gametes of F1 hybrids results in F2 hybrids with chromosomes that vary in their distribution of parental alleles. Unless plastic and non-plastic traits are tightly linked, F2 individuals should vary randomly with respect to these phenotypes. F2 individuals could then be measured for their degree of plasticity relative to the parental populations, and fitness assessed in multiple environments. If plasticity does confer a fitness benefit apart from non-plastic traits, then the phenotypic background for the plastic phenotype should not matter. This method relies on a number of practical considerations (plasticity must be measurable in individuals – that is, plasticity must be reversible) and assumptions (no relationship between plastic and non-plastic traits, no linkage). To our knowledge such an F2 experiment has not been employed.
5.5.2.5 Genomics and Fitness
Novel genomic techniques have increased our ability to detect and measure plastic differences between populations. For instance, researchers can now use gene expression tools (quantitative reverse transcriptase PCR, microarrays, RNA-Sequencing, etc.) in association with common garden experiments to associate experimentally-manipulated transcript abundance with fitness (Rest et al. 2013), measure gene expression for fitness-related traits (Zhou et al. 2012), or compare populations for gene expression profiles (Levine et al. 2011). Gene sequencing can identify alleles associated with fitness-related differences in plasticity (Powers and Schulte 1998), while QTL mapping using microsatellites or Single Nucleotide Polymorphisms can identify regions of the genome associated with divergent plasticities (Ungerer et al. 2003; Gerald et al. 2006; Gutteling et al. 2007; Tétard-Jones et al. 2011). F2 selection experiments could provide compelling associations between genotype, phenotype, and fitness, if QTLs for plasticity can be shown to be associated with F2 survival or fecundity in a common garden. Finally, a genomics perspective permits us to ask questions regarding which genes and genomic structures are likely to facilitate the evolution of adaptive plasticity (Leichty et al. 2012).
5.5.3 Summary
One cannot assume a priori that plasticity confers a fitness advantage relative to non-plastic individuals. Indirect evidence supports the adaptive nature of both plasticity and non-plasticity, but direct measures of fitness offer the most compelling results. This direct evidence has revealed that plasticity may increase fitness under all environments, a subset of environments, or can decrease fitness across environments. Ecological knowledge of the study organism is required to make predictions that can differentiate between these alternatives. Just as ecology cannot be ignored, so the underlying genome cannot be ignored, as plasticity will only evolve if genetic variation is present. Future work in the ecological genomics of plasticity will involve identifying alleles that confer fitness advantages in plastic or non-plastic organisms, and determining the genomic architectures that constrain or encourage the evolution of plasticity.
5.6 Conclusion
Phenotypic plasticity research has undergone a renaissance of sorts in developmental, ecological, and evolutionary biology. Integrating these disciplines with novel genomic tools has allowed researchers to test key predictions regarding the production of plasticity (e.g. the importance of transcriptional vs. proteomic plasticity), the evolution of plasticity (e.g. plasticity as a leader or a follower in evolution), the ecological significance of plasticity (e.g. plasticity genes involved in heritable community traits), and the fitness of plasticity (e.g. relaxed selection as a precondition for adaptive plasticity). Furthermore, the ecological genomics of plasticity has emphasized the complexity and difficulty of elucidating the complete developmental, evolutionary, and ecological story for a single phenotype, even in well-known and well-studied species. Theoretical work currently outpaces data, but the tide is changing. Genomic tools are enabling researchers to understand the significance of plasticity like never before, and the incoming data should stimulate research for years to come.
References
Agrawal A (2001) Phenotypic plasticity in the interactions and evolution of species. Science 294:321–326
Akkas S, Kepenek A, Beklioglu M, Severcan F (2010) Molecular approach to the chemical characterization of fish-exuded kairomone: a Fourier transform infrared spectroscopic study. Aquat Sci 72:71–83
Alpert P, Simms E (2002) The relative advantages of plasticity and fixity in different environments: when is it good for a plant to adjust? Evol Ecol 16:285–297
Andrew RL, Bernatchez L, Bonin A, Buerkle CA, Carstens BC, Emerson BC, Garant D, Giraud T, Kane NC, Rogers SM, Slate J, Smith H, Sork VL, Stone GN, Vines TH, Waits L, Widmer A, Rieseberg LH (2013) A roadmap for molecular ecology. Mol Ecol 22:2605–2626
Angers B, Castonguay E, Massicotte R (2010) Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol Ecol 19:1238–1295
Ashton I, Miller A, Bowman W, Suding K (2010) Niche complementarity due to plasticity in resource use: plant partitioning of chemical N forms. Ecology 91:3252–3260
Aubin-Horth N, Renn S (2009) Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol 18:3763–3780
Aubret F, Shine R (2009) Genetic assimilation and the postcolonisation erosion of phenotypic plasticity in island tiger snakes. Curr Biol 19:1932–1936
Aubret F, Shine R (2010) Fitness costs may explain the post-colonisation erosion of phenotypic plasticity. J Exp Biol 213:735–739
Aubret F, Shine R, Bonnet X (2004) Evolutionary biology: adaptive developmental plasticity in snakes. Nature 431:261–262
Bachmann D, Both S, Bruelheide H, Ding B, Gao M, Härdtle W, Scherer-Lorenzen M, Erfmeier A (2012) Functional trait similarity of native and invasive herb species in subtropical China – environment-specific differences are the key. Environ Exp Bot 83:82–92
Bajić D, Poyatos J (2012) Balancing noise and plasticity in eukaryotic gene expression. BMC Genomics 13:343
Baldwin J (1896) A new factor in evolution. Am Nat 30:441–451
Baldwin J (1902) Development and evolution. MacMillan, New York
Barrett R, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol 23:38–44
Barry M (2002) Progress toward understanding the neurophysiological basis of predator-induced morphology in Daphnia pulex. Physiol Biochem Zool 75:179–186
Beaton M, Hebert P (1997) The cellular basis of divergent head morphologies in Daphnia. Limnol Oceanogr 42:346–356
Beldade P, Mateus R, Keller R (2011) Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol 20:1347–1363
Bell G (2010) Experimental genomics of fitness in yeast. Proc R Soc B 277:1459–1467
Bentz B, Ryan B, Mock K, Pfrender M (2011) Genetic architecture and phenotypic plasticity of thermally-regulated traits in an eruptive species, Dendroctonus ponderosae. Evol Ecol 25:1269–1288
Berrigan D, Scheiner S (2003) Modeling the evolution of phenotypic plasticity. In: DeWitt T, Scheiner S (eds) Phenotypic plasticity: functional and conceptual approaches. Oxford University Press, Oxford
Bock C (2012) Analysing and interpreting DNA methylation data. Nat Rev Genet 13:705–719
Bossdorf O, Richards C, Pigliucci M (2008) Epigenetics for ecologists. Ecol Lett 11:106–115
Bossdorf O, Arcuri D, Richards C, Pigliucci M (2010) Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol Ecol 24:541–553
Bradshaw A (1965) Evolutionary significance of phenotypic plasticity in plants. Adv Genet 13:115–155
Brettner L, Masel J (2012) Protein stickiness rather than number of functional protein-protein interactions predicts expression noise and plasticity in yeast. BMC Syst Biol 6:128
Castillo-Davis C, Mekhedov S, Hartl D, Koonin E, Kondrashov F (2002) Selection for short introns in highly expressed genes. Nat Genet 31:415–418
Charmantier A, McCleery R, Cole L, Perrins C, Kruuk L, Sheldon B (2008) Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320:800–803
Chevin L-M, Lande R (2011) Adaptation to marginal habitats by evolution of increased phenotypic plasticity. J Evol Biol 24:1462–1476
Chevin L-M, Gallet R, Gomulkiewicz R, Holt R, Fellous S (2013) Phenotypic plasticity in evolutionary rescue experiments. Phil Trans R Soc B 368:20120089
Chown S, Jumbam K, Sørensen J, Terblanche J (2009) Phenotypic variance plasticity and heritability estimates of critical thermal limits depend on methodological context. Funct Ecol 23:133–140
Clune J, Ofria C, Pennock R (2007) Investigating the emergence of phenotypic plasticity in evolving digital organisms. In: Almeida e Costa F, Rocha L, Costa E, Harvey I, Coutinho A (eds) Proceedings of the 9th European conference on advances in artificial life. Springer-Verlag, Berlin
Compton T, Rijkenberg M, Drent J, Piersma T (2007) Thermal tolerance ranges and climate variability: a comparison between bivalves from differing climates. J Exp Mar Biol Ecol 352:200–211
Conover D, Schultz E (1995) Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol Evol 10:248–252
Cresswell W, McCleery R (2003) How great tits maintain synchronization of their hatch date with food supply in response to long-term variability in temperature. J Anim Ecol 72:356
Crews D, Gillette R, Scarpino S, Manikkam M, Savenkova M, Skinner M (2012) Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci USA 109:9143–9148
Crispo E (2007) The Baldwin Effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 61(11):2469–2479
Crispo E (2008) Modifying effects of phenotypic plasticity on interactions among natural selection, adaptation and gene flow. J Evol Biol 21:1460–1469
Crispo E, Moore J, Lee-Yaw J, Gray S, Haller B (2011) Broken barriers: human-induced changes to gene flow and introgression in animals: an examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. Bioessays 33:508–518
Dalziel A, Rogers S, Schulte P (2009) Linking genotypes to phenotypes and fitness: how mechanistic biology can inform molecular ecology. Mol Ecol 18:4997–5017
Dawkins R (1976) The selfish gene. Oxford University Press, Oxford
Dawkins R (1982) The extended phenotype. Oxford University Press, Oxford
Day T, Pritchard J, Schluter D (1994) A comparison of two sticklebacks. Evolution 48:1723–1734
De Boer TE, Birlutiu A, Bochdanovits Z, Timmermans MJTN, Dijkstra TMH, Van Straalen NM, Ylstra B, Roelofs D (2011) Transcriptional plasticity of a soil arthropod across different ecological conditions. Mol Ecol 20:1144–1154
De Jong G (1990) Quantitative genetics of reaction norms. J Evol Biol 3:447–468
Debat V, David P (2001) Mapping phenotypes: canalization, plasticity and developmental stability. Trends Ecol Evol 16:555–561
Debes P, Normandeau E, Fraser D, Bernatchez L, Hutchings J (2012) Differences in transcription levels among wild, domesticated and hybrid Atlantic salmon (Salmo salar) from two environments. Mol Ecol 21:2574–2587
Deredge D, Baker J, Datta K, LiCata V (2010) The glutamate effect on DNA binding by Pol I DNA polymerases: osmotic stress and the effective reversal of salt linkage. J Mol Biol 401:223–238
Dettman J, Rodrigue N, Melnyk A, Wong A, Bailey S, Kassen R (2012) Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol Ecol 21:2058–2077
DeWitt T, Scheiner S (2003) Phenotypic variation from single genotypes: a primer. In: DeWitt T, Scheiner S (eds) Phenotypic plasticity: functional and conceptual approaches. Oxford University Press, Oxford
DeWitt T, Sih A, Wilson DS (1998) Costs and limits of phenotypic plasticity. Trends Ecol Evol 13:77–81
Dieckmann U, Doebeli M, Metz J, Tautz D (2004) Adaptive speciation. Cambridge University Press, New York
Diz A, Martínez-Fernández M, Rolán-Alvarez E (2012) Proteomics in evolutionary ecology: linking the genotype with the phenotype. Mol Ecol 21:1060–1080
Eames M, Kortemme T (2012) Cost-benefit tradeoffs in engineered lac operons. Science 336:911–915
Engel K, Tollrian R (2009) Inducible defences as key adaptations for the successful invasion of Daphnia lumholtzi in North America? Proc R Soc B 276:1865–1873
Espinosa-Soto C, Martin O, Wagner A (2011) Phenotypic plasticity can facilitate adaptive evolution in gene regulatory circuits. BMC Evol Biol 11:5
Evans JD, Wheeler DE (2000) Expression profiles during honeybee caste determination. Genome Biol 2:research0001-research0001.6
Feder M, Mitchell-Olds T (2003) Evolutionary and ecological functional genomics. Nat Rev Genet 4:649–655
Findlay G, MacCoss M, Swanson W (2009) Proteomic discovery of previously unannotated rapidly evolving seminal fluid genes in Drosophila. Genome Res 19:886–896
Fitzpatrick B (2012) Underappreciated consequences of phenotypic plasticity for ecological speciation. Int J Ecol 2012:256017
Ford EB (1964) Ecological genetics. Chapman and Hall, London
Fordyce J (2006) The evolutionary consequences of ecological interactions mediated through phenotypic plasticity. J Exp Biol 209:2377–2383
France R (1992) The North American latitudinal gradient in species richness and geographical range of freshwater crayfish and amphipods. Am Nat 139:342–354
Frankenhuis WE, Panchanathan K (2011) Individual differences in developmental plasticity may result from stochastic sampling. Perspect Psychol Sci 6:336–347
Fraser H (2011) Genome-wide approaches to the study of adaptive gene expression evolution. Bioessays 33:469–477
Freeman A, Meszaros J, Byers J (2009) Poor phenotypic integration of blue mussel inducible defenses in environments with multiple predators. Oikos 118:758–766
Fusco G, Minelli A (2010) Phenotypic plasticity in development and evolution: facts and concepts. Phil Trans R Soc B 365:547–556
Garland T Jr, Kelly S (2006) Phenotypic plasticity and experimental evolution. J Exp Biol 209:2344–2361
Gerald J, Lehti-Shiu M, Ingram P, Deak K, Biesiada T, Malamy J (2006) Identification of Quantitative Trait Loci that regulate Arabidopsis root system size and plasticity. Genetics 172:485–498
Ghalambor C, McKay J, Carroll S, Reznick D (2007) Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol 21:394–407
Gibson G, Wagner G (2000) Canalization in evolutionary genetics: a stabilizing theory? Bioessays 22:372–380
Gracey A, Fraser E, Li W, Fang Y, Taylor R, Rogers J, Brass A, Cossins A (2004) Coping with cold: an integrative multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci USA 101:16970–16975
Grangier J, Lester P (2012) Behavioral plasticity mediates asymmetric competition between invasive wasps and native ants. Commun Integr Biol 5:127–129
Greenberg J, Xia J, Zhou X, Thatcher S, Gu X, Ament S, Newman T, Green P, Zhang W, Robinson G, Ben-Shahar Y (2012) Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav 11:660–670
Grether G (2005) Environmental change, phenotypic plasticity and genetic compensation. Am Nat 166:E115–E123
Griffith T, Sultan S (2012) Field-based insights to the evolution of specialization: plasticity and fitness across habitats in a specialist/generalist species pair. Ecol Evol 2:778–791
Guo Y, Lang S, Ellis R (2009) Independent recruitment of F Box genes to regulate hermaphrodite development during nematode evolution. Curr Biol 19:1853–1860
Gutteling E, Riksen J, Bakker J, Kammenga J (2007) Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98:28–37
Hackett J, Sengupta R, Zylicz J, Murakami K, Lee C, Down T, Surani M (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339:448–452
Hahn M, van Kleunen M, Müller-Shärer H (2012) Increased phenotypic plasticity to climate may have boosted the invasion success of polyploid Centaurea stoebe. PLoS One 7:e50284
Hanshew B, Garica T (2012) Invasion of the shelter snatchers: behavioural plasticity in invasive red swamp crayfish Procambarus clarkia. Freshwater Biol 57:2285–2296
Hayden EJ, Ferrada E, Wagner A (2011) Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474:92–95
Hayward S, Murray P, Gracey A, Cossins A (2007) Beyond the lipid hypothesis: mechanisms underlying phenotypic plasticity in inducible cold tolerance. Adv Exp Med Biol 594:132–142
Herrera CM, Bazaga P (2012) Epigenetic correlates of plant phenotypic plasticity: DNA methylation differs between prickly and nonprickly leaves in heterophyllous Ilex aquifolium (Aquifolaceae) trees. Bot J Linn Soc 171:441–452
Hersch-Green E, Turley N, Johnson M (2011) Community genetics: what have we accomplished and where should we be going? Phil Trans R Soc B 366:1453–1460
Hunt B, Ometto L, Wurm Y, Shoemaker D, Yi S, Keller L, Goodisman M (2011) Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc Natl Acad Sci USA 108:15936–15941
Iwasaki WM, Tsuda ME, Kawata M (2013) Genetic and environmental factors affecting cryptic variations in gene regulatory networks. BMC Evol Biol 13:91
Juliano RL, Dixit VR, Kang H, Kim TY, Miyamoto Y, Xu D (2005) Epigenetic manipulation of gene expression: a toolkit for cell biologists. J Cell Biol 169:847–857
Kelly S, Czech P, Wight J, Blank K, Garland T Jr (2006) Experimental evolution and phenotypic plasticity of hindlimb bones in high-activity house mice. J Morphol 267:360–374
Kovach-Orr C, Fussmann G (2012) Evolutionary and plastic rescue in multitrophic model communities. Phil Trans R Soc B 368:20120084
Laforsch C, Tollrian R (2010) Cyclomorphosis and phenotypic changes. In: Likens G (ed) Plankton of inland waters. Academic, San Diego
Lande R (2009) Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J Evol Biol 22:1435–1446
Lande R, Arnold S (1983) The measurement of selection on correlated characters. Evolution 37:1210–1226
Landry C, Oh J, Hartl D, Cavalieri D (2006) Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366:343–351
Lang G, Murray A, Botstein D (2009) The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci USA 106:5755–5760
Langerhans R, DeWitt T (2002) Plasticity constrained: over-generalized induction cues cause maladaptive phenotypes. Evol Ecol Res 4:857–870
Lankau R (2012) Coevolution between invasive and native plants driven by chemical competition and soil biota. Proc Natl Acad Sci USA 109:11240–11245
Lee M, Topper S, Hubler S, Hose J, Wenger C, Coon J, Gasch A (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol 7:514
Leggett H, Benmayor R, Hodgson D, Buckling A (2013) Experimental evolution of adaptive phenotypic plasticity in a parasite. Curr Biol 23:139–142
Lehner B (2010) Conflict between noise and plasticity in yeast. PLoS Genet 6:e1001185
Leichty A, Pfennig D, Jones C, Pfennig K (2012) Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Integr Comp Biol 52:16–30
Levine M, Eckert M, Begun D (2011) Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from eastern Australia. Mol Biol Evol 28:249–256
Levy S, Siegal (2008) Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol 6:e264
Maier T, Schmidt A, Güell M, Kühner S, Gavin A, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7:511
Malakar P, Venkatesh K (2012) Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Appl Micriobiol Biot 93:2543–2549
Martin S, Richier S, Pedrotti M, Dupont S, Castejon C, Gerakis Y, Kerros M, Oberhänsli F, Teyssié J, Jeffree R, Gattuso J (2011) Early development and molecular plasticity in the Mediterranean sea urchin Paracentrotus lividus exposed to CO2-driven acidification. J Exp Biol 214:1357–1368
Matesanz S, Horgan-Kobelski T, Sultan S (2012) Phenotypic plasticity and population differentiation in an ongoing species invasion. PLoS One 7:e44955
Matzek V (2012) Trait values, not trait plasticity, best explains invasive species’ performance in a changing environment. PLoS One 7:e48821
McCairns R, Bernatchez L (2010) Adaptive divergence between freshwater and marine sticklebacks: insight into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution 64(4):1029–1047
McGuigan K, Nishimura N, Currey M, Hurwit D, Cresko W (2011) Cryptic genetic variation and body size evolution in threespine stickleback. Evolution 65(4):1203–1211
Meister P, Schott S, Bedet C, Xiao Y, Rohner S, Bodennec S, Hudry B, Molin L, Solari F, Gasser S, Palladino F (2011) Caenorhabditis elegans Heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol 12:R123
Mithöfer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol 63:431–450
Miyakawa H, Imai M, Sugimoto N, Ishikawa Y, Ishikawa A, Ishigaki H, Okada Y, Miyazaki S, Koshikawa S, Cornette R, Miura T (2010) Gene up-regulation in response to predator kairomones in the water flea Daphnia pulex. BMC Dev Biol 10:45
Moczek A (2012) The nature of nurture and the future of evodevo: toward a theory of developmental evolution. Integr Comp Biol 52:108–119
Moczek A, Sultan S, Foster S, Ledón-Rettig C, Dworkin I, Nijhout H, Abouheif E, Pfennig D (2011) The role of developmental plasticity in evolutionary innovation. Proc R Soc B 278:2705–2713
Molina-Montenegro M, Naya D (2012) Latitudinal patterns in phenotypic plasticity and fitness-related traits: assessing the climatic variability hypothesis (CVH) with an invasive plant species. PLoS One 7:e47620
Molina-Montenegro M, Peñuelas J, Munné-Bosch S, Sardans J (2012) Higher plasticity in ecophysiological traits enhances the performance and invasion success of Taraxacum officinale (dandelion) in alpine environments. Biol Invasions 14:21–33
Morris M, Rogers SM (2013) Overcoming maladaptive plasticity through plastic compensation. Curr Zool 59:526--536
Morris M, Fraser D, Eddington J, Hutchings J (2011) Hybridization effects on phenotypic plasticity: experimental compensatory growth in farmed-wild Atlantic salmon. Evol Appl 4:444–458
Mozdzer T, Megonigal P (2012) Jack-and-master trait responses to elevated CO2 and N: a comparison of native and introduced Phragmites australis. PLoS One 7:e42794
Müller S, Bianchi M, Knapp S (2001) Thermodynamics of HMGB1 interaction with duplex DNA. Biochemistry 40:10254–10261
Neyfakh A, Hartl D (1993) Genetic control of the rate of embryo development: selection for faster development at elevated temperatures. Evolution 47:1625–1631
Nosil P (2012) Ecological speciation. Oxford University Press, New York
Nussey D, Postma E, Gienapp P, Visser M (2005) Selection on heritable phenotypic plasticity in a wild bird population. Science 310:304–306
Orsini L, Andrew R, Eizaguirre C (2013) Evolutionary ecological genomics. Mol Ecol 22:527–531
Padilla D, Adolph S (1996) Plant inducible morphologies are not always adaptive: the importance of time delays in a stochastic environment. Evo Ecol 10:105–117
Pavey S, Collin H, Nosil P, Rogers S (2010) The role of gene expression in ecological speciation. Ann NY Acad Sci 1206:110–129
Pavey S, Bernatchez L, Aubin-Horth N, Landry CR (2012) What is needed for next-generation ecological and evolutionary genomics? Trends Ecol Evol 27:673–678
Peacor S, Allesina S, Riolo R, Pascual M (2006) Phenotypic plasticity opposes species invasions by altering fitness surface. PLoS Biol 4:e372
Pelletier F, Réale D, Garant D, Coltman D, Festa-Bianchet M (2007) Selection on heritable seasonal phenotypic plasticity of body mass. Evolution 61(8):1969–1979
Peñalva-Arana D, Lynch M, Robertson H (2009) The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol 9:79
Pfennig D, Wund M, Snell-Rood E, Cruickshank T, Schlichting C, Moczek A (2010) Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol Evol 25:459–467
Podrabsky J, Somero G (2004) Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J Exp Biol 207:2237–2254
Post E, Forchhammer M (2008) Climate change reduces reproductive success of an Arctic herbivore through trophic mismatch. Phil Trans R Soc B 363:2367–2373
Powers D, Schulte P (1998) Evolutionary adaptations of gene structure and expression in natural populations in relation to a changing environment: a multidisciplinary approach to address the million-year saga of a small fish. J Exp Zool 282:71–94
Price T (2003) The role of phenotypic plasticity in driving genetic evolution. Proc R Soc B 270:1433–1440
Purchase C, Moreau D (2012) Stressful environments induce novel phenotypic variation: hierarchical reaction norms for sperm performance of a pervasive invader. Ecol Evol 2:2562–2571
Queitsch C, Sangster T, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624
Raimondi P, Forde S, Delph L, Lively C (2000) Processes structuring communities: evidence for trait-mediated indirect effects through induced polymorphisms. Oikos 91:353–361
Ranz J, Machado C (2006) Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol 21:29–37
Raser J, O’Shea E (2005) Noise in gene expression: origins, consequences and control. Science 309:2010–2013
Reekie J, Hicklenton P, Reekie E (1994) Effects of elevated CO2 on time of flowering in four short-day and four long-day species. Can J Bot 72:533–538
Refsnider J, Janzen F (2012) Behavioral plasticity may compensate for climate change in a long-lived reptile with temperature-dependent sex determination. Biol Cons 152:90–95
Reimer O, Tedengren M (1996) Phenotypic improvement of morphological defences in the mussel Mytilus edulis induced by exposure to the predator Asterias rubens. Oikos 75:383–390
Renn SCP, Aubin-Horth N, Hofmann HA (2008) Fish and chips: functional genomics of social plasticity in an African cichlid fish. J Exp Biol 211:3041–3056
Rest J, Morales C, Waldron J, Opulente D, Fisher J, Moon S, Bullaughey K, Carey L, Dedousis D (2013) Nonlinear fitness consequences of variation in expression level of a eukaryotic gene. Mol Biol Evol 30:448–456
Richards CL, Bossdorf O, Pigliucci M (2010) What role does heritable epigenetic variation play in phenotypic evolution? BioScience 60:232–237
Richards CL, Rosas U, Banta J, Bhambhra N, Purrugganan MD (2012) Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet 8:e1002662
Roberts SB, Gavery MR (2012) Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol 2:116
Rossitter M (1996) Incidence and consequences of inherited environmental effects. Annu Rev Ecol Syst 27:451–476
Rowntree J, Shuker D, Preziosi R (2011) Forward from the crossroads of ecology and evolution. Phil Trans R Soc B 366:1322–1328
Ruden DM, Xiao L, Garfinkel MD, Lu X (2005) Hsp90 and environmental impacts on epigenetic states: a model for the trans-generational effects of diethylstibesterol on uterine development and cancer. Hum Mol Gen 14:R149–R155
Sardans J, Peñuelas J, Rivas-Ubach A (2011) Ecological metabolomics: overview of current developments and future challenges. Chemoecology 21:191–225
Scheiner S (1993) Genetics and evolution of phenotypic plasticity. Annu Rev Ecol Syst 24:35–68
Scheiner S (2002) Selection experiments and the study of phenotypic plasticity. J Exp Biol 15:889–898
Scheiner S, Holt R (2012) The genetics of phenotypic plasticity. X. Variation versus uncertainty. Ecol Evol 2:751–767
Scheiner S, Lyman R (1991) The genetics of phenotypic plasticity II. Response to selection. J Evol Biol 4:23–50
Scheiner S, Barfield M, Holt R (2012) The genetics of phenotypic plasticity. XI. Joint evolution of plasticity and dispersal rate. Ecol Evol 8:2027–2039
Schlichting C (2008) Hidden reaction norms cryptic genetic variation and evolvability. Ann NY Acad Sci 1133:187–203
Schluter D (2000) The ecology of adaptive radiation. Oxford University Press, New York
Schmitt J, McCormac A, Smith H (1995) A test of the adaptive plasticity hypothesis using transgenic and mutant plants disabled in phytochrome-mediated elongation responses to neighbors. Am Nat 146:937–953
Schrimpf S, Weiss M, Reiter L, Ahrens C, Jovanovic M, Malmström J, Brunner E, Mohanty S, Lercher M, Hunziker P, Aebersold R, von Mering C, Hengartner M (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7:616–627
Schwander T, Leimar O (2011) Genes as leaders and followers in evolution. Trends Ecol Evol 26:143–151
Schwartz T, Bronikowski A (2013) Dissecting molecular stress networks: identifying nodes of divergence between life-history phenotypes. Mol Ecol 22:739–756
Schwarzenberger A, Courts C, von Elert E (2009) Target gene approaches: gene expression in Daphnia magna exposed to predator-borne kairomones or to microcystin-producing and microcystin-free Microcystis aeruginosa. BMC Genomics 10:527
Schweitzer J, Madritch M, Bailey J, LeRoy C, Fischer D, Rehill B, Lindroth R, Hagerman A, Wooley S, Hart S, Whitham T (2008) From genes to ecosystems: the genetic basis of condensed tannins and their role in nutrient regulation in a Populus model system. Ecosystems 11:1005–1020
Scoville A, Pfrender M (2010) Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc Natl Acad Sci USA 107:4260–4263
Seehausen O, van Alphen J, Witte F (1997) Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 277:1808–1811
Seehausen O, Terai Y, Magalhaes I, Carleton K, Mrosso H, Miyagi R, van der Sluijs I, Schneider M, Maan M, Tachida H, Imai H, Okada N (2008) Speciation through sensory drive in cichlid fish. Nature 455:620–626
Shachrai I, Zaslaver A, Alon U, Dekel E (2010) Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol Cell 38:758–767
Simon J, Pfrender M, Tollrian R, Tagu D, Colbourne J (2011) Genomics of environmentally induced phenotypes in 2 extremely plastic arthropods. J Hered 102:512–525
Singh A, Razooky B, Dar R, Weinberger L (2012) Dynamics of protein noise can distinguish between alternate sources of gene-expression variability. Mol Syst Biol 8:607
Slijper E (1942a) Biologic-anatomical investigations on the bipedal gait and upright posture in mammals with special reference to a little goat born without forelegs. I. Proceedings of the Koninklijke Nederlandse Akademie Wetenschappen, vol 45, pp 288–295
Slijper E (1942b) Biologic-anatomical investigations on the bipedal gait and upright posture in mammals with special reference to a little goat born without forelegs. II. Proceedings of the Koninklijke Nederlandse Akademie Wetenschappen, vol 45, pp 407–415
Snell-Rood E (2012) Selective processes in development: implications for the costs and benefits of phenotypic plasticity. Integr Comp Biol 52:31–42
Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM (2003) Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet 33:70–74
Solberg M, Skaala Ø, Nilsen F, Glover K (2013) Does domestication cause changes in growth reaction norms? A study of farmed wild and hybrid Atlantic salmon families exposed to environmental stress. PLoS One 8:e54469
Somero G (2005) Linking biogeography to physiology: evolutionary and acclimatory adjustments of thermal limits. Front Zool 2:1
Somero G (2010) The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J Exp Biol 213:912–920
Sommer RJ, Ogawa A (2011) Hormone signaling and phenotypic plasticity in nematode development and evolution. Curr Biol 21:R758–R766
Srinivasan DG, Brisson JA (2012) Aphids: a model for polyphenism and epigenetics. Genet Res Int 2012:431531
Starrfelt J, Kokko H (2012) Bet-hedging – a triple trade-off between means variances and correlations. Biol Rev 87(3):742–755
Stearns S, Koella J (1986) The evolution of phenotypic plasticity in life-history traits: predictions of reaction norms for age and size at maturity. Evolution 40:893–913
Stern D (2010) Evolution, development and the predictable genome. Roberts and Company Publishers, Greenwood Village
Stern S, Fridmann-Sirkis Y, Braun E, Soen Y (2012) Epigenetically heritable alteration of fly development in response to toxic challenge. Cell Rep 1:528–542
Stoebel D, Dean A, Dykhuizen D (2008) The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178:1653–1660
Strauss S, Lau J, Carroll S (2006) Evolutionary responses of natives to introduced species: what do introductions tell us about natural communities? Ecol Lett 9:357–374
Sunday J, Bates A, Dulvy N (2011) Global analysis of thermal tolerance and latitude in ectotherms. Proc R Soc B 278:1823–1830
Svanbäck R, Schluter D (2012) Niche specialization influences adaptive phenotypic plasticity in the threespine stickleback. Am Nat 180:50–59
Svanbäck R, Pineda-Krch M, Doebeli M (2009) Fluctuating population dynamics promotes the evolution of phenotypic plasticity. Am Nat 174:176–189
Tétard-Jones C, Kertesz M, Preziosi R (2011) Quantitative trait loci mapping of phenotypic plasticity and genotype-environment interactions in plant and insect performance. Philos Trans R Soc Lond B 366:1368–1379
Thibert-Plante X, Hendry A (2011) The consequences of phenotypic plasticity for ecological speciation. J Evol Biol 2:326–342
Tollrian R, Leese F (2010) Ecological genomics: steps towards unraveling the genetic basis of inducible defenses in Daphnia. BMC Biol 8:51
Tomanek L (2008) The importance of physiological limits in determining biogeographical range shifts due to global climate change: the heat-shock response. Physiol Biochem Zool 81:709–717
Tomanek L, Somero G (2002) Interspecific- and acclimation-induced variation in levels of heat-shock proteins 70 (hsp70) and 90 (hsp90) and heat-shock transcription factor-1 (HSF1) in congeneric marine snails (genus Tegula): implications for regulation of hsp gene expression. J Exp Biol 205:677–685
Ungerer M, Halldorsdottir S, Purugganan M, Mackay T (2003) Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165:353–365
Ungerer M, Johnson L, Herman M (2008) Ecological genomics: understanding gene and genome function in the natural environment. Heredity 100:178–183
Utsumi S (2011) Eco-evolutionary dynamics in herbivorous insect communities mediated by induced plant responses. Popul Ecol 53:23–34
Valena S, Moczek AP (2012) Epigenetic mechanisms underlying developmental plasticity in horned beetles. Genet Res Int 2012:576303
Vezzoli M, Castellani P, Corna G, Castiglioni A, Bosurgi L, Monno A, Brunelli S, Manfredi A, Rubartelli A, Rovere-Querini P (2011) High-mobility group box 1 release and redox regulation accompany regeneration and remodeling of skeletal muscle. Antioxid Redox Signal 15:2161–2174
Via S, Lande R (1985) Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39:505–522
Vilaprinyo E, Alves R, Sorribas A (2010) Minimization of biosynthetic costs in adaptive gene expression responses of yeast to environmental changes. PLoS Comput Biol 6:e10000674
Viñuelas J, Kaneko G, Coulon A, Beslon G, Gandrillon O (2012) Towards experimental manipulation of stochasticity in gene expression. Prog Biophys Mol Biol 110:44–53
Waddington C (1953a) Genetic assimilation of an acquired character. Evolution 7:118–126
Waddington C (1953b) The ‘Baldwin effect’, ‘genetic assimilation’ and ‘homeostasis’. Evolution 7:386–387
Waddington C (1956) Genetic assimilation of the bithorax phenotype. Evolution 10:1–13
Wagner A (2007) Energy costs constrain the evolution of gene expression. J Exp Zool 308B:322–324
Wang Y, Wu L, Wang P, Lv C, Yang Z, Tang X (2012) Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res 2012:1–8
Weiss L, Kruppert S, Laforsch C, Tollrian R (2012) Chaoborus and Gasterosteus anti-predator responses in Daphnia pulex are mediated by independent cholinergic and gabaergic neuronal signals. PLoS One 7:e36879
West-Eberhard M (2003) Developmental plasticity and evolution. Oxford University Press, Oxford
West-Eberhard M (2005) Phenotypic accommodation: adaptive innovation due to developmental plasticity. J Exp Zool B 304B:610–618
Whitehead A, Roach J, Zhang S, Galvez F (2012) Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc Natl Acad Sci USA 108:6193–6198
Whitham T, DiFazio S, Schweitzer J, Shuster S, Allan G, Bailey J, Woolbright S (2008) Extending genomics to natural communities and ecosystems. Science 320:492–495
Windig J, De Kovel C, De Jong G (2003) Genetics and mechanics of plasticity. In: DeWitt T, Scheiner S (eds) Phenotypic plasticity: functional and conceptual approaches. Oxford University Press, New York
Wolf J, Brodie E III, Wade M (2003) The genotype-environment interaction and evolution when the environment contains genes. In: DeWitt T, Scheiner S (eds) Phenotypic plasticity: functional and conceptual approaches. Oxford University Press, New York
Woltereck R (1909) Weitere experimentelle untersüchungen über artveränderung speziell über das wesen quantitativer artunterschiede bei daphniden. Verhandlungen de Deutschen Zooligischen Gesellschaft 19:110–172
Wood H, Spicer J, Widdiecombe S (2008) Ocean acidification may increase calcification rates but at a cost. Proc R Soc B 275:1767–1773
Wund M, Baker J, Clancy B, Golub J, Foster S (2008) A test of the ‘flexible stem’ model of evolution: ancestral plasticity genetic accommodation and morphological divergence in the threespine stickleback radiation. Am Nat 172:449–462
Wund M, Valena S, Wood S, Baker J (2012) Ancestral plasticity and allometry in threespine stickleback reveal phenotypes associated with derived freshwater ecotypes. Biol J Linn Soc 105:573–583
Yampolsky L, Glazko G, Fry J (2012) Evolution of gene expression and expression plasticity in long-term experimental populations of Drosophila melanogaster maintained under constant and variable ethanol stress. Mol Ecol 21:4287–4299
Yeh P, Price T (2004) Adaptive phenotypic plasticity and the successful colonization of a novel environment. Am Nat 164:531–542
Yin M, Laforsch C, Lohr J, Wolinska J (2011) Predator-induced defense makes Daphnia more vulnerable to parasites. Evolution 65(5):1482–1488
Zhou X, Tarver MR, Scharf ME (2007) Hexamerin-based regulation of juvenile hormone-dependent gene expression underlies phenotypic plasticity in a social insect. Development 134:601–610
Zhou S, Campbell T, Stone E, Mackay T, Anholt R (2012) Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet 8:e1002593
Acknowledgments
MM held a Vanier Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC), as well as a research allowance from Alberta Innovates Technology Futures (AITF). SMR is funded by an NSERC Discovery Grant and an AITF New Faculty Award. The authors wish to thank two reviewers for their insightful suggestions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Glossary: Some Definitions of Important Terms
- Adaptive plasticity
-
The production of alternative phenotypes (continuous or discrete) by the same genotype across some environmental variable, such that there is a better match between the organism and its environment (Beldade et al. 2011). Alleles that confer plasticity are more likely to spread through a population relative to competing alleles that do not confer plasticity.
- Behavioural plasticity
-
Environmentally-induced alternative behaviors displayed by a single genotype. Behavioral plasticity is difficult to define. Does an organism display behavioral plasticity if it switches from grazing when there are no predators to predator avoidance when predators are present? Or should behavioral plasticity be restricted to a single behavior type (i.e., different foraging tactics for different foods, or different predator avoidance strategies for different predators)? Or is behavior only plastic if one particular behavioral trait is expressed differently when the same environmental variable in manipulated, such as different foraging strategies for a single food under different light conditions, or different predator avoidance strategies for a single predator under different conspecific densities? One’s definition will determine the magnitude of the relationship one finds between behavioral plasticity and other reaction norms.
- Canalization
-
See Robustness.
- Community genetics
-
The study of how genes within a community shape the phenotypes and evolution of other members of the community, and the identification of genes that contribute to heritable community traits.
- Cryptic genetic variation
-
The subset of standing genetic variation that exists in a population but does not affect the phenotype or performance under normal environmental conditions. Upon exposure to a novel environment, this genetic variation produces novel phenotypic variation, and may facilitate adaptation.
- Developmental noise
-
The production of alternative phenotypes by a single genotype under identical environmental conditions (Raser and O’Shea 2005) due to molecular stochasticity in the birth and death rates of transcripts, the effects of low-abundance regulatory proteins, the stickiness of proteins, and random fluctuations in promoter behavior (Raser and O’Shea 2005; Brettner and Masel 2012; Singh et al. 2012).
- Dominant plasticity
-
In niche complementarity, occurs when a superior competitor with high resource use plasticity alters the resources it uses depending on the competitive environment.
- Ecological speciation
-
A theory of speciation in which adaptive phenotypic and genetic divergence, contributing to reproductive isolation, is due to divergent selection (Nosil 2012).
- Environmental robustness (environmental
-
canalization) The production of a stable reaction norm despite environmental perturbations. Non-plastic reaction norms are canalized against at least one environmental variable. A plastic reaction norm can be environmentally canalized if: (1) the reaction norm does not exhibit discontinuous change under extreme environments, or (2) the reaction norm maintains its height and slope in the presence of a second environmental variable.
- Epigenetics
-
The study of environmentally-induced, sometimes heritable modifications to the phenotype, caused by mechanisms other than changes to the underlying DNA sequence (i.e., DNA methylation, histone modification, etc.).
- Epigenome
-
The entire suite of epigenetic modifications that have occurred in a particular cell, tissue, developmental stage, or organism, including the number and placement of methylated sites, the number and nature of histone modifications, etc.
- Flexible stem
-
A model of adaptive phenotypic divergence whereby an initially plastic ancestral population diverges into two populations residing in distinct environments, such that each population expresses opposing ends of a reaction norm. These phenotypes become genetically assimilated, such that plasticity is lost and phenotypic divergence is maintained (West-Eberhard 2003; Wund et al. 2008).
- Gene expression
-
The context-dependent production of gene product, including pre-mRNA, mRNA, microRNA, and protein. Context can include cell type, tissue type, genotype, developmental stage, time, and environment.
- Genetic assimilation
-
The evolved loss of adaptive phenotypic plasticity, such that environmental induction is no longer necessary for the production of the phenotype (Waddington 1953a, b).
- Genetic compensation
-
The evolved loss of maladaptive phenotypic plasticity, resulting in phenotypic similarity (cryptic evolution) between populations living in regular and novel environments (Grether 2005).
- Genetic robustness (genetic robustness)
-
The production of a stable reaction norm despite different genetic backgrounds (Gibson and Wagner 2000). Genotypically distinct individuals that display the same plastic or non-plastic reaction norm are genetically canalized against the alleles that differentiate them. A lack of genetic robustness can be evidenced by changes to the slope or height of the reaction norm.
- Hierarchy of plasticities
-
The production of a plastic or non-plastic macrophenotype due to interactions between numerous underlying reaction norms (Bradshaw 1965).
- Macrophenotype
-
The visible manifestation of numerous underlying phenotypes, sometimes referred to as the “end phenotype” (Beldade et al. 2011).
- Maladaptive plasticity
-
The production of alternative phenotypes (continuous or discrete) by the same genotype across some environmental variable, such that the match between organism and environment is reduced (Ghalambor et al. 2007).
- Model organism
-
Any non-human species that has been readily cultured or raised over many generations in a laboratory setting, for which genomic tools have been developed and applied, and that is used to answer biological questions that can be applied to other species. Examples: Arabidopsis, Drosophila, Daphnia, Mus. The ideal model species for ecological genomics has locally adapted populations, characterized phenotypic and genetic variation, a sequenced genome, a known phylogeny, and is studied by a large community of researchers (Feder and Mitchell-Olds 2003).
- Molecular phenotype
-
Measures of context-specific gene expression or protein behavior (Ranz and Machado 2006; Pavey et al. 2010).
- Molecular plasticity
-
A form of phenotypic plasticity that focuses on environmentally-sensitive gene expression or protein behavior (plasticity in the molecular phenotype).
- Neutral plasticity
-
The production of alternative phenotypes (continuous or discrete) by the same genotype across some environmental variable, that does not contribute positively or negatively to fitness.
- Non-model organism
-
Any species that has not been readily incorporated into biological research in the last several decades. Basic biological information, including genomic information, is often lacking for these organisms, although genomic tools may be developed and used.
- Non-plasticity
-
A reaction norm with a slope of zero. Sometimes referred to as environmental robustness (Gibson and Wagner 2000). Non-plasticity may be adaptive, maladaptive, or neutral, relative to competing alleles that confer plasticity.
- Phenotypic accommodation
-
A source of adaptive phenotypic novelty, in which a genetically- or environmentally-induced change to a phenotype during development is accommodated through plastic changes in other phenotypes (West-Eberhard 2005).
- Phenotypic capacitor
-
Any phenotype that can suppress phenotypic variation that would otherwise be expressed via developmental noise, microenvironmental variation, and genotypic variation (Queitsch et al. 2002; Levy and Siegal 2008).
- Phenotypic plasticity
-
The environmentally sensitive production of alternative phenotypes by a single genotype (DeWitt and Scheiner2003).
- Plastic compensation
-
The production of phenotypic similarity between populations living in regular and novel environments, due to plasticity in some compensating phenotype. Without plasticity in the compensating phenotype, maladaptive plasticity would generate phenotypic divergence between populations. Usually comes with a cost to some other phenotype and may mask the existence of maladaptive plasticity (Morris and Rogers 2013).
- Plasticity-mediated population extinction
-
(PMPE) The unsuccessful colonization of a new environment due to phenotypic plasticity induced by the new environment (Morris and Rogers 2013).
- Plasticity-mediated population persistence
-
(PMPP) The successful colonization of a new environment due to phenotypic plasticity induced by the new environment (Baldwin 1896; Pavey et al. 2010).
- Proteome
-
The full complement of proteins present within a particular context (see gene expression).
- Proteomic plasticity
-
A form of molecular plasticity that focuses on environmentally-sensitive protein abundance.
- Reaction norm
-
A function of all possible phenotypic states across some environmental gradient.
- Robustness (Canalization)
-
The production of a stable reaction norm despite genetic, environmental, or stochastic perturbations. Both plastic and non-plastic reaction norms can display robustness (Waddington 1953a, b).
- Symmorphosis
-
The theory that biological structures match their functional requirements, without unnecessarily exceeding those requirements. This includes the idea that the components of a system will not exceed in possible performance their weakest unit.
- Standing genetic variation (SGV)
-
Genetic variation that exists at a single locus in natural populations (Barrett and Schluter 2008). One form of SGV, cryptic genetic variation (CGV), is not apparent until exposed by novel environments (Schlichting 2008).
- Stochastic robustness (Stochastic robustness)
-
The production of a stable reaction norm despite stochasticity (developmental noise) in transcript abundance, protein activity, etc.
- Transcriptional plasticity
-
A form of molecular plasticity that focuses on environmentally-sensitive mRNA transcript abundance.
- Transcriptome
-
The full complement of mRNA present within a particular context (see gene expression).
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Morris, M., Rogers, S.M. (2014). Integrating Phenotypic Plasticity Within an Ecological Genomics Framework: Recent Insights from the Genomics, Evolution, Ecology, and Fitness of Plasticity. In: Landry, C., Aubin-Horth, N. (eds) Ecological Genomics. Advances in Experimental Medicine and Biology, vol 781. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-7347-9_5
Download citation
DOI: https://doi.org/10.1007/978-94-007-7347-9_5
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-7346-2
Online ISBN: 978-94-007-7347-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)