Abstract
Icosahedral viruses exhibit elegant pathways of capsid assembly and maturation regulated by symmetry principles. Assembly is a dynamic process driven by consecutive and genetically programmed morphogenetic interactions between protein subunits. The non-symmetric capsid subunits are gathered by hydrophobic contacts and non-covalent interactions in assembly intermediates, which serve as blocks to build a symmetric capsid. In some cases, non-symmetric interactions among intermediates are involved in assembly, highlighting the remarkable capacity of capsid proteins to fold into demanding conformations compatible with a closed protein shell. In this chapter, the morphogenesis of structurally simple icosahedral viruses, including representative members of the parvoviruses, picornaviruses or polyomaviruses as paradigms, is described in some detail. Icosahedral virus assembly may occur in different subcellular compartments and involve a panoplia of cellular and viral factors, chaperones, and protein modifications that, in general, are still poorly characterized. Mechanisms of viral genome encapsidation may imply direct interactions between the genome and the assembly intermediates, or active packaging into a preformed empty capsid. High stability of intermediates and proteolytic cleavages during viral maturation usually contribute to the overall irreversible character of the assembly process. These and other simple icosahedral viruses were pioneer models to understand basic principles of virus assembly, continue to be leading subjects of morphogenetic analyses, and have inspired ongoing studies on the assembly of larger viruses and cellular and synthetic macromolecular complexes.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Icosahedral capsid
- Triangulation number
- Assembly intermediate
- Protein contacts
- Nuclear translocation
- Protein folding
- Nucleation
- Packaging
- Encapsidation
- Virus factory
- Maturation cleavage
1 Introduction
All viral entities have a capsid, which in structurally simple viruses is built up from one or a few types of protein subunits. At late stages of the intracellular phase of their life cycle (see Chap. 1) viruses perform capsid assembly, a process by which the structural capsid protein (CP) subunits are joined by maximal hydrophobic contacts and/or non-covalent interactions (and occassionally covalent bonds) to construct the viral particle. This process is essential for viruses to mature (become infectious) and release progeny, and in the simple icosahedral viruses proceeds by strict principles of genetic economy and symmetry (see Chap. 2). This chapter reviews the main stages in the assembly and genome encapsidation of small or medium sized viruses with a relatively simple icosahedral architecture, exemplified by three distinct virus models: (i) Adeno-associated virus (AAV) and minute virus of mice (MVM), as respective representative members of the Dependovirus and Parvovirus genera of the Parvoviridae, a family of single-stranded (ss) DNA viruses with a T = 1 capsid (25 nm in diameter) assembling in the nucleus; (ii) Poliovirus, genus Enterovirus of the Picornaviridae, RNA(+) viruses with a pseudoT = 3 capsid (30 nm in diameter) assembling in the cytoplasm; and (iii) Polyomavirus and Simian Virus (SV40), members of the Polyomavirus genus of the Polyomaviridae, double-stranded (ds) DNA viruses with an all-pentamers T = 7d capsid (45 nm in diameter) assembling in the nucleus. In these so-called “simple” icosahedral viruses, assembly occurs through an orchestrated pattern of interactions of the capsid subunits to form complexes or assembly intermediates, whose composition and conformation usually change along the process. The structural dynamics undergone by the assembly intermediates must also fulfill another important function, which is to traffic within the infected cell towards the compartment where the genome is being replicated. It is the accumulation of assembly intermediates at a specific compartment what triggers genome encapsidation and maturation, allowing the virus to finally propagate in nature.
2 Icosahedral Capsids: Symmetry and Genetic Regulation
2.1 Structural Principles in Icosahedral Capsid Assembly
In simple icosahedral viruses, the capsid is formed by many copies of one or a few protein subunits that assemble by making multiple contacts to build a hollow shell of proper size and symmetry. The regular icosahedron is formed by a defined number of copies of a capsid building block (CBB), which can be built up by a single CP subunit, by several identical CPs, or by non-identical CPs. The CBBs are related by two-fold (2×), three-fold (3×), and five-fold (5×) symmetry axes, and CPs within the CBBs establish regular interactions with their neighbors depending on their position in relation to these icosahedral axes. The Caspar and Klug (1962) theory explained how some multiples of 60 identical subunits could be arranged with similar (quasi-equivalent) interactions, according to the rule T = h2 + hk + k2, where h and k are integers and T is called the triangulation number (see Chap. 2 for a detailed explanation of capsid icosahedral symmetry and quasi-equivalence).
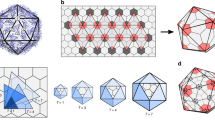
In the members of the Parvoviridae, the capsid is a T = 1 perfect icosahedron, formed by a total of 60 CPs (termed viral proteins, VPs, in this and some other virus families), which include two to three variant types (VP1, VP2 and VP3) with identical amino acid sequence and fold except for short streches of sequences at the C- or N-termini, which are intrinsically disordered and are not observed in the X-ray structures of the virus particles (Fig. 10.1a). The topology of the capsid surface differs among the parvoviruses due to the characteristic prominence of peptide loops and spikes at the 3x symmetry axes, and the depth and contour of the depression surrounding the 5x axes [1, 2, 7–10]. In Poliovirus and related picornaviruses the protein shell is built up of 60 copies of a fundamental subunit (the protomer) composed of three different proteins (termed VP1, VP2 and VP3) which are not related in amino acid sequence but have a similar fold, and a small extended polypeptide (VP4). VP4 is located at the inner surface of the protein shell and remains covalently linked to the N-terminus of VP2 until the final stages of virus assembly and maturation. The interactions between the VP1 subunits around the 5x axes are not equivalent to those engaging the alternating VP2 and VP3 subunits in the 3x axes, accounting for the features of the surface of the capsid (Fig. 10.1c), including the conspicuous protrusion of the VP1 subunits at the 5x axes [3, 4]. In polyomaviruses such as SV40, the icosahedral capsid is formed by 360 identical CP subunits of a single protein (termed VP1), whose arrangement does not follow the quasi-equivalence rules of Caspar and Klug, as the basic structural elements (capsomers) are 72 pentamers displayed in a T = 7d surface lattice ([5]; Fig. 10.1e). Each pentamer contains in addition one copy of either of two other proteins (termed VP2 and VP3), which share most of their amino acid sequences. The capsomers located around each 5x axis are referred to as the pentavalent pentamers, and those capsomers arranged around each of the 3x axis are referred to as the hexavalent pentamers.
Atomic structure of icosahedral virus particles and capsid subunits whose assembly is described in this chapter. (a) Structure of Parvovirus MVM (p and i strains; [1, 2]). Different colours distinguish the subunits surrounding the 5x axes (pentagon) from those interdigitated at the 3x axes (triangle). (b) Folding of the VP1 and VP2 subunits in the MVM structure. Note the prominent loops projecting away from the capsid surface. (c) Structure of Poliovirus, a picornavirus [3, 4]. The VP1 subunits around the 5x axes (pentagon), and the alternating VP2 and VP3 subunits around the 3x axes (hexagon) are respectively shown in green, red and violet. (d) Folding of the VP1 subunit in the capsid of Poliovirus. The β-strands (arrows) forms two antiparallel sheets juxtaposed in a wedgelike structure. (e) Structure of the SV40 virus showing the organization of the VP1 pentamers [5]. The 5x-coordinated pentamers are represented in gray and the VP1 subunits of the pentamers in hexameric arrays are represented in different colors. (f) folding of the VP1 subunit as observed in the SV40 capsid. The β-barrel (jellyroll) is radial to the capsid surface. The figure was prepared using the Pymol program (http://www.pymol.org/) and the VIPER resources [6], using the atomic coordinates of MVM (1MVM, 1Z1C), Poliovirus (2PLV), and SV40 (1SVA), deposited in the Protein Data Bank (PDB)
Although the parvovirus, poliovirus and polyomavirus capsid proteins are not related in amino acid sequences, the core of all these proteins are folded in their capsids as a β-sheet structure termed β-barrel jelly roll (or Swiss-roll β-barrel, or an eight-stranded antiparallel β-barrel), a wedge-shaped structure comprising two antiparallel β-sheets. The topology of all these capsid subunits is, thus, similar (Fig. 10.1b, d and f). The major structural differences between them are in the loops that connect the strands, which are particularly prominent in parvoviruses (Fig. 10.1b), and in the N- and C- terminal segments that extend from the central β-barrel domains. The remarkable similarity of these β-barrel jelly rolls among viral proteins that do not share primary amino acid sequences, and belong to different families of viruses with unrelated biological properties, suggests that it may represent one of the few structural solutions allowing proteins to be packaged in icosahedral capsids, or be a testimony of a common ancestral evolutionary history (see also Chaps. 2 and 7).
2.2 Synthesis of the Capsid Subunits: Setting the Assembly Scenario
For successful assembly, the synthesis of the capsid proteins must be tightly regulated during the virus life cycle, in order to satisfy at least three key requirements: (i) quantity, viruses must induce the accumulation of high amounts of capsid proteins in the host cells, in order to compete with the vast amount and diversity of preexisting cellular proteins; (ii) timing, capsid proteins synthesis must reach maximum levels by the time the viral genome is being replicated, so it can be efficiently packaged; and (iii) stoichiometry, capsid subunits must be synthesized at the proper ratio to ensure assembly of an infectious viral particle. Genetic regulation of viral gene expression is therefore crucial for a successful assembly process. Mechanisms controlling viral gene expression involve multiple networks which are out of the scope of this book, so only a brief outline of those involved in assembly are mentioned below.
In Poliovirus (see [11] for a review of its life cycle), the genomic RNA is released in the cytoplasm by the incoming virion, and translated as a single open reading frame to produce a very large polyprotein, which undergoes cotranslational cis-cleavage by the viral protease 2A. This cleavage releases from the N-terminus the precursor polyprotein myristoyl-P1, which contains the CP sequences. In this and related picornaviruses, CPs are synthesized to high levels by the combination of the translational competence of the genomic RNA with an effective host protein shut-off induced by viral proteases, whereas the proper protein stoichiometry results from the cleavage of a common precursor.
In parvoviruses, polyomaviruses and other DNA viruses, the stoichiometry of capsid proteins is regulated mainly at the level of splicing of their messenger RNAs. In the parvovirus MVM for instance, site-directed mutagenesis at the minor splicing sites, or independent cDNA cloning, allows to obtain genomic clones expressing either VP protein [12]. VP2 alone can form a capsid which can encapsidate the viral genome, but VP1 is necessary for the infectivity of the particles due to specific domains residing at its N-terminal segment [13, 14]; and (Fig. 10.3c). A 1:5 ratio of VP1:VP2 is found for the soluble synthesized proteins, as well as in the assembled capsid, and preserving this ratio is critically important for an ordered assembly avoiding protein aggregates [16]. In polyomaviruses VP1, VP2, and VP3 levels in the infected cells are regulated by alternative splicing from a common trascript, occurring soon after viral DNA replicative intermediates accumulate. In these small DNA viruses protein shut-off is not a major mechanisn to counteract host protein synthesis, but the nuclear accumulation of protein products prior assembly (see below) facilitates their interactions at certain nuclearly confined enviroments.
3 Capsid Building Blocks and Assembly Intermediates
The capacity of the viral structural proteins to form capsids ultimately result from their folding and self-assembly properties, which are conferred by the encoded amino acid sequences. Virus capsids are assembled from CBBs which may be CP monomers or, in many cases, CP oligomers. These stable oligomeric CBBs may be considered as the first stable intermediates of the capsid assembly process. The use of in vitro assembly systems in combination with diverse theoretical approaches has allowed the investigation of fundamental principles of virus capsid self-assembly starting from CBBs. The theory of capsid self-assembly is outside the aims of this chapter, which is dedicated to the description of assembly processes in different viruses from in vitro and in vivo experimental evidences supported by structural and functional studies. The reader is referred to Chap. 1 for a brief overview, and to Chap. 19 for a detailed description on the thermodynamic and kinetic aspects of the assembly of simple virus capsids. These studies are generally consistent with experimental observations on the assembly of very simple virus capsids, frequently carried out in controlled in vitro experiments. The models support assembly as a nucleated cooperative process in which CP or CBB concentration is critical, with a lag phase reflecting the time required to build up an assembly line of intermediate structures. The intermediates are expected to be at very low concentration, but this steady state of intermediates is required for efficient assembly in any stepwise reaction.
3.1 Structure of CBBs and Assembly Intermediates
The stepwise assembly pathway has been well characterized in Poliovirus (and other picornaviruses including human rhinovirus and foot-and-mouth disease virus), as discrete intermediates are stable enough to be isolated. A common strategy that many viruses adopt to build blocks for capsid assembly is to initiate assembly while the structural units are linked into a polyprotein precursor. In Poliovirus, the first intermediate of the assembly pathway is an inmature structural unit (unprocessed protomer) formed by a folded polyprotein (termed P1). P1 contains three structural domains. These domains are split apart from each other upon cleavage at specific sites in the linker sequences by the viral 3CDpro protease. The result is a processed protomer (5S protomer) which sediments as a 5S particle and is formed by one copy each of VP0, VP3, and VP1, which correspond to the cleaved P1 domains (Fig. 10.2a). It is unclear when the β-barrel of these proteins core fold, but unprocessed P1 of picornaviruses is recognised by panels of virus-neutralising antibodies elicited against discontinuous epitopes of mature virions, strongly suggesting that the domains in unprocessed P1 are already folded like the mature CPs in the assembled capsid. The structures of the isolated unprocessed or processed P1 protomers have not been solved for any picornavirus yet; however, in the virus capsid structure VP1, VP2 and VP3 form an intricate network of intermolecular interactions among the surfaces of their β-barrel domains, which must greatly stabilize the protomer and contribute to the early stages of the assembly. VP4 remains covalently linked to VP2 in the VP0 protein until virus assembly is completed and maturation occurs (see below).
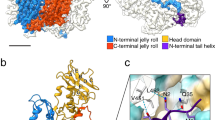
Structural models of assembly intermediates in icosahedral viruses. (a) Poliovirus P1 polyprotein precursor showing the four proteins forming the heterometric structural unit (protomer). Cleavage by 3CDpro protease yields the 5S assembly intermediate. (Adapted from Flint et al. (2009) Principles of Virology, ASM Press, with permission). (b) Structure of the VP1-VP2 assembly intermediate of polyomavirus. VP2 is shown in red and the three VP1 monomers that form contacts with VP2 in green (middle) and blue (left and right). (c) Schematic representation of the VP1/VP2 interaction in this complex. VP2 (red) enters in the VP1 pentamer (black) from the base, continuing to the upper part of the conical depression. It then loops back to interact specifically with the inner face of VP1, forming a hairpin-like structure. (b, c: Reprinted from [17], with permission). (d) Structure of the VP1 pentamer in the SV40 capsid. (e) Structure of the VP trimer in the MVM capsid. Residues involved in NLM function (see below) are highlighted. (d, e: prepared using the Pymol program (http://www.pymol.org/) and the 1SVA and 1MVM respective atomic coordinates deposited in the PDB)
The 5S precursor is followed in the assembly line by the 14S pentamer (Fig. 10.4a), which is formed by oligomerization of five 5S protomers, and is stabilized by extensive protein-protein interactions and by others mediated by myristate chains incorporated in the five VP0 N-termini. These multiple interactions determine a molecular interlocking of the five protomers in the pentamer, conferring high stability and directionality to the whole assembly pathway.
In polyomaviruses as well as in parvoviruses, the CP subunits interact in the cytoplasm before nuclear capsid formation. The composition of the assembly intermediates is characteristic for each virus system and plays a role of paramount importance in the correct timing and spatial ordering during assembly. A relatively stable cytoplasmic assembly intermediate seems to be a common need for these viruses completing capsid assembly and maturing within the nucleus. In Polyomavirus and SV40, stable pentamers of VP1 are formed in the cytoplasm. Each VP1 pentamer binds either one VP2 or one VP3 protein that becomes allocated in the axial cavity of the pentamer (Fig. 10.2b), and the VP1-VP2/3 complex is stabilized by strong hydrophobic interactions [17]. The contacts between subunits significantly alter the configuration of the VP1 pentamer, as demonstrated by changes in epitope accesibility and by direct structural insights obtained from the crystal structure (Fig. 10.2b, c). In SV40, transient disulphide bridges are established intramolecularly, and subsequently intermolecularly, as the monomers assemble into pentamers, which facilitate the folding and interdigitation of structural elements [19]. The non-covalent interactions and covalent bonds collectively conform a stable cytoplasmic CP pentameric complex (Fig. 10.2d), which is the major assembly intermediate in these viruses.
In parvoviruses, taking the murine MVM as a reference model, trimers of VP subunits assemble in the cytoplasm. The VP1 (82 kDa) and VP2 (63 kDa) proteins synthesized at a VP1:VP2 1:5 M ratio assemble into two types of trimers (Fig. 10.2e), which are produced in stoichiometric amounts. The larger trimer (200 kDa) is a heterotrimer formed by one VP1 and two VP2 subunits, whereas the smaller (180 kDa) is a VP2-only homotrimer [16]. In the formation of the cytoplasmic trimer, the VP2 protein may act as a scaffolding factor assisting VP1 to acquire a proper folding, as deletion mutants of the VP1-specific region undergo extensive ubiquitination degradative reaction that can be significantly prevented by co-expression of VP2 [14]. The assembly of VP cytoplasmic trimers for these viruses was structurally supported first by the higher stability of the trimer (measured by the buried surface area on oligomer formation) as compared to putative dimers or pentamers of CP subunits, due to the multiple contacts in the intertwined loops of the subunits around the 3x symmetry axes (Fig. 10.1a). Furthermore, crosslinking experiments and mutations disrupting the intertrimer interfaces in the MVM capsid allowed the isolation of stable trimers (see below).
3.2 Intracellular Traffic of CBBs and Assembly Intermediates
For capsid formation, CBBs or other assembly intermediates must accumulate in the subcellular compartment where the viral genome replicates, and at the right time. In some viral systems, the capsid subunits must traffic from the site of synthesis in the cytoplasm to the assembly compartment. This traffic is directed by protein signals in CBBs or intermediates that are accessible to the transport machineries of the cell. The nature of the signals and the configuration of the intermediate exposing them, which will determine the transport route accessed by the intermediate and its fate in the cell, are thus key elements in the viral assembly process.
In polyomaviruses and parvoviruses, the viral genome is replicated in the nucleus, and the CPs of these viruses are therefore karyophilic polypeptides that traverse the nuclear membrane. Translocation of proteins into the nucleus imposes two restrictions, firstly by the need of nuclear localization sequences (NLS) to access the cellular transport machinery, and secondly by the size of the complex to be transported, which cannot exceed 25–30 nm, the functional diameter of the aperture in the nuclear pore complex (NPC) [20], a supramolecular structure embedded in the nuclear envelope (Fig. 10.3a). The exchange of macromolecules across the NPC is mostly mediated by proteins of the importin β (karyopherin β) superfamily, which comprises importins and exportins. The NLS are, in the so-called conventional configuration [21], single or bipartite stretches of basic amino acids that access, generally upon direct binding to a protein adaptor, the importin α/β transport pathway [22]. However, many karyophilic proteins harbor non-classical NLS that may bind karyopherin β1 directly, or access the alternative karyopherin β2/transportin import pathway. A current active area of research is the identification of the transport routes accessed by the viral proteins in specific cell hosts. Functional conventional NLS are found in the major VP1 as well as in the minor VP2/3 subunits of Polyomavirus [23, 24] and SV40 [25] (Fig. 10.3b). It is remarkable the sequence conservation among the structural proteins of these viruses at the NLS domains, compared to the high sequence divergence in the rest of these proteins. In polyomaviruses the VP1-VP2/3 cytoplasmic complex is translocated through the NPC by the functional cooperation of the multiple NLS displayed by the VP subunits [26].
Nuclear localization sequences in polyomavirus and parvovirus capsid proteins. (a) Basic architecture of the nuclear pore complex (NPC). The central ring is illustrated with the filament facing the cytoplasm and the basket protruding into the nucleus. (b) Examples of identified NLS in the VP proteins of Polyomavirus and SV40. (c) Two conserved domains with NLS activity identified in the VP1 N-terminal sequence of several parvoviruses: MVM, parvovirus H1 (H1-PV), CPV, feline parvovirus (FPV), mink enteritis virus (MEV), porcine parvovirus (PPV). (d) Configuration of the NLM in the capsid subunits of MVM and conservation of the NLM sequence in related parvoviruses. Basic residues contributing to nuclear targeting of assembly intermediates are shown in bold. (Adapted from [15] with permission)
In MVM, used as a representative molecular model of the Parvoviridae, the two types of VP trimers are major CBBs, or stable assembly intermediates, translocating across the nuclear membrane. The protein subunits within the trimer cooperate for nuclear transport, as both VP1 and VP2 proteins genetically depleted of functional nuclear transport sequences can be co-transported into the nucleus by expressed intact subunits [14]. Indeed VP1 and VP2 carry independent NLS and efficiently target the nucleus of transfected cells when singly expressed [12]. VP1 harbours two conventional NLS at its N-terminal specific region (Fig. 10.3c), which function independently. These NLS are required for nuclear translocation of the expressed VP1 subunits in MVM [14] and in canine parvovirus, (CPV) [27], and also for the MVM virion to initiate infection [14], suggesting that they are exposed out of the virus shell during the cell entry process. Similar separate regions with basic amino acids essential for assembly and infectivity were identified in the capsid proteins of AAV [28]. In addition, both VP1 and VP2 (the major capsid-forming polypeptide) contain, in their common folded sequence, a structured domain with nuclear targeting capacity (named NLM) [15]. In the assembled capsid the NLM is localized in the amphipatic ß-strand I at the inner capsid surface (Fig. 10.3d). The NLM is the only functional nuclear targeting sequence identified in VP2. The structured NLM, highly conserved in many members of the Parvoviridae, shows, when displayed in the capsid structure (Fig. 10.2e), all its charged basic amino acids placed within the side of the strand facing the solvent at the interior surface of the capsid, and the hydrophobic amino acids oriented towards the protein core. This structuration of the NLM probably occurs only upon folding and trimerization of the VPs, which may occur concomitantly. Folding and oligomerization of the capsid subunits probably occur in the cytoplasm, leading to trimeric assembly intermediates acquiring nuclear transport competence. The structuration of the two types of trimers translocating across the NPC may exert a quality control role in the virus morphogenetic flow, as misfolded subunits (not exposing the NLM) or oligomers with aberrant structures and protein composition would not access the nucleus [15, 16].
4 Forming the Capsid
4.1 Contacts and Structural Changes in CBBs and Assembly Intermediates
The CBBs or initial stable intermediates of capsid assembly may be found in an assembly-incompetent state, incapable of establishing productive interactions between them to accommodate the final configuration of the capsid [29]. Thus, the assembling oligomers must frequently undergo conformational changes during the late stages of the assembly pathway. In Poliovirus and other picornaviruses, the similar β-barrel folding of the VP1, VP2 and VP3 proteins that facilitate their interactions to form the 60 structural subunits (5S protomers) and the 14S pentameric CBBs (Fig. 10.4a), also favours the subsequent assembly of these latter intermediates into a complete viral particle [30]. For this, the extensive interactions among the β-barrels of adjacent proteins help to form a rather rigid protein shell with a dense network of intersubunit interactions. Inside the capsid, a network of additional protein contacts stabilizes the mature particle. These contacts are largerly contributed by the long (40–80 residues) N-terminal arms of the three VP subunits, which have a similar path inside the capsid in different picornaviruses, although their primary amino acid sequence is drastically different. These interactions are most extensive at the 5x axes, where the N-termini of five VP3 molecules are arranged in a tubelike parrallel β-sheet. The conformation of these arms is ordered only after capsid assembly, so it is difficult to evaluate the contribution of the interactions involving these arms in the final particle stability based on structural data only.
Interactions among assembly intermediates to form the capsid. (a) Assembly of the 5S protomer into a 14S pentamer in Poliovirus. The protomer is not identical to the icosahedral asymmetric unit in the capsid. (b) Contacts between capsomers in SV40. Four pentamers assembled in the SV40 capsid are shown. Each subunit of the pentamers projects an arm that makes different contacts with the jellyroll of the subunit in another pentamer. (Adapted from [5], with permission). (c) Capsid formation in Parvovirus: a VP trimer is represented; some residues involved in major inter-trimer contacts of MVM capsid and required for capsid assembly are shown as spacefill models and coloured violet. (Adapted from [18], with permission). (d) Sedimentation analysis in sucrose gradients of the K153A mutant of the MVM capsid proteins. This mutant lacks a side chain that is important for establishing critical contacts between MVM trimers during capsid assembly. The sedimentation position of complete capsids, trimers, and VP monomers are indicated. (Adapted from [16], with permission)
In members of the Parvoviridae, capsid formation in the natural infection starts in the nucleus. Indeed the nucleolus was identified as the subcellular compartment where AAV assembly inititates, colocalizing with the non-structural replicative viral Rep proteins; subsequently, capsid accumulation spreads across the entire nucleus [31]. In MVM, as the assembly intermediates (mainly trimers) accumulate in the nucleus, the nucleation reaction is triggered and multiple non-covalent interactions are established between amino acids localized at the edges of the binding trimers. A few evolutionary conserved residues involved in presumably strong intertrimer contacts were found to be necessary for capsid assembly (Fig. 10.4c). These residues buried a large hydrophobic surface upon trimer association, or formed buried intertrimer hydrogen bonds or salt bridges [18]. Assembly intermediates other than the CBBs are difficult to isolate in parvoviruses, presumably because of the high efficiency of the assembly reaction in which intermediates are highly transient and accumulate at very low levels, in agreement with theoretical studies on the assembly of simple virus capsids (see Chap. 19). A genetic approach to trap unstable assembly intermediates based on the introduction of disruptive mutations at some of the residues that in the capsid are involved in contacts between MVM trimers yielded high amounts of trimers, without evidence of any other larger intermediate [16]. These trimers could be isolated by sedimentation in sucrose gradients (Fig. 10.4d), and were competent in a nuclear transport assay performed in permeabilized cells [32], indicating that their configuration properly expose the nuclear transport sequences, and thus mutations precluded just the final capsid formation stage.
The acquisition of competence for assembly by MVM trimers involves conformational changes that have been indirectly detected. An MVM-induced monoclonal antibody recognizing a discontinuous epitope located at each 3x axis of the capsid (which corresponds to the center of each trimer) failed to react with non-assembled trimers accumulated in the cell nucleus or isolated in vitro [16, 32]. Trimers must therefore change their conformation during nuclear capsid assembly, and the process may be triggered by some external factor and/or phosphorylation of the subunits (see below). These induced conformational rearrangements would reorganize some residues located at the vertex of the thee-fold axes, creating a new epitope.
In the viral particle of Polyomavirus and SV40, all the capsomers are pentamers, even though the capsid shell is built by subunits arranged in an icosahedral T = 7 lattice. Therefore the architecture of this virus does not fullfil the arrangement of pentameric and hexameric capsomers predicted by the quasiequivalent theory for capsids with T >1 values ([33]; see Sect. 10.2.1). However the SV40 capsid is stable enough, and this is accomplished in part by virtue of unique intercapsomers bonds established during assembly by the C-terminal domain of the VP1 protein (Fig. 10.4b). This domain acts as a connecting arm which, under six different configurations, makes contacts between the pentavalent pentamers (around each 5× axis) and the hexavalent pentamers (around each 3× axis). The different types of contacts in which the arms are involved act as multiple clamps holding the subunits together, and thus ensuring the stability of the capsid shell [5].
The VP1 protein of Polyomavirus can self-assemble into pentamers in heterologous cell expression systems [34], and the purified pentamers can associate to form capsid-like assemblies (VLPs) that become stabilized at low ionic strength by calcium. Interestingly, these VLPs assemble without the minor virion protein components, VP2 and VP3, suggesting that the non-equivalently related subunits of the penta- and hexavalent capsomeres may spontaneously switch their bonding specificity during assembly. VP1-only capsids however do not represent the physiological assembly pathway observed in the natural virus infections, in which the assembly of the VP1 pentamers is driven upon regular interactions involving VP2 and VP3 with the viral genome DNA (see below), and empty capsids are thought to be minor abortive assembly by-products.
4.2 Compartments, Factors, and Protein Modifications Influencing Capsid Formation
Icosahedral capsid assembly is influenced by environmental conditions in the cell and also by multiple molecular factors (in addition to the VP protein themselves) that may be encoded by the cells or by the virus genomes. These factors may act at specific subcellular compartments, and their contribution may critically determine in many cases the efficiency of the assembly process in vivo. Some of these factors have been identified in several viral systems, although multiple experimental evidences suggest that many others remain to be identified. For instance in Poliovirus, the rate of assembly of the structural proteins in vitro is reduced by at least two orders of magnitude compared to the rate observed in infected cells, and the empty capsids formed showed altered conformation, unless the reaction is primed by 14S pentamers isolated from infected cells. This experiment implies that proper folding, interactions, and/or modifications of the proteins forming the pentameric 14S intermediate, are essential for assembly to proceed successfully. A candidate for this function is the cellular Hsp70 chaperone, which associates with the P1 polyprotein precursor during its folding to form the 5S structural unit.
The cellular modulations of virus assembly may involve complex signalling pathways. For parvoviruses, capsid assembly during natural infections occurs with high efficiency in the nuclear compartment of infected host cells. However virus-like particles (VLPs) devoid of nucleic acid may be formed in the cytoplasm of heterologous expression systems (e.g. recombinant baculovirus expressing VP2 of MVM in insect cells) at low efficiency [35, 36]. This distinct assembly efficiency may be accounted, at least in part, by post-translational modifications of the capsid subunits. In MVM-infected cells, VPs and native capsid become extensively post-translationally modified by phosphorylation [37], whereas VLPs purified from heterologous insect cells were not phosphorylated [32]. In spite of the absence of modifications, these VLPs showed a 3-D structure identical to the native capsid and virus [2, 35], indicating that phosphorylation is not important for the icosahedral T = 1 ordering. The 2D-tryptic phosphopeptides analysis of native MVM capsid subunits resulted in a complex pattern of phosphoserine and phosphothreonine residues which was different for VP1 compared to VP2 [37]. In the host cell systems studied, the phosphorylation of the VP subunits of MVM was mainly catalyzed by the cytoplasmic activity of the Raf-1 kinase of the MAPK signalling pathway [32], and this modification was crucial for the acquisition of nuclear transport competence by the trimers. Phosphorylation by cellular kinases may be a general strategy to connect capsid assembly with host cell physiology, ensuring a spatially and timely regulated process maximizing virus yield.
As in parvoviruses, in Polyomavirus the efficiency of nuclear assembly is regulated by phosphorylation of the capsid subunits. Polyomavirus is highly tumorigenic in mouse, but host range transforming (hr-t) mutants of this virus defective in tumour induction are blocked in virion assembly when infecting non-permissive cells [38], although viral DNA and capsid proteins are synthesized to wild type levels. In purified Polyomavirus particles the several VP1 species identified by 2-D electrophoresis are generated by acetylation and phosphorylation of threonine and serine residues of the initial translation product. The hr-t mutants failed to assemble the complete (240S) viral particle, correlating with the lack of acidic forms of VP1 [39]. These acidic forms resulted from phosphorylation of threonine residues, and at least one of the phosphotreonines was shown to be essential for the encapsidation of the viral minichromosome [45]. In SV40, phosphate groups are added in natural infections on serine and threonine residues flanking the NLS of the VP proteins. Phosphorylation was shown to act on the activity of these NLS indirectly regulating the nuclear accumulation of VPs and virus assembly.
An example of a factor encoded by the viral genome favouring assembly was identified in studies with the parvovirus AAV. Capsid assembly in AAV begins in the nucleolus, and spreads throughout the nucleus at later stages of infection. In addition to the three capsid proteins, VP1, VP2 and VP3, the cap gene also encodes, by using an alternative open reading frame, the assembly-activating protein (AAP) that is essential for capsid assembly [31]. The AAP factor targets newly synthesized capsid proteins to the nucleolus, and becomes stabilized upon co-expression of the capsid protein VP3 of the same virus serotype, or from an AAV serotype of the same assembly group. The assembly-promoting activity of AAP is mediated by interaction between two hydrophobic domains in the N-terminal region of the molecule with the C-terminus of the VP proteins, which forms the capsid protein interface at the 2× symmetry axes [40]. AAP seems to act as a scaffolding factor able to change the conformation of non-assembled VP molecules.
5 Genome Encapsidation and Virus Maturation
5.1 Poliovirus Cytoplasmic Maturation
Pathways of genome packaging adopted by some icosahedral viruses have been, and continue to be a matter of debate, as it is quite difficult in many cases to distinguish between “concerted assembly”, in which the capsid is formed as a result of the ordered association of the protein subunits with the genome, from the “sequential assembly” that occurs when the genome is encapsidated into a preformed capsid, which may require an active stage (see Chap. 12 for detailed descriptions of mechanisms of encapsidation referred mostly to structurally complex viruses). Figure 10.5a illustrates this dilema in Poliovirus. In this system, most evidences support a “concerted” assembly of the 14S pentamer condensing around the RNA genome. Alternatively, an empty procapsid containing 60 copies of the VP0-VP3-VP1 structural unit could be transiently required at low concentrations for packaging. The apparent equilibrium between pentamers and an empty capsid of low stability makes it difficult to demonstrate whether the RNA is encapsidated by association with the pentamers or becomes inserted into the capsid [11].
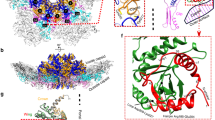
Genome pakaging and virus maturation. (a) Late stages of Poliovirus assembly and virion maturation cleavage [30]. (b) Organization of the Polyomavirus factory. Electron microscopy of plastic-embedded thin sections of the nucleus of mouse fibroblasts showing tubular structures adjacent to a virus cluster (black arrowhead, “full” tubular structure; white arrowhead, full virion; black arrow, “empty” tubular structure; white arrow, empty virion). (c) Spherical virions and tubular structures seen in the nucleus of Polyomavirus-infected cells showing a lighter and well-arranged density at their periphery (white arrowhead) corresponding to capsid protein density, and a dense core (black arrowhead) suggesting DNA. (b, c: Adapted from [41]). (d) Integrative model of major stages in parvovirus assembly and maturation: I. VP capsid subunits assemble into trimers in the cytoplasm (top image). II. Inmature empty capsids (60S) form in the nucleus (top center). III. Genome packaging reaction (bottom center). IV. Mature viral particle (110S) (bottom image)
Whichever encapsidation pathway is dominant, the maturing Poliovirus virion must undergo a number of modifications prior to becoming infectious. Maturation involves lipid modification and proteolytic cleavage at a specific site, making assembly an irreversible process. Along the final stages of assembly, a molecule of the fatty acid myristate is added post-translationally to the N-terminus of each VP4 subunit. This lipid mediates the interaction of the β-sheet formed by VP3 N-termini with a second β-sheet structure containing strands contributed by both VP4 and VP1 molecules. This feature of the capsid does not form until final stages of the maturation, when proteolytic processing liberates VP2 and VP4 from their precursor VP0, and this reaction is associated with a significant increase in the stability of the viral particle, priming it for entry into a new host cell.
5.2 Polymorphic Nuclear Maturation of Polyomaviruses
To form the SV40 and Polyomavirus virions, the VP1-pentamers become associated with single copies of a minor capsid protein (either VP2 or VP3) and, once imported into the nucleus, interact with the replicating DNA. Although the virus structure is known to atomic resolution, how the virions mature in the nucleus during productive infections is only partly understood. In a stepwise model, the capsid proteins would be sequentially added to and arranged on the viral DNA, resulting in its condensation and packaging to form the virion. The process implies multiple molecular recognition events of the viral genome by the CP subunits, in which DNA sequences near the viral origin of replication and the T-antigen (major replicating non-structural Polyomavirus protein) play important functions. Several DNA-binding domains (DBD) localized in all three VP proteins contribute to the packaging process. Major DBD of VP2 and VP3 were found in their C-terminal segments, whereas the DBD of VP1 was localized overlapping with the N-terminal bipartite NLS [42]. This VP1 N-terminal sequence of 15 amino acids is not visible in the virion crystal due to structural disorder, but it likely extends into the virion core to interact with the viral DNA.
The dynamics of maturation of the Polyomavirus and SV40 virions is not precisely known yet, but most evidences suggest a polymerization of the CP subunits onto the viral genome, acting as a scaffold. Although the intermediates are not well defined, the final nuclear assembly process seems to proceed through large, polymorphic structures that may serve as virus factories (Fig. 10.5b, c) (see Chap. 14 for a description of virus factories). Electron microscopy reveals tubular structures in the nucleus adjacent to clusters of virions. They share an organization consisting of a protein shell surrounding an electron dense DNA core, suggesting that they are the main factories able to yield mature virions budding from their ends, although the resolution mechanism is unknown.
5.3 Viral Genome Encapsidation in a Pre-formed Parvovirus Capsid
The final stages of parvovirus virions assembly and maturation are a subject of current active research, but in the best known cases it proceeds through the formation of large amounts of empty capsids in the nucleus, which serve as preformed substrate for active genome encapsidation. In MVM for example, empty capsids are first detected accumulated in the nucleus of synchronized cells preceding the accumulation of DNA-filled virions. A consensus model for parvovirus maturation, based on the available current data obtained in the AAV and MVM systems [43, 44] is outlined in Fig. 10.5d, although many details of the ssDNA packaging mechanism are still poorly understood. The reaction proceeds through a “packaging stage” in which the major non-structural proteins, NS1 in the Parvovirus genus and Rep 78/52 in AAV, mediate the association of the empty capsid with DNA replicative intermediates. These multifunctional proteins harbor helicase, DNA binding, and endonuclease activities that are critical for genome replication, and form oligomers in vitro in the presence of ATP. A Rep/NS1 hexameric packaging motor is probably formed at the five-fold symmetry axes, injecting the ssDNA genome through the channel of the capsid, which acts as the portal for genome encapsidation. For the packaging reaction the helicase activity of the non-structural proteins is essential. Genome encapsidation operates in the 3′ to 5′direction, mediated by specific contacts with the resolving hairpins and ATP hydrolysis. Encapsidation is coupled to DNA replication, ending as the capsid is filled with the complete ssDNA genome, which establishes regular contacts with some internal capsid residues and become icosahedrally ordered along certain sequences. The Rep/NS1 subunits may remain bound to the genome outside of the viral particle. The reader is referred to Chap. 12 for comparisons with equivalent mechanisms of dsDNA and dsRNA packaging into preformed bacteriophage capsids.
6 Perspectives and Conclusions
The assembly of simple icosahedral viruses proceeds through elegant genetically encoded and virus-specific pathways, which direct the self-association of the synthesized asymmetric protein subunits into a symmetric viral capsid shell. The general mechanism of capsid assembly involves first the oligomerization of CP subunits into early stable intermediates, usually of one major type per virus, which constitute the building blocks (CBBs). Capsid assembly then proceeds along a pathway regulated by ordered interactions between activated CBBs, most likely through a cascade of second-order reactions (see Chap. 19). The general architecture and the organization of the capsid subunits in the intermediates resembles those in the final capsid. However, the assembly process implies further structural rearrangements within and between the capsid subunits, including the establishment of multiple inter-protein non-covalent interactions, protein cleavages and covalent bonds (in some cases), and intracellular traffic of the assembly intermediates, prior to their final condensation into a closed container. Assembly is a dynamic process, implying that many of these interactions among subunits may have a transient morphogenetic role, and will not be preserved in the assembled virion. Functional capsid assembly cannot be understood without concomitant or later packaging of the genome, a complex process that viruses solve through molecular recognition patterns between the replicating genomes and either the assembly intermediates or a preformed empty capsid (see Chap. 12). During the final stages of morphogenesis of many viruses, cleavage of the viral particle leads to a mature infectious virion (see Chap. 13 for a description of maturation strategies in different viruses, including structurally complex bacteriophages). Virion morphogenesis in the cell, even for simple viruses, is frequently mediated by additional viral and or cellular factors, and may involve specific cellular factories (see Chap. 14). The efficiency of assembly critically determines some important features of viral fitness, such as the virus yield per host cell, or the acquisition of enough stability to prevail in natural enviroments.
The intensive research on small icosahedral viruses over the past decades have provided detailed information on the atomic structure of viral particles, and on the overall organization and mechanisms of assembly in vitro and in the host cells. Despite these advances, our knowledge on some crucial steps of the assembly of even structurally simple viruses is still poor or unclear in many respects, as exemplified by the following unsolved problems: the detailed structure of assembly intermediates in picornaviruses and parvoviruses; the cellular factors and transporters regulating VP traffic in the cell; the enzimology of genome encapsidation in parvoviruses; the processes of protein-protein and protein-nucleic acid recognition leading to the maturation of Poliovirus and Polyomavirus; or the resolution of the mature virions from the Polyomavirus factories. Ongoing research on these and other subjects related with morphogenesis promises exciting insights into essential aspects of the structural biology of simple icosahedral viruses in the near future.
Abbreviations
- AAP:
-
assembly-activating protein
- AAV:
-
adeno-associated virus
- CBB:
-
capsid building block
- CP:
-
capsid protein
- CPV:
-
canine parvovirus
- DBD:
-
DNA-binding domains
- H1-PV:
-
parvovirus H1
- hr-t:
-
host range-transforming
- FPV:
-
feline parvovirus
- MEV:
-
mink enteritis virus
- MVM:
-
minute virus of mice
- NLM:
-
nuclear localization motif
- NLS:
-
nuclear localization sequence
- NPC:
-
nuclear pore complex
- PPV:
-
porcine parvovirus
- SV40:
-
simian virus 40
- VLP:
-
virus-like particle
- VP:
-
viral protein
- 5x:
-
five-fold axis
- 3x:
-
three-fold axis
- 2x:
-
two-fold axis.
References and Further Reading
Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG (1998) Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369–1381
Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M (2005) Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol 79:10931–10943
Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Ruecker RR (1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 307:145–153
Hogle JM, Chow M, Filman DJ (1985) Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358–1365
Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC (1991) Structure of simian virus 40 at 3.8-A resolution. Nature 28:278–284
Natarajan P, Lander GC, Shepherd CM, Reddy VS, Brooks CL III, Johnson JE (2005) Exploring icosahedral virus structures with VIPER. Nat Rev Microbiol 3:809–817
Tsao J, Chapman MS, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith TJ, Rossmann MG, Compans RW, Parrish CR (1991) The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456–1464
Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS (2002) The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A 99:10405–10410
Gurda BL, Parent KN, Bladek H, Sinkovits RS, DiMattia MA, Rence C, Castro A, McKenna R, Olson N, Brown K, Baker TS, Agbandje-McKenna M (2010) Human bocavirus capsid structure: insights into the structural repertoire of the parvoviridae. J Virol 84:5880–5889
Kaufmann B, Simpson AA, Rossmann MG (2004) The structure of human parvovirus B19. Proc Natl Acad Sci U S A 101:11628–11633
Racaniello VR (2001) Picornaviridae: the viruses and their replication. In: Knipe D, Howley P (eds) Fields virology. Lippincot Williams & Sons, New York, pp 685–722
Tullis GE, Lisa RB, Pintel DJ (1993) The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol 67:131–141
Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P (2001) A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell 1:291–302
Lombardo E, Ramírez JC, García J, Almendral JM (2002) Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J Virol 76:7049–7059
Lombardo E, Ramirez JC, Agbandje-McKenna M, Almendral JM (2000) A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J Virol 74:3804–3814
Riolobos L, Reguera J, Mateu MG, Almendral JM (2006) Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J Mol Biol 357:1026–1038
Chen XS, Stehle T, Harrison SC (1998) Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J 17:3233–3240
Reguera J, Carreira A, Riolobos L, Almendral JM, Mateu MG (2004) Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc Natl Acad Sci USA 101:2724–2729
Li PP, Nakanishi A, Clark SW, Kasamatsu H (2002) Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc Natl Acad Sci USA 99:1353–1358
Raices M, D’Angelo MA (2012) Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 13:687–699
Robbins J, Dilworth SM, Laskey RA, Dingwall C (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623
Mattaj IW, Englmeier L (1998) Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67:265–306
Chang D, Haynes JI, Brady JN, Cosigli RA (1992) Identification of a nuclear localization sequence in the polyomavirus capsid protein VP2. Virology 191:978–983
Moreland RB, Garcea RL (1991) Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology 185:513–518
Ishii N, Minami N, Chen EY, Medina AL, Chico MM, Kasamatsu H (1996) Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J Virol 70:1317–1322
Ishii N, Nakanishi A, Yamada M, Macalalad MH, Kasamatsu H (1994) Functional complementation of nuclear targeting-detective mutants of simian virus 40 structural proteins. J Virol 68:8209–8216
Vihinen-Ranta M, Wang D, Weichert WS, Parrish CR (2002) The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J Virol 76:1884–1891
Grieger JC, Snowdy S, Samulski RJ (2006) Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J Virol 80:5199–5210
Johnson JE (1996) Functional implications of protein-protein interactions in incosahedral viruses. Proc Natl Acad Sci USA 93:27–33
Hogle JM (2002) Poliovirus cell entry: common structural themes in viral cell entry. Annu Rev Microbiol 56:677–702
Sonntag F, Schmidt K, Kleinschmidt JA (2010) A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 107:10220–10225
Riolobos L, Valle N, Hernando E, Maroto B, Kann M, Almendral JM (2010) Viral oncolysis that targets Raf-1 signaling control of nuclear transport. J Virol 84:2090–2099
Caspar DLD, Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol XXVII:1–24
Salunke DM, Caspar DL, Garcea RL (1986) Self-assembly of purified polyomavirus capsid protein VP1. Cell 46:895–904
Hernando E, Llamas-Saiz AL, Foces-Foces C, McKenna R, Portman L, Agbandje-McKenna M, Almendral JM (2000) Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 267:299–309
Yuan W, Parrish CR (2001) Canine parvovirus capsid assembly and differences in mammalian and insect cells. Virology 279:546–557
Maroto B, Ramírez JC, Almendral JM (2000) Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J Virol 74:10892–10902
Garcea R, Benjamin T-L (1983) Host range of trasnforming gene of polyoma virus plays a role in virus assembly. Proc Natl Acad Sci USA 80:3613–3617
Garcea RL, Ballmer-Hofer K, Benjamin TL (1985) Virion assembly defect of polyomavirus hr-t mutants: underphosphorylation of major capsid protein VP1 before viral DNA encapsidation. J Virol 54:311–316
Naumer M, Sonntag F, Schmidt K, Nieto K, Panke C, Davey NE, Popa-Wagner R, Kleinschmid JA (2012) Properties of the AAV assembly activating protein AAP. J Virol 86:13038–13048
Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL (2012) Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog 8:1–15
Li PP, Nakanishi A, Shum D, Sun PC, Salazar AM, Fernandez CF, Chan S-W, Kasamatsu H (2001) Simian virus 40 VP1 DNA-binding domain is functionally separable form the overlapping nuclear localization signal and is required for effective virion formation and full viability. J Virol 75:7321–7329
King JA, Dubielzig R, Grimm SW, Kleinschmidt JA (2001) DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20:3282–3291
Plevka P, Hafenstein S, Li L, D’Abramo A, Cotmore SF, Rossmann MG, Tattersall P (2011) Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis. J Virol 85:4822–4827
Li M, Garcea RL (1994) Identification of threonine phosphorylation sites on the polymavirus major capsid protein VP-1: relationship to the activity of middle T antigen. J Virol 68:320–327
Further Reading
Carrasco L, Almendral JM (eds) (2006) Virus patógenos. Hélice and Fundación BBVA, Madrid
Flint SJ, Enquist LW, Racaniello VR, Skalka AM (2009) Principles of virology, 3rd edn. ASM Press, Washington, DC
Kerr JK, Cotmore SF, Bloom ME, Linden RM, Parrish CR (eds) (2006) Parvoviruses. Hodder Arnold, London
Harrison SC (2006) Principles of virus structure. In: Knipe DM, Howley PM (eds in chief) Fields virology, 5th edn. Lipincott Williams and Wilkins, Philadelphia, p 59–98
Reddy VS, Johnson JE (2005) Structure-derived insights into virus assembly. Adv Virus Res 64:45–68
Zlotnick A, Fane BA (1997) Mechanisms of icosahedral virus assembly. In: Agbandje-McKenna M, McKenna R (eds) Structural biology. RSC Publishing, Cambridge, pp 180–202
Acknowledgements
I gratefully acknowledge the collaborative work of Mavis Agbandje-Mckenna (University of Florida) on solving parvovirus MVMp capsid structure to atomic resolution, of Mauricio G. Mateu (CBMSO, Universidad Autónoma de Madrid) on MVM assembly, and of Thomas L. Benjamin (Harvard Medical School, Massachussets) on aspects related to Polyomavirus assembly. The many inspiring conversations along the years with regular participants to the “Virus Assembly” FASEB meeting (Vermont Academy, USA), in particular with Peter Tattersall (Yale University, Connecticut), are also deeply acknowledged. This review stands from the experience gained by the author on Parvovirus assembly thanks to the enthusiastic work of Eva Hernando, Eleuterio Lombardo, Beatriz Maroto, Juan C. Ramírez, Laura Riolobos, and Noelia Valle. Support on figures design by Jon Gil-Ranedo and Jorge Sánchez is also acknowledged. Research in JM Almendral’s laboratory is currently supported by grant SAF 2011-29403 from the Spanish Ministerio de Ciencia e Innovación.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Almendral, J.M. (2013). Assembly of Simple Icosahedral Viruses. In: Mateu, M. (eds) Structure and Physics of Viruses. Subcellular Biochemistry, vol 68. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-6552-8_10
Download citation
DOI: https://doi.org/10.1007/978-94-007-6552-8_10
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-6551-1
Online ISBN: 978-94-007-6552-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)