Abstract
A color image-based segmentation method for segmenting skin lesions is proposed in this paper. This proposed methodology mainly includes two parts: First, a combination of scale-invariant and semantic mathematic model is utilized to classify different pixels. Second, a strategy based on skeleton corner point’s extraction is proposed in order to extract the seed points for the skin lesion image. By this method, the skin slices are processed in series automatically. As a result, the lesions present in the skin can be segmented clearly and accurately. The proposed algorithm is trained and tested for 360 skin slices in order to evaluate the accuracy of segmentation. Overall accuracy of the proposed method is compared with existing conventional techniques. An average missing pixel rate of 3.02 % and faulting pixel rate or 2.36 % has been obtained for segmenting the skin lesion images.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Melanoma is the most deadly form of skin cancer, with an estimated 76,690 people being diagnosed with melanoma and 9480 people dying of melanoma in the United States in 2013 [1]. In the United States, the lifetime risk of getting melanoma is 1 in 49 [1]. Melanoma accounts for approximately 75 % of deaths associated with skin cancer [2]. It is a malignant tumor of the melanocytes and usually occurs on the trunk or lower extremities [3]. Recent trends found that incidence rates for non-Hispanic white males and females were increasing at an annual rate of approximately 3 % [4]. If melanoma is detected early, while it is classified at Stage I, the 5-year survival rate is 96 % [5]; however, the 5-year survival rate decreases to 5 % if the melanoma is in Stage IV [5]. With the rising incidence rates in certain subsets of the general population, it is beneficial to screen for melanoma in order to detect it early. To reduce costs of screening melanoma in the general population, development of automated melanoma screening algorithms have been proposed.
Human body visualization research includes cutting the sequences of body slices, developing the computerized methods with the overall goal of reconstructing the 3D virtual models of the human body. With the help of these 3D virtual models, the human organs can be hidden selectively and viewed from any side. The virtual human models can be implemented for risky experiments instead of real human body. In summary, the visible human project (VHP) has a lot of applications and significant value for human society. This ambitious scheme has caused attentions of many countries. By the end of 2012, the United States, South Korea, China, and Japan had all begun their own Visible Human projects. Researchers all over the world are now planning to append physical and chemical attributes to the 3D Visible. If new technology by which the live human body can be scanned to build personalized 3D virtual models is invented, the Virtual Human Project will have a more promising future [1–3].
Image segmentation is a critical stage of the cancer detection. That is because errors at this stage will inevitably lead to later bigger problems. In order to actualize the precise and accurate descriptions of inner structure of the human body, immense volumes of raw data are collected. Therefore, a rapid and accurate method should be utilized to segment the cross-sectional photographs of the Visible Human.
Currently, a wide variety of color image segmentation methods have been proposed. Among them, the k-means method and fuzzy c-means method are accurate and practical [6, 7]. However, considering the large data set of Visible Human, their high computational complexity will lead to considerable time cost. Color image segmentation methods based on region-growing can be implemented easily. The result is also acceptable. However, the seed points should be manually picked. It is hard to imagine picking out seed points for each picture. So does the graph cut method whose manual mode will be tedious for the large-scale image datasets [8, 9]. The color structure code (CSC) segmentation algorithm can tolerate the sensitivity of threshold, but it is easy to be affected by the noise [10–13]. The segmentation algorithm based on fuzzy connectedness can well mark the target region which is not easy to be accurately defined due to the fuzzy edge, and this algorithm is not sensitive to the noise. But there is a lot of iterative calculation algorithm which will seriously reduce the speed of the segmentation [14–17]. The support vector machine (SVM)-based segmentation algorithm can achieve the automatic segmentation in some degree, but it need too much time for acquiring the accurate space information during the training. What is more, the segmentation speed is not fast enough. So it cannot realize the real-time and efficient segmentation [18]. Another kind of segmentation methods is the interactive image segmentation methods which can manually extract the regions of interest. Most importantly, these methods are designed only for a single-layer color image, and they cannot be directly utilized as the serialized slice image segmentation method for the immense datasets of the Visible Human. The cross-sectional photographs of Visible Human are serialized, coherent, and slowly changing. Taking these features into account, an automatic-serialized color image segmentation method is proposed in this paper. The purpose of sequential and automatic segmentation in 3D color image spaces with lower time cost and higher accuracy can be achieved.
The rest of the paper is organized as follows: Sect. 2 presents the computation method of color similarities between two pixels and the automatic seed points extraction method for serialized slices. Section 3 presents experimental segmentation results for the primary organs of Visible Human. Concluding remarks are given in Sect. 4.
2 Materials and Methods
2.1 Selection of Color Feature
There are many color similarity measure methods. For example, Minkowski distance measure, Canberra distance measure, and cylindrical distance metric [19]. They all regard the color as feature and try to group the pixels with a similar color in the image. However, too much time is wasted on the calculations during the color space transformation to decouple brightness and hue. These calculations will be a bad influence when I am dealing with thousands of images in the Virtual Human Project.
The most widely used color space is RGB model. RGB is an abbreviation of red-green-blue. The three fundamental components of RGB color space are all influenced by brightness, which make it not perfectly good at color discrimination. Besides RGB color space, many other color spaces are designed based on RGB color space. By some calculations, the RGB color space can be turned into other spaces, such as HSI color space [20]. Some of these spaces are specially designed for color image processing. They mimic human visual perception in a more accurate way. HSI space is a typical representative of them. H means hue (the difference of color). S means saturation (the gradation of color). I represents intensity (the degree of light and shade of color). Human is more sensitive to brightness rather than hue. HSI space shows the brightness and hue explicitly and separately. So, HSI space is more common-used on color image processing. Although HSI space and RGB space are two expressions of the same thing, the transformation between them is very complicated. Therefore, it will surely improve the speed of the segmentation method if it does not need the color space transformation procedure (RGB → HIS) but can accord with the color vision perception characteristics of human eyes (HSI space).
2.2 Computation of Color Similarities Between Two Pixels
In this paper, a color similarity based segmentation algorithm which was proposed by Shikai Wang was utilized. This method is accurate and robust with low computational complexity [21]. A scale-invariant and semantic mathematic model is utilized to classify different pixels. This method can compute the color similarity between two RGB pixels of a color image in real time. Color space transformation is avoided in this method, so the computation time will be further reduced. However, this similarity reflects the characteristics of the HSI color space, which is what the most notable feature of this method.
Given two pixels f and g in the RGB color space: (R0, G0, B0), (R1, G1, B1).
Let
Then the proposed algorithm is computed as follows:
The proposed model considers both brightness and hue and does not perform color space transformation. The proposed method, which represents the level of similarity, ranges from 0 to 1. When the scale-invariant and semantic mathematic model is equal to 1, it means two color pixels are same. When the proposed model is equal to 0, it means two color pixels are absolutely different. After the seed point is picked out, (R0, G0, B0) is obtained. So a threshold should be chosen to help us determine whether the ambient pixels in the slice are similar to the seed point or not. Experiment results indicate that this threshold can be found by several tests. Once confirmed, the threshold will be valid and invariant in the whole segmentation process of an image sequence.
2.3 Automatic Seed Points Extraction for Serialized Slices
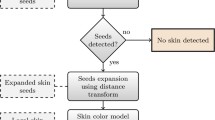
In the traditional image segmentation algorithm based on region-growing, the seed points must be clicked manually. To pick out seed points in each slice image is exhausting. In this paper, the research goal is achieving an automatic and serialized segmentation of skin lesions in the image sequence. Automatic image segmentation requires less manual intervention to some extent. The automation degree of the segmentation algorithm is limited by setting the seed points. In other words, before the next image is to be segmented, the corresponding seed points should be provided automatically rather than manually.
The next image to be segmented must be different from the current image. However, the changes are small and predictable. For instance, current target regions are merged into one region or split into more regions. In these cases, more seed points should be provided in order to make sure at least one seed point in each separate region. Because of the characteristics of the visible skin dataset, the contours of the target regions in adjacent images are similar. So after extracting the target regions of the current image, the skeletons of target regions can be computed and stored. And then, an effective method to further extract the feature points of the skeletons to generate the seed points is needed. As a typical feature point extraction method, Harris operator has the advantage of simple calculation, uniform distribution, and stable performance. Therefore, the Harris corners of skeleton will be utilized as the seed points of the next image. The detailed process is as follows:
The current image which has already been segmented is regarded as a binary image. The pixel belongs to the target region is stored as 1, while the others are stored as 0. Zhang-Suen Thinning algorithm is utilized to extract the skeleton of the segmented region [22]. Because the skeleton can reflect topological features and is close to the center of the region, the seed points can be obtained from the skeleton pixels instead of checking each pixel. This new strategy is better than my previous method [23].
Harris corners are the points where the curvature has a sudden change [24]. The edges of the segmented regions are frequently changing. However, the Harris corners of the skeleton are not. These corner points are characteristic of the most unchangeable local features owned by the segmented regions. In other words, the extracted corner points will land in the target regions of next slice for certain. So Harris corners can be seed points of the adjacent image. As a consequence, the automatic seed points extraction method proposed in this paper is possible.
Algorithm for the proposed serialized segmentation method is as follows:
-
(1)
Set the color similarity threshold T;
-
(2)
Select a number of pixels manually in the target region, and set each of them as an initial seed pixel (x 0, y 0);
-
(3)
Set pixel (x 0, y 0) as the center, and obtain the eight neighborhood pixels (x i , y i ) (i = 0, 1, …., 10);
-
(4)
Compute the color similarity between (x i , y i ) and (x 0, y 0) by the combination of scale-invariant and semantic mathematic model;
-
(5)
If the color similarity between pixel (x i , y i ) and pixel (x 0, y 0) is larger than threshold T, pixel (x i , y i ) and pixel (x 0, y 0) can be grouped into a same region. Pixel (x i , y i ) will be pushed into a stack;
-
(6)
If the stack is not empty, iterate each pixel and regard one as the pixel (x 0, y 0). Return to step (3). When the stack is empty, the growing process for the target region segmentation of the current color slice ends. If current slice is the last image, the whole segmentation process ends;
-
(7)
In the obtained target regions, compute the seed pixels (number ≥ 1) for the next slice by utilizing the skeleton corner point method;
-
(8)
For the next slice, each seed pixel is regarded as (x 0, y 0). Then turn to step (3).
3 Results and Discussion
Accuracy of segmentation has been calculated in order to determine the performance of the proposed segmentation algorithm. In this section, the results of this segmentation methodology in terms of quality evaluation through image display and experimental analysis are presented. The system is validated through accuracy metrics for finding satisfactory segmentation results. The performances of various segmentation techniques have been analyzed and compared with the results obtained for segmenting skin lesion images. The segmented outputs of the proposed model are shown in Table 1.
In this paper, methods of calculating the missing pixel rate P m and faulting pixel rate P f are used to evaluate the segmentation quality of different algorithms.
where G is the pixel set of reference segmentation result (Ground truth) and S is the set of pixels present in the original image. The experimental results are shown in Table 2. From the tables, it can be seen that the missing pixel rate of the color structure code method is most serious and the faulting pixel rates of the first three segmentation methods are similar. The missing pixel rate and the faulting pixel rate of this method are all lower than other three methods.
In terms of time, the proposed method shows satisfactory results. The experiments were performed on an ordinary dual-core personal computer. The time cost of different methods is shown in Table 3. From the table, it can be observed that the overall speed of this proposed method is faster than other methods.
4 Conclusions
In this paper, a novel lesion segmentation algorithm using the concept of combination of scale-invariant and semantic mathematic model is proposed. A scale-invariant and semantic mathematic model is introduced based on the seeded points of normal skin and lesion textures. The images are divided into numerous smaller regions and each of those regions are classified as lesion or skin. The entire proposed framework is tested by using various skin lesion images as the input to the proposed segmentation algorithm. The proposed algorithm is compared with other conventional segmentation algorithms, including three algorithms designed for color images. The proposed framework produces the highest segmentation accuracy using manually segmented images as ground truth. From the experimental results, it can be observed that the proposed method is very effective for the segmentation of skin lesions with relatively consistent color.
References
Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (2013) SEER cancer statistics review, 1975–2010. National cancer institute, Bethesda, MD, USA, technology reports
Jerants AF, Johnson JT, Sheridan CD, Caffrey TJ (2000) Early detection and treatment of skin cancer. Am Family Phys 62(2):1–6
Public Health Agency of Canada (2013) Melanoma skin cancer. http://www.phac-aspc.gc.ca/cd-mc/cancer/melanomaskincancer-cancerpeaumelanome-eng.php
Jemal A, Saraiya M, Patel P, Cherala SS, Barnholtz-Sloan J, Kim J, Wiggins CL, Wingo PA (2011) Recent trends in cutaneous melanoma incidence and death rates in the united states, 1992–2006. J Am Acad Dermatol 65(5):S17.e1–S17.e11
Freedberg KA, Geller AC, Miller DR, Lew RA, Koh HK (1999) Screening for malignant melanoma: a cost-effectiveness analysis. J Am Acad Dermatol 41(5, pt. 1):738–745
Lim YW, Lee SU (1990) On the color image segmentation algorithm based on the thresholding and the fuzzy c-means techniques. Pattern Recogn 23(9):935–952
Siang Tan K, Mat Isa NA (2011) Color image segmentation using histogram thresholding–fuzzy C-means hybrid approach. Pattern Recogn 44(1):1–15
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905
Felzenszwalb PF, Huttenlocher DP (2004) Efficient graph-based image segmentation. Int J Comput Vision 59(2):167–181
Priese L, Sturm P. Introduction to the color structure code and its implementation [[EB/OL]. doi:10.1.1.93.3090. http://citeseerx.ist.psu.edu/viewdoc/summary?
Lia H, Gua H, Hana Y, Yang J (2010) Object-oriented classification of high-resolution remote sensing imagery based on an improved colour structure code and a support vector machine. Int J Remote Sens 31(6):1453–1470
Priese L, Rehrmann V, Schian R, Lakmann R, Bilderkennen L (1993) Traffic sign recognition based on color image evaluation [C]. In: Proceedings IEEE intelligent vehicles symposium ‘93, pp 95–100
von Wangenheim A, Bertoldi RF, Abdala DD, Richter MM, Priese L, Schmitt F (2008) Fast two-step segmentation of natural color scenes using hierarchical region-growing and a color-gradient network. J Braz Comput Soc 14(4):29–40
Udupa JK, Samarasekera S (1996) Fuzzy connectedness and object definition: theory, algorithms, and applications in image segmentation. Graph Models Image Process 58(3):246–261
Udupa JK, Saha PK (2003) Fuzzy connectedness and image segmentation. Proc IEEE 91(10):1649–1669
Saha PK, Udupa JK, Odhner D (2000) Scale-based fuzzy connected image segmentation: theory, algorithms, and validation. Comput Vis Image Underst 77:145–174
Yu Z, Bajaj CL (2002) Normalized gradient vector diffusion and image segmentation [C]. In: Proceedings of the 7th European conference on computer vision (ECCV 2002), pp 517–530
Cyganek B (2008) Color image segmentation with support vector machines: applications to road signs detection. Int J Neural Syst 18(4):339–345
Ikonomakis N (2000) Color image segmentation for multimedia applications. J Intell Robot Syst 28:5–20
Tao W, Jin H, Zhang Y (2007) Color image segmentation based on mean shift and normalized cuts. IEEE Trans Syst Man Cybern 37(5):1382–1389
Wang S (2009) Color image segmentation based on color similarity [C]. In: IEEE international conference on computational intelligence and software engineering, pp 1–4
Zhang TY, Suen CY (1984) A fast parallel algorithm for thinning digital patterns. Commun ACM 27(3):236–239
Liu B, Li H, Xianyong Jia X, Zhao ZL, Zhao Q, Zhang H (2014) A simple method of rapid and automatic color image segmentation for serialized Visible Human slices. Comput Electr Eng 40(3):870–883
Harris C, Stephens MJ (1988) A combined corner and edge detector [C]. Alvey vision conference, pp 147–152
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer India
About this paper
Cite this paper
Faizal Khan, Z. (2016). Automated Segmentation of Skin Lesions Using Seed Points and Scale-Invariant Semantic Mathematic Model. In: Suresh, L., Panigrahi, B. (eds) Proceedings of the International Conference on Soft Computing Systems. Advances in Intelligent Systems and Computing, vol 397. Springer, New Delhi. https://doi.org/10.1007/978-81-322-2671-0_21
Download citation
DOI: https://doi.org/10.1007/978-81-322-2671-0_21
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-2669-7
Online ISBN: 978-81-322-2671-0
eBook Packages: EngineeringEngineering (R0)