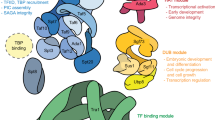
Abstract
Sin3, a global transcription regulator, acts as a molecular scaffold for complex assembly and also as a molecular adapter bridging HDAC (histone deacetylase complex) with an astonishingly large and diverse group of DNA-binding transcription factors and chromatin-binding proteins. Most of the protein-protein interactions are achieved through six conserved domains of Sin3 that include four paired amphipathic helices (PAH 1–4), one histone deacetylase interaction domain (HID) and one highly conserved region (HCR). These PAH domains, though, are structurally homologous to one another having different amino acid sequences which result in differential protein-protein interactions. The PAH domains also show conformational flexibility under different physiological conditions such as pH and temperature and conformational heterogeneity upon interaction with different proteins. Interaction with large groups of transcription factors and co-repressor complexes is also possible due to the fact that Sin3 shows structural modification such as phosphorylation, myristoylation, ubiquitination, and SUMOylation as well as structural allostery upon interaction with proteins. Till now, only N-terminal region of Sin3 has been characterized, which makes it more important to characterize C-terminal region of Sin3 so as to find and understand its interacting partners and their role in Sin3-mediated gene regulation. Future studies should also be directed towards understanding the regulation of Sin3 protein under different physiological conditions and modulation of its biological activity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Sin3 complex
- Protein-protein interaction
- Allosteric regulation
- Post-translation modification
- Co-repressor
- Ubiquitination
1 Introduction
Sin3, a global transcription regulator, helps to regulate many biological functions including nucleosome remodelling, DNA methylation, cell proliferation and apoptosis. Sin3 does not bind to DNA but is a scaffold protein that helps the transcription of various genes by interacting with different transcription factors, forming Sin3 complex (Grzenda et al. 2009). The core complex of Sin3 consists of eight components in humans: Sin3, SAP18, SAP30, HDAC1, HDAC2, RbAp46, RbAp48 and SDS3 (McDonel et al. 2009). Presence of different sub-complexes has also been reported in different organisms; these sub-complexes contain additional components besides the components present in a core complex. Sin3 interacts with numerous transcriptional factors through its six distinct yet conserved domains that include the four imperfect repeats of paired amphipathic helices (PAH 1–4), histone deacetylase interaction domain (HID) and highly conserved region (HCR) (Grzenda et al. 2009). These PAH domains, though distinct in sequence, are structurally homologous to one another and recognize unique sets of proteins. NMR and X-ray crystallographic studies of individual PAH domains as well as PAH domains complexed with DNA-binding domains and co-repressor complex revealed that each PAH domain folds in a distinct way to interact with the proteins. There are a number of other factors which are responsible for making PAH domains an ideal region of Sin3 for interacting with large numbers of transcription factors. Presence of different splice variants and isoforms of Sin3 in humans and various other organisms further increases the flexibility of Sin3 protein to recruit a large set of proteins (Ayer et al. 1995). Interaction of Sin3 with the proteins indirectly via co-repressor complex adds another level of complexity to Sin3 functions. Post-translation modifications of Sin3 such as phosphorylation, myristoylation, ubiquitination and SUMOylation help in maintaining protein-protein stability and proper regulation of genes. Structural variation in domains of Sin3 and allosteric regulation of Sin3 further increase the flexibility and conformational heterogeneity of Sin3 protein, making it an ideal regulator of gene expression.
2 Sin3 Complex
Sin3 is a large protein which is thought to function as a molecular scaffold due to presence of several protein-protein interaction domains. It helps in the assembly of co-repressor complex and as a molecular adapter to bridge components of the complex with DNA-bound repressors (Sheeba et al. 2007; Silverstein and Ekwall 2005). Chromatographic data of components of Sin3 complex suggested that there are different Sin3 complexes and these are variable in their structural components. Certain proteins are found to be conserved between them and thus referred to as “core complex” while some have additional proteins. The mammalian Sin3 complex analogous to Sin3/RPD3 complex of yeast comprises of at least seven polypeptides besides Sin3, including HDACs HDAC1 and HDAC2, two histone-binding proteins RbAp46 and RbAp48 (Rb-associated polypeptides) and three Sin3-associated polypeptides SAP18, SAP30 and SAP45/Sds3. Human family with sequence similarity 60 member A (FAM60A) protein, a new member of Sin3 core complex, has been discovered recently (Smith et al. 2012). Other protein components might vary between different sub-complexes. p33ING1b is one such example as it is thought to associate with only certain species of Sin3/HDAC complex (Campos et al. 2004). Similarly, Sin3A, an isoform of Sin3 present in mammals, contains five extra polypeptides besides components of core complex which includes three SAP polypeptides SAP25, SAP130, SAP180 and one RBP1 (Rb-binding protein 1) protein (Xie et al. 2011). In the years following the elucidation of the core complex, a number of other associated proteins were uncovered, including BRMS1, CpG methylated binding protein (MeCP2) and ING1/2 (Grzenda et al. 2009; Nan et al. 1998; Zhang et al. 2014). The ability to support different compositions within the complex may be yet another way to expand the functional flexibility of Sin3 complex.
In the core complex, Sin3 is the platform for protein interaction, most importantly the enzymatic activity of HDAC1 and HDAC2. RbAp46 and RbAp48 are able to bind histone H4 and H2A and might thus function in stabilizing the interaction between co-repressor complex and chromatin. HDAC1 and HDAC2 bind to conserved HID region on Sin3 and provide the enzymatic activity to the complex. RbAp46 and RbAp48 that bind to nucleosomal histones are involved in chromatin modelling such as histone acetylation, nucleosome remodelling and nucleosomal assembly (Spencer and Davie 1999). Sin3-associated proteins (SAP) provide structural support and stabilize the complex. Brief functions of different components of Sin3 complex present in different organisms are listed in Table 1.1.
3 Structural Overview
The basic structure of Sin3 is evolutionary conserved from yeast to mammals. It contains four paired α-helices known as PAH (paired amphipathic helix) domains separated by less conserved amino acids forming the spacer region. There are also two other conserved protein interaction domains, HID (histone interacting domain) located between PAH3 and PAH4 regions and HCR (highly conserved region) situated C-terminally to PAH4 (Fig. 1.1). The PAH domains are meant for protein-protein interaction; HID interacts with HDACs and many other components of Sin3/HDAC complex and HCR, recently identified as another protein interacting domain (Grzenda et al. 2009). The current evidence advocates Sin3 as a modular protein where PAH1-3 are earmarked for interactions with different transcription factors while the HID and PAH4 domains primarily serve scaffolding function by interacting with other components/sub-units of the repressor complex. PAH1-3 domains form pre-folded binding modules on full-length Sin3, like a beads-on-a-string model (Le Guezennec et al. 2006). Unlike the other PAH domains, PAH4 most likely does not fold as a four-helix bundle but instead adapts a distinct fold (van Ingen et al. 2006). Out of the four Sin3 PAH domains, the tertiary structures of only mSin3A and mSin3B PAH1 and PAH2 domains have been determined by NMR and X-ray crystallography (Kumar et al. 2011; Nomura et al. 2005; Sahu et al. 2008). The fundamental structures of both PAH1 and PAH2 are similar; however, the helices formed in PAH1 are shorter than that of PAH2 (Sahu et al. 2008). The Sin3A PAH2 domain homodimerizes and exists in unfolded state, but Sin3B PAH2 domain is monomeric and is fully folded while PAH1 exists in homodimeric form in both isoforms. Thus, PAH2 domain shows conformational heterogeneity in two isoforms of Sin3 (Kumar et al. 2011). Structures of PAH1, PAH2 and PAH3 complex with SID domain of interacting partners have also been determined by NMR and X-ray crystallography (Sahu et al. 2008; Swanson et al. 2004). PAH2 domains of mSin3A and mSin3B hold a wedged four-helix bundle structure associated with the Sin3-interacting domain (SID) of Mad1, a transcription factor involved in cell proliferation and differentiation in mammalian cells. PAH2 domain complex with the Mad1 SID adopts an amphipathic α- helix whereas PAH1 holds a rather globular four-helix bundle structure with a semi-ordered C-terminal tail on interaction with the NRSF/REST repressor domain (Nomura et al. 2005). Solution structure of PAH3 and SAP30 complex showed that PAH3 forms a canonical hydrophobic cleft and a discrete surface formed largely by the PAH3 α2, α3 and α3′ helices. Sin3 interaction domain (SID) of SAP30 binds to PAH3 via a tripartite structural motif, including a C-terminal helix that targets the canonical PAH hydrophobic cleft while two other helices and an N-terminal extension target a discrete surface of the PAH3 domain formed by α helices (α2, α3 and α3′) (Xie et al. 2011).
The yeast Sin3 protein: Contains 1,536 amino acids and has six regions that are highly conserved throughout evolution. The PAH (paired amphipathic helix) domains appear to be protein-interaction domains, separated by spacer. The HID (Histone interacting domain) region between PAH3 and PAH4 interacts with HDACs and many of the other core components of the Sin3/ HDAC complex. The HCR (Highly conserved region) was recently identified as another protein-interaction domain that resides on C-terminal region of Sin3 protein
In spite of substantial structural homology and similarity between PAH1 and PAH2 domains of Sin3 in various organisms, the two domains recognize different sequence motifs, thereby enabling differential target specificity. However, PAH3 domains share relatively low levels of sequence identity with PAH1 and PAH2 domains (25 and 16 %, respectively), yet these domains are structurally homologous to one another (Xie et al. 2011). Recent studies from our lab have shown that the three PAH domains, although showing homology in the tertiary structure, respond differentially to the environmental signals (pH and temperature). We found that PAH2 and PAH3 domains of Sin3B largely exist in unfolded state and are thermodynamically unstable in nuclear pH condition with respect to physiological pH while the structural identity of PAH1 remains unaltered at both the pH values (Tauheed Hasan et al. 2015). Based on these studies, we assume that there exists a flexibility and conformational heterogeneity in structure of Sin3 which provides additional new surfaces for protein-protein interactions.
4 Isoforms of Sin3
The basic protein structure of Sin3 is highly conserved among eukaryotes from yeast to mammals and has a varied number of isoforms (Fig. 1.2). Saccharomyces cerevisiae has only one isoform of Sin3 while Schizosaccharomyces pombe has three isoforms Pst1, Pst2 and Pst3, each encoded by a separate gene. Study on three paralogues of fission yeast (Sch. Pombe) revealed that these isoforms have originated due to gene duplication. Pst1 is most closely related to Sin3 of budding yeast (S. cerevisiae), suggesting that gene duplication occurred early in the evolution process. Pst2 appears to have arisen from Pst1 gene duplication during evolution of fission yeast. Pst3 seems to have arisen from duplication very early in evolution (Benedik et al. 1999). In Drosophila, three isoforms of Sin3 (Sin3 187, Sin3 190 and Sin3 220) are reported, which are produced by alternate splicing, having different C-terminals. Sin3 220 is expressed in proliferating cells whereas Sin3 187 is expressed in differentiated tissues. The adult females and embryos contain Sin3 190. Thus, each isoform expresses in different cells and tissues and regulates different sets of genes and therefore can perform non-overlapping functions. Multiple Sin3 isoforms have also been reported in three species of mosquitoes which are conserved at splice sites and have a similar protein sequence to that of Drosophila Sin3 187 and Sin3 220 (Sharma et al. 2008).
In mammals, two isoforms of Sin3 are present, Sin3A and Sin3B, which are encoded by two separate genes (Ayer et al. 1995). These isoforms are results of gene duplication. The mouse Sin3A is more closely related to human Sin3A than to mouse Sin3B. Mammalian Sin3A and Sin3B proteins are approximately 57 % identical throughout the length of their polypeptide chains with the highest degree of homology localized in the PAH and HID suggesting that the duplication of Sin3 in mammals occurred before speciation (Alland et al. 1997; Silverstein and Ekwall 2005; Yang et al. 2000). The two isoforms also differ in the length of their polypeptide chain. Sin3B has a shorter amino acid tail prior to PAH1 with respect to Sin3A. Both Sin3A and Sin3B are widely expressed, frequently within the same cells and tissues, and interact with common as well as distinct transcriptional repressors and complexes. Splice variants of both the isoforms have also been reported in mice and humans. One example is mouse Sin3BSF (Sin3 B short form), in which the first 327 amino acids are identical to those present in long-form mSin3B (Sin3BLF) but are followed by a unique 19-residue stretch in the C-terminus, resulting in a truncated mSin3B with intact PAH1/2 domains but lacking the remainder of the C-terminus portion, including PAH3 and HID. It is envisaged that the splice variant forms perform differential function in same types of cells and tissues. Sin3BSF provides a more attenuated and reversible type of regulation on the basal transcriptional apparatus, while mSin3BLF acts as a nucleosomal condenser to provide more effective multi-level gene repression (Alland et al. 1997; Nair and Burley 2006). Xenopus Sin3 has one isoform which shows homology to Sin3A, revealing duplication of genes before divergence to vertebrates (Silverstein and Ekwall 2005). Isoform of Sin3 in C. elegans shows a unique structural hierarchy, having only one PAH domain in contrast to four PAH domains of Sin3 in other eukaryotes. This PAH domain shows 50 % sequence similarity with PAH1 domain of mammalian Sin3A. Thus, PAH domain of C. Elegans is structurally and functionally more evolved than Sin3 of higher organisms (Wysocka et al. 2003).
Purification and characterization of the components of the Sin3 complex by our group have shown that these isoforms contain different subsets of protein on their core complex. The different complexes show some common sub-unit whereas other sub-units are specific to each complex (Table 1.2). The mechanism of regulation of genes is also different in these complexes. For example, in yeast (S. cerevisae), two distinct complexes, large Rpd3L and small Rpd3S, are present. Although sub-units such as Sin3, Rpd3 and Ume1 are shared by both the complexes, Dep1, Pho23, Rxt1, Rxt2, Sds3 and Sap30 are specifically present in Rpd3L complex while Rco1 and Eaf3 are unique sub-units of Rpd3S complex (Keogh et al. 2005). The mechanism of repression for each complex is also distinct; the Rpd3L complex represses gene expression by binding to the promoter through DNA-binding factors or other co-repressors. In contrast, the Rpd3S-mediated trans-repression is complex and involves recognition of methylated H3K36 by chromo-domain of Eaf3 and subsequent recruitment of core complex. This in turn brings about deacetylation of nucleosomes of the transcribed genes. Furthermore, during transcription elongation, Rpd3S complex enhances histone deacetylation thereby preventing transcription initiation from the intragenic cryptic promoters (Li et al. 2007). A similar repression mechanism has also been observed in S. pombe, wherein Pst1 and Pst2 form two distinct complexes (Complex I and Complex II). Complex I contains Pst1 and Sds3, and complex II encompasses Pst2, Alp13, Cph1 and Cph2 as distinct subunits whereas Clr6 and Prw1 are common to both the complexes (Nicolas et al. 2007). Likewise in Drosophila, Sin3 187 and Sin3 220 isoforms form two discrete complexes that are localized on distinct regions of polytene chromosome (Spain et al. 2010). Recently, a homologue of Rpd3S complex comprising of Sin3B, HDAC1, Mrg15 and Pf1 was reported in mammals. This complex mediates restoration of repressed chromatin structure at specific actively transcribing loci approximately 1 kb downstream of transcription start site (Jelinic et al. 2011). The above-mentioned examples of unique regulatory functions for each Sin3 isoform justify the concept of gene duplication to achieve complete functional plasticity during evolution.
5 Functional Aspect of Sin3A and Sin3B
Human Sin3A (hSin3A) is more homologous to mouse Sin3A (mSin3A), suggesting that divergence of Sin3A and Sin3B from Sin3 gene occurred before speciation during evolution (Halleck et al. 1995; Silverstein and Ekwall 2005). Interestingly, both Sin3A and Sin3B are expressed ubiquitously and are often present in same types of cells and tissues (Silverstein and Ekwall 2005). Since both Sin3A and Sin3B are evolutionary conserved, at least some of the functional aspects of each isoform must be specific and unique. Differential expression and interaction with distinct protein components would provide extensive flexibility both during embryonic development and normal cellular homeostasis. Studies have indicated that Sin3A is essential in early embryonic development of eukaryotes whereas Sin3B is essential for late embryonic development (Brunmeir et al. 2009; McDonel et al. 2009). It is also reported that Sin3A and Sin3B target similar as well as unique sets of genes. DNA-binding transcription factors like KLF, Mad1, REST and ESET are observed to interact with both isoforms. On the one hand, transcription regulators SMRT and MeCP2 associate with mSin3A while the master regulator of MHC II, CIITA mediates IFN-γ induced repression of collagen type I gene transcription via HDAC2/Sin3B (Kong et al. 2009; Nagy et al. 1997; Nan et al. 1998). Sin3B was associated with repression of subsets of E2F target genes while both Sin3A and Sin3B interact with p53 and regulate its target gene repression. Sin3B also plays an essential role in promoting the cell cycle via the E2F–Rb pathway (Bansal et al. 2011; Blais and Dynlacht 2007; Kadamb et al. 2013). In fact, Sin3B-deficient mice confirmed the role of mSin3B in cell cycle exit control and repression of E2F target genes in vivo, along with a role in the differentiation of erythrocytes and chondrocytes. Sin3B is upregulated during oncogenic stress signalled by Ras overexpression and that expression of Sin3B was required for cellular senescence (David et al. 2008). In contrast, removal of Sin3A did not affect cell cycle progression; it instead promoted apoptosis of breast cancer cells MCF7. These observations reinforce the idea that each isoform of Sin3 has several unique targets in the cell (Hurst 2013).
6 Sin3-Protein Interactions
Sin3 targets chromatin either through direct interaction with DNA-binding transcription factors or indirectly through another adaptor molecule, known as co-repressor complex. A large number of transcription factors interact with Sin3 through its six conserved domains that include the four PAH1-4, HID and HCR. NMR and crystallographic structural data available so far for Sin3 suggest that N-terminal region of Sin3, which includes PAH1-3 domains of Sin3, shows conformational heterogeneity whereas the structure of C-terminal region of Sin3 (HID, PAH4 and HCR) is highly stable. This may be one of the reasons due to which PAH1-3 regions of Sin3 can interact with a large number of transcription factors, whereas very few transcription factors interact with C-terminal region of Sin3. Other factors such as difference in sequence of PAH domains, recognition of different SID domains (Sin3 interacting domain) by PAH domains, etc. make each PAH domain a unique region, capable of interacting with diverse transcription factors and co-repressors to regulate transcription. Thus, Sin3 acts as a master scaffold to provide a platform for the assembly of numerous transcription factors and co-repressors. Besides this, multiple domains within Sin3 serve as excellent protein-binding interfaces which in turn make Sin3 to be suitable for multiple protein interactions.
6.1 Interaction with DNA-Binding Proteins
The PAH domains of Sin3 are specialized for interaction with DNA-binding domains. These PAH domains are structurally homologous to one another, yet they recognize different sets of transcription factors or DNA-binding proteins due to the fact that these PAH domains have different amino acid sequences which recognize different sequence motifs of transcription factors to bring out transcription repression. Figure 1.3 shows CLUSTAL-Omega-guided multiple sequence alignment of PAH1, PAH2 and PAH3 of Sin3.
Majority of proteins so far identified to interact with sin3 interact with its PAH2 domain. Although PAH2 domain is the closest relative of the PAH1 domain having 30 % sequence identity and 54 % sequence similarity, only a few interacting partners are known to interact with PAH1 in contrast to PAH2, suggesting dissimilar modes of engaging targets. However, PAH3 domains share relatively low levels of sequence identity with the PAH1 and PAH2 domains having 25 % sequence identity and 45 % sequence similarity with PAH1 and 16 % sequence identity and 48 % sequence similarity with PAH2 domains respectively, yet it recognizes larger sets of proteins than PAH1 (Le Guezennec et al. 2006; Moehren et al. 2004; Sahu et al. 2008). Previous studies have also shown that PAH1 amino acid residues Val7, Leu14 and Lys39 play an important role in the specificity between PAH1 and SID domains whereas PAH2 residues Phe7, Val14 and Gln39 are important in determining the specificity between PAH2 and SID domains (Le Guezennec et al. 2006). Similar observations are lacking for PAH3 domains. Mutation in these amino acid sequences may decrease in binding specificity of SID (Sin3 interacting domain) domains.
Moreover, each PAH domain of Sin3 recognizes different sequence motifs on DNA-binding proteins of Sin3 which is also known as SID domain (Sin3 interacting domain). Initial attempts to identify a SID consensus sequence for Mad family members in PAH2 revealed the following degenerate sequence: φφZZφφXAAXXφnXXn with X for any non-proline residues, φ for bulky hydrophobic residues and n for negatively charged residues. Other studies have reported the SID of SP1-like members with PAH2 as φφXAAXXφ. Similarly in case of PAH1, LXXLL consensus sequence was identified as SID of SAP25, a component of the Sin3 complex. Thus by performing a yeast two-hybrid screening of a peptide aptamer library, a small repertoire of peptides interacting specifically either with PAH1 or PAH2 domain of mSin3B was identified, which is listed in Table 1.3 (Le Guezennec et al. 2006).
Structural flexibility and conformational heterogeneity of PAH domains are also two of the major factors which help in recruiting different sets of DNA-binding proteins onto the PAH domains of Sin3. NMR and X-ray crystallographic structures of PAH1 and PAH2 have shown that these are structurally independent and fold in a different way to interact with the SID domains of the peptides (Guezennec 2006; Swanson et al. 2004). In the complex, PAH1 holds a left-handed four-helix bundle structure followed by a semi-ordered C-terminal tail; however, PAH2 domain adopts a wedge-shaped helical conformation in the complex engaging the cleft through a non-polar surface (Nomura et al. 2005). Essentially, the architecture of the four helices of PAH1 is similar to the corresponding structure of the PAH2 domains, except that they are shorter (Zhang et al. 2001). There also exists a structural heterogeneity in orientation of PAH domains on binding with different transcription factors. Mad1 and HBP1 trans-repression domain binds through a helical structure to the hydrophobic cleft of mSin3A PAH2, but the HBP1 helix binds PAH2 in a reversed orientation relative to Mad1 (Swanson et al. 2004).
Presence of different isoforms and splice variants of Sin3 in various organisms further add to the structural complexity and heterogeneity of PAH domains to interact with diverse binding partners. Though PAH2 domains of both mSin3A and Sin3B proteins interact with their targets in an analogous manner, the non-interactive forms of these two paralogues differ in their structure. The PAH2 domain of apo-mSin3A homodimerizes and exists in unfolded state, but apo-mSin3B PAH2 domain is monomeric and is fully folded (Kadamb et al. 2013).
Structures of PAH3 complex with SAP30 have been recently characterized, which suggests that PAH3 domain also forms a four-helix structure similar to PAH1 and PAH2 domains of Sin3 on interaction with SID domain of SAP30 (Xie et al. 2011). However, the individual structure of PAH3 domain is yet to be characterized by NMR and X-ray crystallography. Recent study from our lab suggests that PAH3 and PAH2 domains of Sin3B largely exist in unfolded state whereas PAH1 exists in folded state in nuclear pH conditions. The flexibility in the structure of PAH2 and PAH3 at nuclear pH condition allows them to interact with large numbers of binding partners (Tauheed Hasan et al. 2015). Transcription factors have also been reported to interact with two different regions of Sin3 thus giving the next level of complexity in the interaction of proteins. Table 1.7 shows proteins interacting with two different regions of proteins.
Few transcription factors have also been shown to interact with regions of Sin3 not predicted to form amphipathic helices such as HCR, HID and PAH4. The PAH4 domain of Sin3 is highly conserved throughout in the length of their polypeptide in different organisms and exists in a folded state and thus does not show any structural heterogeneity (Cowley et al. 2004; Kadamb et al. 2013). Only one interacting partner has been known so far in case of PAH4 domain. Like PAH4 domain, HID and HCR regions of Sin3 are also highly conserved in different organisms. Thus, it is believed that N- terminal of Sin3, which includes PAH1-3, is involved in interacting with different DNA-binding transcription factors, whereas C-terminal part which includes HID, PAH4 and HCR is involved in the scaffolding function of Sin3 by serving as interaction site for other subunits of the co-repressor complex.
The first transcription factor characterized as interacting with Sin3 was Mad1. The Mad1 repressor belongs to a family of four proteins (Mad1, Mxi1, Mad3 and Mad4) that are thought to antagonize the transcriptional activation, proliferation-promoting and transformation functions of the oncoprotein Myc and thereby acts as tumour suppressor (Pang et al. 2003). Solution structures of the PAH2 domain complexed to the Sin3 interacting domain (SID) of Mad1 showed that the complex is folded as a ‘wedged helical bundle’, in which the PAH2 domain adopts a four-helix bundle conformation in which the α-helix of the SID is inserted (Brubaker et al. 2000). Till now, numerous binding partners have been discovered that interact with Sin3 for transcription regulation. A brief list of the proteins interacting with different domains of Sin3 are given in Tables 1.4, 1.5, 1.6, 1.7 and 1.8.
Earlier, Sin3 was thought to be a transcription repressor, but many transcription factors are able to positively regulate gene expression. The first report of Sin3 as an activator was published in the 1990s wherein it was found to activate GAM3, a gene encoding an extracellular glucoamylase in yeast, through interaction with STA1 (transcription factor) (Yoshimoto et al. 1992). Another interesting example is the heat stress-induced Sin3-mediated gene activation (CTT1, ALD3, PNS1 and TPS1) (Ruiz-Roig et al. 2010). Further studies are required to understand the mechanism of Sin3-mediated gene regulation.
6.2 Interaction of Proteins with Members of Sin3 Core Complex and Co-repressor Complex
Other targeting peptides interact with Sin3 in a more indirect manner to bring transcription regulation. They may interact through one or several members of the core complex, or indirectly via special adapter molecules called co-repressors. Sin3-associated proteins (SAPs) are important members of core complex that play a vital role in interaction with DNA-binding proteins. Binding with transcription factors results in conformational change and altered binding of SAP with PAH domains of Sin3 and consequent tethering of SAP onto the promoter of DNA. Since SAP also directly binds to HDAC, the SAP-Sin3-HDAC complexed with DNA results in histone deacetylation and thus repression of the genes (Zhang et al. 1998). The first member of SAP protein discovered to interact with transcription factors is SAP30, which specifically interacts with Sin3A isoform (Laherty et al. 1998). SAP 30 interacts with transcription factors such as papillomavirus binding factor (PBF), a nuclear-cytoplasmic shuttling factor which represses the cell growth. On interaction with PBF, SAP 30 is recruited on PAH3 domain of Sin3A to bring about transcription regulation (Sichtig et al. 2007). Solution structure of SAP30-PAH3 complex revealed that SID domain of SAP30 binds to PAH3 via a tripartite structural motif, including a C-terminal helix that targets the canonical PAH hydrophobic cleft while two other helices and an N-terminal extension target a discrete surface formed largely by the PAH3 α2, α3 and α3′ helices (Xie et al. 2011). SAP protein is also recruited directly by PAH domains onto the promoter of transcription regulation machinery to bring about transcription regulation. For example, SAP30 and SAP25 of core complex function as DNA-binding proteins that specifically bind to the promoter region of genes. Solution NMR structure of SAP25 and PAH1 domain shows that SAP25, highly unstructured, except for portions of the SID interacting segments, adopts a helical conformation. SAP25 binds through an amphipathic helix to a predominantly hydrophobic cleft on the surface of PAH1 (Sahu et al. 2008). Thus, SAP25 and SAP30 adopt a different fold to get recruited by PAH1 and PAH3 domains of Sin3 to bring about transcription regulation. Other members of the Sin3 complex recruited directly or indirectly by different PAH domains of Sin3 are listed in Table 1.9.
Co-repressors typically serve as link between the DNA-binding proteins and Sin3/HDAC complex. However, co-repressors may augment transcriptional silencing through their intrinsic repressor activities, or even by recruiting auxiliary functionality. Nevertheless, addition of co-repressor to Sin3/HDAC complex alters the properties of the complex, sometimes drastically. The best studied co-repressors are associated with nuclear receptors: nuclear receptor co-repressor (N-CoR) and neural restrictive silencer factor (NRSF)/repressor element 1 silencing transcription factor (REST). These co-repressors provide a link between the Sin3/ HDAC complex and nuclear receptors, a family of transcription factors that regulate gene expression in a ligand-dependent manner. Nuclear receptors recruit HAT and HDAC complexes in a context-dependent manner and thus facilitate both gene activation and repression without dissociating from the DNA (Alland et al. 1997; Nomura et al. 2005; Torchia et al. 1998). Thus, by simply changing its interaction partners, a transcription factor is able to modulate the transcriptional activity of genes.
In mammals, the N-terminal repressor domain of NRSF/REST interacts with PAH1 domain of Sin3B and represses neuronal gene expression in non-neuronal tissues. The NMR structures of the complex suggest that PAH1 holds left-handed, four-helix bundles with a semi-ordered C-terminal tail associated with a hydrophobic short α-helix of NRSF/REST. The NRSF/REST short helix is sandwiched between α1 and α2 helices of PAH1 and positioned at an angle of about 55° relative to the α2 helix. While in case of nuclear hormone receptor co-repressor (N-CoR), the vital structure of the four helices of PAH1 is similar to that of corresponding structure of the PAH1-NRFS/REST domains, but the affinity of binding of N-CoR to the PAH1 domain is low. One of the reasons could be that C-terminal region of N-CoR contains fewer hydrophobic amino acid residues than the NRSF/REST helix (Nomura et al. 2005; Wolffe 1997). Thus, for strong binding to a repressor, PAH1 preferentially requires a short α-helix with a large number of hydrophobic amino acid residues within the repressor. The conformation flexibility in structure of PAH domains binds co-repressors with different binding affinities thereby providing additional flexibility for regulating the promoter activity. For some transcription factors, special adapters have been developed that can mediate or stabilize the contact between these proteins and Sin3/HDAC. For instance, the adapter molecule RBP1 interacts with the nuclear phosphoprotein Rb and also serves to recruit Sin3/HDAC by interacting with the core component SAP30 and Rb protein (Lai et al. 2001).
7 Altered Structure of Sin3 Protein
7.1 Alteration in Structure Due to Post-Translation Modification of Sin3 Protein
The Sin3/HDAC co-repressor complex can be recruited by a large number of DNA-binding transcription factors or co-repressors, thereby requiring a precise and coordinated mechanism to achieve specific and timely regulation of transcription. Although poorly investigated, post-translational modifications in Sin3 protein play a very important role in fine-tuning of its regulation. Sin3 protein contains several potential sites for post-translation modifications such as phosphorylation, myristoylation, ubiquitination and SUMOylation (Bansal et al. 2011).
7.1.1 Phosphorylation
Protein phosphorylation is a post-translational modification of proteins in which a serine, threonine or a tyrosine residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Sin3 phosphorylation by tyrosine kinase at key tyrosine residue helps to regulate protein stability through PAH domain-mediated protein-protein interactions (Bansal et al. 2011). For example, under normal conditions, levels of p53 (tumor suppressor protein) are tightly regulated by MDM2-mediated degradation. However, in case of cellular stress such as DNA damage, the levels of p53 increase significantly due to its stable interaction with phosphorylated sin3A. Interaction of p53 with Sin3A effectively masks the MDM2-binding motif and protects p53 from proteasome-mediated degradation in a manner independent of MDM2 (Bansal et al. 2011; Honda et al. 1997). Thus, under conditions of cellular and/or genotoxic stress, phosphorylation of Sin3A can mediate stabilization of its interacting partner proteins.
7.1.2 Myristoylation
Myristoylation is an irreversible protein lipidation modification where a myristoyl group, derived from myristic acid, is covalently attached by an amide bond to the alpha-amino group of an N-terminal glycine residue. Myristic acid is a 14-carbon saturated fatty acid (14:0) with the systematic name of n-Tetradecanoic acid (Farazi et al. 2001). In Saccharomyces cerevisiae, myristoylation status of Sin3 is used for regulation of Opi1 transcription factor which regulates biosynthesis of phospholipids. During glucose starvation, Sin3 gets myristoylated and recruits Opi1 transcription factors, which results in activation of Ino2p and Ino4p genes. Ino2p and Ino4p are basic helix–loop–helix (bHLH) transcription factors that form heterodimeric complexes which bind to inositol-choline response elements (ICRE, also known as UASINO). In presence of inositol, binding of Ino2p and Ino4p represses the biosynthesis of phospholipid (Chen et al. 2007). Thus, Saccharomyces cerevisiae can overcome glucose starvation by minimizing energy utilization by shutting down phospholipid biosynthesis.
7.1.3 Ubiquitination
Ubiquitination, a post-translational modification where ubiquitin (a small regulatory protein that has been found in almost all tissues of eukaryotic organisms) is attached to a substrate protein. The addition of ubiquitin can affect proteins in many ways: it can signal for their degradation via the proteasome, alter their cellular location, affect their activity and promote or prevent protein interactions (Peng et al. 2003). Ubiquitination is carried out in three main steps: activation, conjugation and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s), respectively (Kadamb et al. 2013). The result of this sequential cascade binds ubiquitin to lysine residues on the protein substrate via an isopeptide bond or to the amino group of the N-terminus via a peptide bond.
Sin3 protein gets attached to a novel ubiquitin ligase, RING finger protein 220 (RNF220), and gets transported to cytoplasm where proteosomal degradation takes place. Although the mechanism is not clear, it is postulated that RNF220 acts as an E3 ubiquitin ligase for Sin3B and can promote its ubiquitination (Kong et al. 2010). Sin3 protein is thought to be a nuclear protein where it brings about transcriptional regulation of various genes. Thus, not only can ubiquitination of Sin3 protein alter gene regulation, but its movement to cytoplasm may provide it with novel functions inside the cell.
7.1.4 SUMOylation
SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress and progression through the cell cycle. SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO (Small Ubiquitin-like Modifier) is not used to tag proteins for degradation. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on the target protein (Pungaliya et al. 2007).
Recent studies indicated that SUMOylation of Sin3 helps in stabilization of interaction between retinoblastoma-binding protein 1 (RBP1) and SAP 30 in Sin3/HDAC complex. RBP1 is a cellular protein that interacts with the pocket of retinoblastoma protein (pRB) and appears to be an important factor in the repression of E2F-dependent transcription by RB family of proteins. pRB appears to function principally as a transcriptional repressor that blocks E2F-dependent transcription, thus regulating entry into the S phase of the cell cycle (Binda et al. 2006). This cellular proliferation pathway is deregulated in a vast majority of cancers. Thus, Sin3 SUMOylation may play an important role in suppression of cell proliferation.
7.2 Modification of Sin3 Structure Due to Protein-Protein Interaction
The Sin3 has a dual role, functioning not only as a molecular scaffold for complex assembly but also as a molecular adapter, bridging HDACs with an astonishingly large and diverse group of DNA-binding transcription factors and chromatin-binding proteins. Most of the protein-protein interactions are achieved through six conserved domains of Sin3 that include four paired amphipathic helices (PAH 1–4), one HID and one highly conserved region (HCR). However, most of them interact with PAH domains of Sin3, especially with PAH2 and PAH3. Although there is a high degree of similarity between the PAH domains, yet PAH domains recognize different sequence motifs thereby exhibiting high degrees of specificity for their target. The PAH domains mediate specific protein–protein interactions, most likely through their independent associations with various repressors. Upon interaction with the transcription factors, these PAH domains adopt diverse conformational folds that help PAH domain to recruit different binding partners. Of the four Sin3 PAH domains, the tertiary structures of only PAH1 and PAH2 domains have been determined by NMR.
The second copy of the PAH domain (PAH2) is the site for interactions with numerous factors of which some are listed in Table 1.5. Structural studies have revealed that the PAH2 domain of mammalian Sin3 exhibits conformational heterogeneity that enables it to interact with diverse protein targets. PAH2 domain of mSin3B protein interacts with its targets in a manner similar to that of mSin3A, but these two paralogues differ in their non-interactive forms. The apo-mSin3A PAH2 domain homodimerizes and exists in unfolded state, but apo-mSin3B PAH2 domain is monomeric and is fully folded (Kadamb et al. 2013). Solution structures of the PAH2 domain complexed to the Sin3 interacting domain (SID) of Mad1 have shown that the complex is folded as a ‘wedged helical bundle’, in which the PAH2 domain adopts a four-helix bundle conformation into which the α- helix of the SID is inserted (van Ingen et al. 2006). However, the HMG box-containing repressor HBP1, which targets several cell cycle-specific and differentiation-specific genes, was shown to interact with the PAH2 domain. The structure of this complex showed a reversed orientation of the SID relative to the Mad1 complex, while maintaining the overall fold. The reversal of this helix orientation is correlated with a reversal in the SID sequence motif (Swanson et al. 2004). Furthermore, molecular dynamic simulation has suggested that TIEG2 SID binds PAH2 in a different orientation than the Mad SID, suggesting multiple ways to interact with PAH2 domain.
By contrast, less is known about the mode of interaction and recruitment of the Sin3 proteins via the PAH1 domain. The solution structures of the apo-mSin3A PAH1 domain adopts the canonical left-handed four-helix bundle fold while the SAP25 SID in the mSin3A PAH1 complex is largely unstructured, except for some segments that adopt a helical conformation (Sahu et al. 2008). However, PAH1 holds a rather globular four-helix bundle structure with a semi-ordered C-terminal tail on interaction associated with the NRSF/REST repressor domain (Nomura et al. 2005). Although the PAH2 domain is the closest relative of the PAH1 domain, the domains exhibit distinct patterns of sequence conservation, suggesting dissimilar modes of engaging targets.
However, the folded structure of PAH3 and PAH4 domains of Sin3 has not been solved yet. More recently, some of the interacting partners of PAH3 domain have been identified (see Table 1.6), suggesting that PAH3 domain of Sin3 also exhibits conformational plasticity upon interacting with proteins. Till now, only one interacting partner of PAH4 has been identified (Table 1.8), suggesting that this domain may not exhibit conformational flexibility. Thus, structural and dynamic plasticity in Sin3 PAH domains, upon protein –protein interaction, helps Sin3 to interact with large numbers of transcription factors.
8 Structural Allostery
Structural allostery refers to the biological phenomenon where ligand/effector binding or energetic perturbation at one molecular site results in structure or activity changes at a second distinct site. The site where the effector binds is termed as the allosteric site and is different from the protein’s active site. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change. Effectors that enhance the protein’s activity are referred to as allosteric activators, whereas those that decrease the activity of proteins are called allosteric inhibitors (Hilser et al. 2012).
Allostery in the structure of Sin3 protein helps to recruit unique sets of proteins onto different sub-domains of Sin3 to bring transcription regulation. As discussed earlier in this chapter, Sin3 acts as a scaffold, which provides platforms to various proteins and complexes to coordinate the transcription regulation of diverse genes. Structural allostery in Sin3 shows two types of modulation as under.
8.1 Allosteric Modulation
Allosteric modulation of a receptor results from the binding of allosteric modulators at a different site (a ‘regulatory site’) from that of the endogenous ligand (an ‘active site’) and enhances or inhibits the effects of the endogenous ligand. Under normal circumstances, it acts by causing a conformational change in a receptor molecule, which results in a change in the binding affinity of the ligand (Fig. 1.4) (del Sol et al. 2009; Hilser et al. 2012).
8.1.1 Positive Modulation
Positive allosteric modulation (also known as allosteric activation) occurs when the binding of one ligand enhances the attraction between substrate molecules and other binding sites (Fig. 1.4a) (Hilser et al. 2012). HDAC1 (histone deacetylase complex 1) and HDAC2 (histone deacetylase complex 2) bind to the highly conserved HID region of Sin3, bring allosteric change in the structure of PAH2 domain of Sin3B and form a ternary complex. The ternary complex interacts with Mad/Myc proteins (allosteric activation) that is able to direct trichostatin A (TSA, a HDAC inhibitor) sensitive repression to luciferase under the control of a Myc-binding element (Nan et al. 1998).
8.1.2 Negative Modulation
Negative allosteric modulation (also known as allosteric inhibition) occurs when the binding of one ligand decreases the affinity for substrate at other active sites (Fig. 1.4b) (Hilser et al. 2012). Sin3/HDAC complex undergoes an allosteric conformational shift upon binding with reduced nicotinamide adenine dinucleotide (NADH) which in turn regulates the interaction of CtBP (C-terminal-binding protein) with REST co-repressor complex. High NADH levels cause CtBP to interact weakly with REST (negative modulation or allosteric inhibition, resulting in de-repression of transcription. However, low NADH levels allow CtBP to bind strongly with REST co-repressor complex and shut down transcription. Since the bulk of nuclear NADH is thought to be in equilibrium with cytoplasmic NADH and is derived in large part through glycolysis, lower rates of glycolysis result in reduced NADH, increased CtBP binding to REST and more repression of REST target genes (Chinnadurai 2002; Ooi and Wood 2007). This mechanism was exploited to control pro-epileptic gene expression in the kindling model using glycolytic inhibitors.
Structural allostery in Sin3 also helps in governing protein-protein interaction stability by which they can carry gene regulation. Phosphorylation of Sin3 by tyrosine kinase at key tyrosine residues induces conformational change in the structure of PAH2 domain. Upon phosphorylation, PAH2 domain forms α-helical pocket which interacts with SID domain of p53 (tumor suppressor protein). This interaction between SID domain of p53 and Sin3 complex alleviates proteasome-mediated degradation in a manner independent of MDM2 (Zilfou et al. 2001).
Similarly, NRSF/REST is a Kruppel-type zinc finger protein that mediates transcriptional repression through the association of its C-terminal repressor domain (RD-2) with CoREST/HDAC complex and N-terminal repressor domain (RD-1) with Sin3/HDAC complex. The N-terminal domain of NRSF/REST is folded into a unique hydrophobic α-helix which on interaction with Sin3 induces allosteric modification in unstructured C-terminal region of PAH1 domain such that the latter adopts a left-handed four-helix bundle structure. This in turn stabilizes the binding of PAH1 to RD-2 domain of NRSF/REST. Both RD-1 and RD-2 interact directly with TATA-binding protein (TBP) via a chromatin-independent mechanism (Nomura et al. 2005). Several neurological diseases such as Down syndrome, medullo-blastoma and Huntington’s disease are related with dysregulation of NRSF/REST and its target genes (Song et al. 2014).
9 Conclusion
Sin3 plays dual roles as a scaffold protein as well as adapter protein both in higher and lower eukaryotes. Due to the conformational diversity and structural flexibility of its different sub-domains, it is able to recruit a large number of DNA-binding proteins and co-repressor complexes. Various processes such as allosteric regulation, protein-protein interactions and post-translation modifications contribute towards allosteric structure and conformational diversity of Sin3. The flexible nature of Sin3 structure helps to interact with various proteins and facilitates regulation of diverse cellular processes inside the cell such as growth, differentiation and senescence as well as oncogenic transformation in pathological conditions.
10 Future Prospective
In spite of active research on the structure of Sin3 protein, the NMR structure of only PAH1 and PAH2 domains have been solved. Sin3 is able to recruit a large number of transcription factors and co-repressors due to flexible natures of different sub-domains. It will be imperative to decipher the function of different motifs present in the C-terminal regions of Sin3 so as to characterize and understand its interacting partners and their role in Sin3-mediated gene regulation. Future studies should also be directed towards understanding the regulation of Sin3 protein under different physiological conditions and modulation of its biological activity.
References
Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA (1997) Role for n-cor and histone deacetylase in sin3-mediated transcriptional repression. Nature 387:49–55
Ayer DE, Lawrence QA, Eisenman RN (1995) Mad-max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor sin3. Cell 80(5):767–776
Bansal N, Kadamb R, Mittal S, Vig L, Sharma R, Dwarakanath BS, Saluja D (2011) Tumor suppressor protein p53 recruits human sin3b/hdac1 complex for down-regulation of its target promoters in response to genotoxic stress. PLoS One 6(10), e26156
Benedik MJ, Ekwall K, Choi J, Allshire RC, Levin HL (1999) A new member of the sin3 family of co-repressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol Cell Biol 19(3):2351–2365
Binda O, Roy J-S, Branton PE (2006) Rbp1 family proteins exhibit sumoylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence. Mol Cell Biol 26(5):1917–1931
Blais A, Dynlacht BD (2007) E2f-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 19(6):658–662
Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, Eisenman RN, Radhakrishnan I (2000) Solution structure of the interacting domains of the mad-sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell 103(4):655–665
Brunmeir R, Lagger S, Seiser C (2009) Histone deacetylase 1 and 2-controlled embryonic development and cell differentiation. Int J Dev Biol 53(2):275
Campos EI, Chin MY, Kuo WH, Li G (2004) Biological functions of the ING family tumor suppressors. Cell Mol Life Sci 61(19–20):2597–2613
Chen M, Hancock LC, Lopes JM (2007) Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochimica et Biophysica Acta (BBA)-Mol Cell Biol Lipids 1771(3):310–321
Chinnadurai G (2002) Ctbp, an unconventional transcriptional co-repressor in development and oncogenesis. Mol Cell 9(2):213–224
Coulson JM (2005) Transcriptional regulation: cancer, neurons and the rest. Curr Biol 15(17):R665–R668
Cowley SM, Kang RS, Frangioni JV, Yada JJ, DeGrand AM, Radhakrishnan I, Eisenman RN (2004) Functional analysis of the mad1-msin3a repressor-co-repressor interaction reveals determinants of specificity, affinity, and transcriptional response. Mol Cell Biol 24(7):2698–2709
David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, DePinho RA (2008) Specific requirement of the chromatin modifier msin3b in cell cycle exit and cellular differentiation. Proc Natl Acad Sci 105(11):4168–4172
del Sol A, Tsai C-J, Ma B, Nussinov R (2009) The origin of allosteric functional modulation: multiple pre-existing pathways. Structure 17(8):1042–1050
Espinas M, Canudas S, Fanti L, Pimpinelli S, Casanova J, Azorin F (2000) The gaga factor of drosophila interacts with sap18, a sin3a-associated polypeptide. EMBO Rep 1(3):253–259
Farazi TA, Waksman G, Gordon JI (2001) The biology and enzymology of proteinn-myristoylation. J Biol Chem 276(43):39501–39504
Fleischer TC, Yun UJ, Ayer DE (2003) Identification and characterization of three new components of the msin3a co-repressor complex. Mol Cell Biol 23(10):3456–3467
Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T (2000) The isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by ume6p. Cell 103(3):423–433
Grzenda A, Lomberk G, Zhang J-S, Urrutia R (2009) Sin3: master scaffold and transcriptional co-repressor. Biochimica et Biophysica Acta (BBA)-Gene Reg Mech 1789(6):443–450
Guezennec XSl. 2006. A journey inside sin3 and mi-2/nurd co-repressor complexes: A structural and biochemical perspective. [Sl: sn].
Guidez F, Ivins S, Zhu J, Soderstrom WS, Zelent A (1998) Reduced retinoic acid-sensitivities of nuclear receptor co-repressor binding to pml-and plzf-rarî ± underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91(8):2634–2642
Halleck MS, Pownall S, Harder KW, Duncan A, Jirik FR, Schlegel RA (1995) A widely distributed putative mammalian transcriptional regulator containing multiple paired amphipathic helices, with similarity to yeast sin3. Genomics 26(2):403–406
Hasan T, Ali M, Saluja D, Singh LR (2015) pH might play a role in regulating the function of paired amphipathic helices domains of human Sin3B by altering structure and thermodynamic stability. Biochemistry (Moscow) 80(4):424–432
He Y, Imhoff R, Sahu A, Radhakrishnan I (2009) Solution structure of a novel zinc finger motif in the sap30 polypeptide of the sin3 co-repressor complex and its potential role in nucleic acid recognition. Nucleic Acids Res 37(7):2142–2152
Hilser VJ, Wrabl JO, Motlagh HN (2012) Structural and energetic basis of allostery. Annu Rev Biophys 41:585
Honda R, Tanaka H, Yasuda H (1997) Oncoprotein mdm2 is a ubiquitin ligase e3 for tumor suppressor p53. FEBS Lett 420(1):25–27
Hug BA, Lazar MA (2004) Eto interacting proteins. Oncogene 23(24):4270–4274
Hui Ng H, Bird A (2000) Histone deacetylases: silencers for hire. Trends Biochem Sci 25(3):121–126
Hurlin PJ, Queva C, Eisenman RN (1997) Mnt, a novel max-interacting protein is coexpressed with myc in proliferating cells and mediates repression at myc binding sites. Genes Dev 11(1):44–58
Hurst DR (2013) Metastasis suppression by brms1 associated with sin3 chromatin remodeling complexes. Cancer Metastasis Rev 31(3–4):641–651
Jelinic P, Pellegrino J, David G (2011) A novel mammalian complex containing sin3b mitigates histone acetylation and rna polymerase ii progression within transcribed loci. Mol Cell Biol 31(1):54–62
Kaczynski J, Cook T, Urrutia R (2003) Sp1-and kruppel-like transcription factors. Genome Biol 4(2):206
Kadamb R, Mittal S, Bansal N, Batra H, Saluja D (2013) Sin3: insight into its transcription regulatory functions. Eur J Cell Biol 92(8):237–246
Kadosh D, Struhl K (1997) Repression by ume6 involves recruitment of a complex containing sin3 co-repressor and rpd3 histone deacetylase to target promoters. Cell 89(3):365–371
Keogh M-C, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ (2005) Cotranscriptional set2 methylation of histone h3 lysine 36 recruits a repressive rpd3 complex. Cell 123(4):593–605
Kong X, Fang M, Li P, Fang F, Xu Y (2009) Hdac2 deacetylates class ii transactivator and suppresses its activity in macrophages and smooth muscle cells. J Mol Cell Cardiol 46(3):292–299
Kong Q, Zeng W, Wu J, Hu W, Li C, Mao B (2010) Rnf220, an e3 ubiquitin ligase that targets sin3b for ubiquitination. Biochem Biophys Res Commun 393(4):708–713
Kumar GS, Xie T, Zhang Y, Radhakrishnan I (2011) Solution structure of the msin3a pah2-pf1 sid1 complex: a mad1/mxd1-like interaction disrupted by mrg15 in the rpd3s/sin3s complex. J Mol Biol 408(5):987–1000
Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Role of the sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33ing1. Mol Cell Biol 22(3):835–848
Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun J-M, Mullen T-M, Davie JR, Rose DW, Glass CK (1998) Sap30, a component of the msin3 co-repressor complex involved in n-cor-mediated repression by specific transcription factors. Mol Cell 2(1):33–42
Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge M-C, Tsai S-C, Seto E, Zhang Y, Kuzmichev A (2001) Rbp1 recruits the msin3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol 21(8):2918–2932
Le Guezennec X, Vermeulen M, Stunnenberg HG (2006) Molecular characterization of sin3 pah-domain interactor specificity and identification of pah partners. Nucleic Acids Res 34(14):3929–3937
Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL (2007) Combined action of phd and chromo domains directs the rpd3s hdac to transcribed chromatin. Science 316(5827):1050–1054
McDonel P, Costello I, Hendrich B (2009) Keeping things quiet: roles of nurd and sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 41(1):108–116
Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR (2004) Breast cancer metastasis suppressor 1 (brms1) forms complexes with retinoblastoma-binding protein 1 (rbp1) and the msin3 histone deacetylase complex and represses transcription. J Biol Chem 279(2):1562–1569
Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro CL, Messali S, Zollo M, Ledbetter DH (1997) Rox, a novel bhlhzip protein expressed in quiescent cells that heterodimerizes with max, binds a non-canonical e box and acts as a transcriptional repressor. EMBO J 16(10):2892–2906
Moehren U, Dressel U, Reeb CA, Väisänen S, Dunlop TW, Carlberg C, Baniahmad A (2004) The highly conserved region of the co-repressor sin3a functionally interacts with the co-repressor alien. Nucleic Acids Res 32(10):2995–3004
Nagy L, Kao H-Y, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM (1997) Nuclear receptor repression mediated by a complex containing smrt, msin3a, andhistone deacetylase. Cell 89(3):373–380
Nair SK, Burley SK (2006) Structural aspects of interactions within the Myc/Max/Mad network. CTMI 302:123–143, Springer
Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998) Transcriptional repression by the methyl-cpg-binding protein mecp2 involves a histone deacetylase complex. Nature 393(6683):386–389
Nawaz Z, Baniahmad C, Burris TP, O’Malley BW, Stillman DJ, TsaiTsaiTsai M-J (1994) The yeast sin3 gene product negatively regulates the activity of the human progesterone receptor and positively regulates the activities of gal4 and the hap1 activator. Mol Gen Genet MGG 245(6):724–733
Nicolas E, Yamada T, Cam HP, FitzGerald PC, Kobayashi R, Grewal SIS (2007) Distinct roles of hdac complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14(5):372–380
Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y (2005) The neural repressor nrsf/rest binds the pah1 domain of the sin3 co-repressor by using its distinct short hydrophobic helix. J Mol Biol 354(4):903–915
Ooi L, Wood IC (2007) Chromatin crosstalk in development and disease: lessons from rest. Nat Rev Genet 8(7):544–554
Pang Y-P, Kumar GA, Zhang J-S, Urrutia R (2003) Differential binding of sin3 interacting repressor domains to the pah2 domain of sin3a. FEBS Lett 548(1):108–112
Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21(8):921–926
Pungaliya P, Kulkarni D, Park H-J, Marshall H, Zheng H, Lackland H, Saleem A, Rubin EH (2007) Topors functions as a sumo-1 e3 ligase for chromatin-modifying proteins. J Proteome Res 6(10):3918–3923
Romm E, Nielsen JA, Kim JG, Hudson LD (2005) Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem 93(6):1444–1453
Ruiz-Roig C, Viéitez C, Posas F, Nada E (2010) The rpd3l hdac complex is essential for the heat stress response in yeast. Mol Microbiol 76(4):1049–1062
Sahu SC, Swanson KA, Kang RS, Huang K, Brubaker K, Ratcliff K, Radhakrishnan I (2008) Conserved themes in target recognition by the pah1 and pah2 domains of the sin3 transcriptional co-repressor. J Mol Biol 375(5):1444–1456
Sharma V, Swaminathan A, Bao R, Pile LA (2008) Drosophila sin3 is required at multiple stages of development. Dev Dyn 237(10):3040–3050
Sheeba CJ, Palmeirim I, Andrade RP (2007) Chick hairy1 protein interacts with sap18, a component of the sin3/hdac transcriptional repressor complex. BMC Dev Biol 7(1):83
Shi X, Garry DJ (2012) Sin3 interacts with foxk1 and regulates myogenic progenitors. Mol Cell Biochem 366(1–2):251–258
Shiio Y, Rose DW, Aur R, Donohoe S, Aebersold R, Eisenman RN (2006) Identification and characterization of sap25, a novel component of the msin3 co-repressor complex. Mol Cell Biol 26(4):1386–1397
Sichtig N, Karfer N, Steger G (2007) Papillomavirus binding factor binds to sap30 and represses transcription via recruitment of the hdac1 co-repressor complex. Arch Biochem Biophys 467(1):67–75
Silverstein RA, Ekwall K (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet 47(1):1–17
Smith KT, Sardiu ME, Martin-Brown SA, Seidel C, Mushegian A, Egidy R, Florens L, Washburn MP, Workman JL (2012) Human family with sequence similarity 60 member a (fam60a) protein: a new subunit of the sin3 deacetylase complex. Mol Cell Proteomics 11(12):1815–1828
Song Z, Zhao D, Zhao H, Yang L (2014) Nrsf: An angel or a devil in neurogenesis and neurological diseases. J Mol Neurosci 1–14
Spain MM, Caruso JA, Swaminathan A, Pile LA (2010) Drosophila sin3 isoforms interact with distinct proteins and have unique biological functions. J Biol Chem 285(35):27457–27467
Spencer VA, Davie JR (1999) Role of covalent modifications of histones in regulating gene expression. Gene 240(1):1–12
Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I (2004) Hbp1 and mad1 repressors bind the sin3 co-repressor pah2 domain with opposite helical orientations. Nat Struct Mol Biol 11(8):738–746
Torchia J, Glass C, Rosenfeld MG (1998) Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol 10(3):373–383
van Ingen H, Baltussen MAH, Aelen J, Vuister GW (2006) Role of structural and dynamical plasticity in sin3: the free pah2 domain is a folded module in msin3b. J Mol Biol 358(2):485–497
Vietor I, Vadivelu SK, Wick N, Hoffman R, Cotten M, Seiser C, Fialka I, Wunderlich W, Haase A, Korinkova G (2002) Tis7 interacts with the mammalian sin3 histone deacetylase complex in epithelial cells. EMBO J 21(17):4621–4631
Viiri KM, Heinonen TYK, Mäki M, Lohi O (2009) Phylogenetic analysis of the sap30 family of transcriptional regulators reveals functional divergence in the domain that binds the nuclear matrix. BMC Evol Biol 9(1):149
Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller H-J (2001) The negative regulator opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor sin3 and the transcriptional activator ino2. Mol Microbiol 41(1):155–166
Wang J, Saunthararajah Y, Redner RL, Liu JM (1999) Inhibitors of histone deacetylase relieve eto-mediated repression and induce differentiation of aml1-eto leukemia cells. Cancer Res 59(12):2766–2769
Wolffe AP (1997) Sinful repression. Nature 387(6628):16–17
Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W (2003) Human sin3 deacetylase and trithorax-related set1/ash2 histone h3-k4 methyltransferase are tethered together selectively by the cell-proliferation factor hcf-1. Genes Dev 17(7):896–911
Xie T, He Y, Korkeamaki H, Zhang Y, Imhoff R, Lohi O, Radhakrishnan I (2011) Structure of the 30-kda sin3-associated protein (sap30) in complex with the mammalian sin3a co-repressor and its role in nucleic acid binding. J Biol Chem 286(31):27814–27824
Yang X-J, Seto E (2008) The rpd3/hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9(3):206–218
Yang Q, Kong Y, Rothermel B, Garry D, Bassel-Duby R, Williams R (2000) The winged-helix/forkhead protein myocyte nuclear factor î2 (mnf-î2) forms a co-repressor complex with mammalian sin3b. Biochem J 345:335–343
Yochum GS, Ayer DE (2001) Pf1, a novel phd zinc finger protein that links the tle co-repressor to the msin3a-histone deacetylase complex. Mol Cell Biol 21(13):4110–4118
Yoshimoto H, Ohmae M, Yamashita I (1992) The saccharomyces cerevisiae gam2/sin3 protein plays a role in both activation and repression of transcription. Mol Gen Genet MGG 233(1–2):327–330
Yuan J, Tirabassi RS, Bush AB, Cole MD (1998) The C. elegans mdl-1 and mxl-1 proteins can functionally substitute for vertebrate mad and max. Oncogene 17(9):1109–1118
Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D (1998) Sap30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell 1(7):1021–1031
Zhang J-S, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R (2001) A conserved α-helical motif mediates the interaction of sp1-like transcriptional repressors with the co-repressor msin3a. Mol Cell Biol 21(15):5041–5049
Zhang Y, Akinmade D, Hamburger AW (2005) The erbb3 binding protein ebp1 interacts with sin3a to repress e2f1 and ar-mediated transcription. Nucleic Acids Res 33(18):6024–6033
Zhang Y, Ye L, Tan Y, Sun P, Ji K, Jiang WG (2014) Expression of breast cancer metastasis suppressor-1, brms-1, in human breast cancer and the biological impact of brms-1 on the migration of breast cancer cells. Anticancer Res 34(3):1417–1426
Zilfou JT, Hoffman WH, Sank M, George DL, Murphy M (2001) The co-repressor msin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol Cell Biol 21(12):3974–3985
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer India
About this chapter
Cite this chapter
Hasan, T., Saluja, D. (2015). Structural Allostery and Protein–Protein Interactions of Sin3. In: Singh, L.R., Dar, T.A., Ahmad, P. (eds) Proteostasis and Chaperone Surveillance. Springer, New Delhi. https://doi.org/10.1007/978-81-322-2467-9_1
Download citation
DOI: https://doi.org/10.1007/978-81-322-2467-9_1
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-2466-2
Online ISBN: 978-81-322-2467-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)