Abstract
Due to the vast number of medical technologies and equipments, the medical images are growing at a rapid rate. This directs to retrieve efficient medical images based on visual contents. This paper proposed the magnetic resonance imagining (MRI) scan image retrieval system using co-occurrence matrix-based texture features. Here, the principal component analysis (PCA) is applied for optimized feature selection to overcome the difficulties of feature vector creation with Haralick’s texture features. Then, K-means clustering and Euclidean distance measure are used to retrieve best MRI scan images for the query image in medical diagnosis. The experimental results demonstrate the efficiency of this system in clusters accuracy and best MRI scan image retrieval against using all the fourteen familiar Haralick’s texture features.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Content-based image retrieval (CBIR) is a technique in which different visual contents have been measured to search and retrieve images from the mass amount of image databases based on the input image. The content-based medical image retrieval (CBMIR) systems are medical domain-specific search engine for medical image databases, which indexing and retrieving medical images according to their visual contents such as texture, shape, and other information [1–3].
The significance of new technologies such as X-ray radiography, ultrasound, computed tomography (CT), magnetic resonance imagining (MRI), and picture archiving and communication systems (PACS) has resulted in an explosive growth in the number of medical images stored in the database. Medical images classifying, indexing, and retrieval in manual methods are very expensive and time consuming. This will lead various systems for storage, organization, indexing, and retrieval of the medical images.
The most important objective of the CBMIR system is to retrieve the images from the huge volume of medical databases with high accuracy by performing feature extraction, classification, and similarity measure process. So the retrieved images are used for various medical diagnostic purposes.
Generally, the medical image database contains a lot of texture-based information capable for retrieval purpose. This paper proposed the MRI scan image retrieval system in two parts of the human body such as the spine and brain using co-occurrence matrix [4]-based texture features with principal component analysis (PCA) [5] feature selection transformation, K-means clustering [6], and Euclidean distance measure [7]. The accuracy, precision, and recall rate of this system are high compared with using all fourteen Haralick’s texture features [8].
The next section of the paper describes related works of the system. The brief discussion on the proposed work and Haralick’s texture features is given in Sects. 3 and 4. Sections 5 and 6 explain about feature selection and K-means clustering. Section 7 deals the image retrieval. Section 8 shows experiments and results. In Sect. 9, the conclusion of the work with future prospects is given.
2 Related Works
Medical images have become a key investigation tool for medical diagnosis. With the growth of medical databases, new applications committed to statistical analysis of medical data have appeared. There are many existing systems that provide different methods and algorithms for CBMIR. The most important intention of all these systems is to prove the improvement of results so as to give support to the doctors and radiologists in diagnosis of treatments.
In [9], they described a medical image retrieval system using low-level features and high-level semantic features with 90 % of precision and recall rate. Here, medical images were segmented into several sub-images using fuzzy C-mean clustering algorithm and extracting 3 gray-level features using color moments. Then, the sub-images were changed to binary image, and seven shape features and four texture features were extracted using co-occurrence matrix. Then, the genetic algorithm was used to select optimal features, and the text information in the medical image was chosen for the semantic content of the report of radiologists.
Selvarani and Annadurai [10] illustrated a system by combining low-level content features and high-level semantic features for medical image retrieval. The semantic information was extracted from the DICOM header which was used to perform the initial search. This pre-filtering of the images reduced the number of images to be searched. Then, texture features and shape features were found by Gabor filter and the fixed block resolution format. Image retrieval is performed by Euclidean distance measure. This system reduced the time taken to search the entire medical image database. Also, the average precision rate of 80 % is achieved.
Horsthemke et al. [11] explained two different texture feature-based CBMIR systems. The first system can be used to provide context-sensitive tools for computer-aided diagnosis with pixel-level co-occurrence matrices. The second system can be used directly as a computer-aided diagnosis system for case-based and evidence-based medicine with pixel-level and global-level co-occurrence matrices.
Zhang and Zhu [12] proposed a method using co-occurrence matrix to extract texture feature and edge histogram to extract shape feature of medical images. Then, Euclidean distance was used for medical image retrieval. Results of experimentation showed that the system had a recall rate about 90 % and applied to medical image retrieval with promising effect. In [13], the authors presented an evaluation of the diagnosis of dementia using texture analysis of brain MRI with Gabor wavelets and further classified by the back propagation network. Here, three different types of texture features were extracted: The first had the gray-level co-occurrence matrix (GLCM) features, the second had the Haralick’s features, and the third had Gabor wavelet-based Haralick’s features. From the comparison of the average efficiency, the statistical features extracted from Gabor wavelets provided better efficiency of 97 % than the other two methods.
Prasad and Krishna [14] evaluated the performance of two statistical methods of texture features proposed by Haralick’s and Tamura for retrieving similar cases for CT scan brain images. To speed up the search process, selected features were extracted and indexed using hash structure. The Euclidean distance measure was used for similarity measurement. Both the methods were compared based on precision and recall. Tamura features were found to provide better retrieval results for CT scan brain images. In our previous work [15], the performance measures for spine MRI scan image retrieval proved that texture features (black-white symmetry, geometric symmetry, degree of direction, orientation features and central symmetry) based on the texture spectrum were somewhat good compared to Haralick’s texture features (contrast, angular second moment, coarseness, entropy) and the combination of both features of image retrieval was the best.
3 Proposed Work
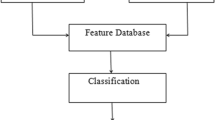
The block diagram of the proposed CBMIR system is shown in Fig. 1 in which two parts of the human body MRI scan images such as the spine and brain are used to construct the training data set.
In our work, we find GLCM to each MRI scan image in the training image database. Then, fourteen Haralick’s textures feature vector values are extracted as feature components, and PCA feature selection transformation is used to create an optimized database.
After that, using k-means clustering, the training images are clustered by means of the selected texture feature-based database. When a testing MRI scan image is submitted, the same texture feature extraction and feature vector value construction process have been applied to obtain the feature vector values for the testing image. Next, for similarity comparison between the query MRI scan image and the clustered MRI scan images, a Euclidean distance function is used. The closest Euclidean distance values for the query image are ranked, and best MRI scan images are retrieved for medical diagnosis.
4 Haralick’s Texture Features
Since we are interested in the statistical approach, we make use of the most suitable Haralick’s features. The major advantage of using the texture attributes is obviously their simplicity. The most common features used in practice are the measures derived from GLCM. These features have been widely used in the analysis, classification, and interpretation of medical images. The following fourteen Haralick’s texture features are extracted for the training MRI scan images.
-
1.
Angular second moment: \( \sum\limits_{i} {\sum\limits_{j} p } (i,j)^{2} \)
-
2.
Contrast: \( \sum\limits_{n = 0}^{{N_{g} - 1}} {n^{2} } \left\{ {\sum\limits_{i = 1}^{{N_{g} }} {\sum\limits_{j = 1}^{{N_{g} }} {p(i,j)} } } \right\},\left| {i - j} \right| = n \)
-
3.
Correlation: \( \frac{{\sum\nolimits_{i} {\sum\nolimits_{j} {(i,j)p(i,j) - \mu_{x} \mu_{y} \mu } } }}{{\sigma_{x} \sigma_{y} }} \)
-
4.
Sum of squares: variance: \( \sum\limits_{i} {\sum\limits_{j} {(1 - \mu )^{2} } } p(i,j) \)
-
5.
Inverse difference moment: \( \sum\limits_{i} {\sum\limits_{j} {\frac{1}{{1 + (i - j)^{2} }}p(i,j)} } \)
-
6.
Sum average: \( \sum\limits_{i = 2}^{{2N_{g} }} {ip_{x + y} (i)} \)
-
7.
Sum variance: \( \sum\limits_{i = 2}^{{2N_{g} }} {(i - f_{8} )^{2} } p_{x + y} (i) \)
-
8.
Sum entropy: \( - \sum\limits_{i = 2}^{{2N_{g} }} {p_{x + y} (i)\log \{ p_{x + y} (i)\} = f_{8} } \)
-
9.
Entropy: \( - \sum\limits_{i} {\sum\limits_{j} {p(i,j)\log (p(i,j))} } \)
-
10.
Difference variance: \( \sum\limits_{i = 0}^{{N_{g} - 1}} {i^{2} } p_{x - y} (i) \)
-
11.
Difference entropy: \( - \sum\limits_{i = 0}^{{N_{g} - 1}} {p_{x - y} (i)\log \{ p_{x - y} (i)\} } \)
-
12.
Information measure of correlation 1: \( \frac{HXY - HXY1}{{\hbox{max} \{ HX, HY\} }} \)
-
13.
Information measure of correlation 2: \( (1 - \exp [ - 2(HXY2 - HXY)])^{{{1 \mathord{\left/ {\vphantom {1 2}} \right. \kern-0pt} 2}}} \)
where,
$$ \begin{aligned} {\text{HXY}} & = - \sum\limits_{i} {\sum\limits_{j} {p(i,j)\log (p(i,j))} } , \\ {\text{HXY1}} & = - \sum\limits_{i} {\sum\limits_{j} {p(i,j)\log \{ p_{x} (i)p_{y} (j)\} } } , \\ {\text{HXY2}} & = - \sum\limits_{i} {\sum\limits_{j} {p_{x} (i)p_{y} (j)\log \{ p_{x} (i)p_{y} (j)\} } } \\ \end{aligned} $$ -
14.
Maximum correlation coefficient: \( Q(i,j) = \sum\limits_{k} {\frac{p(i,k)p(j,k)}{{p_{x} (i)p_{y} (k)}}}. \)
Figure 2 shows the sample Haralick’s feature extraction output screen of our proposed work.
5 Feature Selection
We are using PCA as a feature selection algorithm. PCA is useful when we have obtained features on large number of attributes and believe that there is some redundancy in those features. In our case, redundancy means that some of the features are correlated with one another, possibly because they are measuring the same construct. Because of this redundancy, it should be possible to reduce the observed attributes into a smaller number of principal components (artificial attributes) that will account for most of the variance in the observed attributes.
After extracting fourteen texture features, the database is normalized using the z-transform and rescales the feature values. Then, using PCA transformation, we have selected five best features (i.e.) ASM, entropy, inverse difference moment, inertia, and correlation for MRI scan image retrieval using variance as 0.95. So, instead of using fourteen Haralick’s texture features, we are using only five texture features for best MRI image retrieval.
6 K-means Clustering
K-means clustering is one of the simplest unsupervised learning algorithm that solves the clustering problem. The procedure follows a simple and easy way to classify a given data set through ‘K’ number of clusters. In this work, given 1,250 normal and abnormal MRI scan images as training images in a 5-dimensional metric space, determine a partition of the images into maximum 20 clusters and 100 iterations, such that the images in a cluster are more similar cases to each other than two images in different clusters.
We initialize 20 clusters by arbitrarily selecting one image to represent each cluster. Each of the remaining images is assigned to a cluster, and the clustering criterion is used to calculate the cluster mean. These means are used as the new cluster points, and each image is reassigned to the cluster that it is most similar to. This continues until there is no longer change when the clusters are recalculated.
7 Image Retrieval
The Euclidean distance is calculated between the query image and the clustered images. If x i and y i are 2D feature vectors of the clustered training images and query image, respectively, then the distance measure is defined as,
The calculated distances are sorted in increasing order and display the first N images as the best similar MRI scan images for medical treatment. The sample output screen for MRI brain image retrieval is shown in Fig. 3.
8 Experiments and Results
This method is implemented on a computer system using Java as the programming language and MS Access as the backend. In this work, we used around 1,250 MRI scan images as a training set and 100 MRI scan images as testing set in BMP format with the size of 256 × 256 as a database. Two parts of the human body MRI scan images such as 900 spine and 450 brain images are used. The nature of clustering of the system is to cluster the normal and abnormal human body MRI scan images for spine and brain using 1,250 training data set. The effectiveness of the K-means clustering algorithm can be measured by accuracy, sensitivity, and specificity.
The K-means clustering algorithm gave a test accuracy of 85.2 % while using fourteen Haralick’s texture features. The proposed PCA-based feature selection transformation with five Haralick’s features gave the test accuracy of 95.6 %. Figure 4 shows the empirical receiver operating characteristic curves in support of various cutoff points with a false-positive rate on the X-axis and true-positive rate on the Y-axis.
The effectiveness of the proposed method can be measured by precision and recall, which are often referred together since they measure the different aspects of the system performance. The results are given in the following Fig. 5.
9 Conclusion
In this paper, we have proposed an efficient MRI scan image retrieval system using PCA-based optimized Haralick’s texture features. The experimental results demonstrate that the proposed method has the best accuracy, precision, and recall rate than usual Haralick’s texture feature-based MRI scan image retrieval methods. We have planned to extend our work with all types of human body scan images.
References
H. Muller, N. Michoux, D. Bandon, A. Geissbuhler, A review of content-based image retrieval systems in medical applications: clinical benefits and future directions. Int. J. Med. Inform. 73, 1–23 (2004)
X.S. Zhou, S. Zillner, M. Moeller et al., Semantics and CBIR: a medical imaging perspective, in ACM Conference on Content-based Image and Video Retrieval (2008) pp. 571–580
C.B. Akgül, D.L. Rubin, S. Napel, C.F. Beaulieu et al., Content-based image retrieval in radiology: current status and future directions. J. Dig. Imag. 24(2), 208–222 (2011)
B. Ramamurthy, K.R. Chandran, Content based medical image retrieval with texture content using gray level co-occurrence matrix and K-means clustering algorithms. J. Comput. Sci. 8(7), 1070–1076 (2012)
U. Sinha, H. Kangarloo, Principal component analysis for content-based image retrieval. Radiographics 22, 1271–1289 (2002)
G.N. Lee, H. Fujita, K-means clustering for classifying unlabelled MRI data, in IEEE Conference of the Australian Pattern Recognition Society on Digital Image Computing Techniques and Applications (2007) pp. 92–98
S. Ayyachamy, V.S. Manivannan, Distance measures for medical image retrieval. Int. J. Imag. Syst. Technol. 23(1), 9–21 (2013)
R. Haralick, K. Shanmugam, I. Dinstein, Textural features for image classification. IEEE Trans. Syst. Man. Cybern. 3(6), 610–621 (1973)
H. Shao, W.C. Cui, H. Zhao, Medical image retrieval based on visual contents and text information, in IEEE International Conference on Systems (2004) pp. 1098–1103
A.G. Selvarani, S. Annadurai, Medical image retrieval by combining low level features and Dicom features, in IEEE International Conference on Computational Intelligent and Multimedia Applications (2007), pp. 587–589
W. Horsthemke, D. Raicu, J. Furst, Task-oriented medical image retrieval, in MICCAI Work Shop Proceedings (2007) pp. 31–44
P. Zhang, H. Zhu, Medical image retrieval based on co-occurrence matrix and edge histogram, in IEEE conference on multimedia technology (2011) pp. 5434–5437
T.R. Sivapriya, V. Saravanan, P. Ranjit Jeba Thangaiah, Texture analysis of brain MRI and classification with BPN for the diagnosis of dementia. Eng. Inf. Technol. Commun. Comput. Inf. Sci. 20(4), 553–563 (2011)
B.G. Prasad, A.N. Krishna, Statistical texture feature-based retrieval and performance evaluation of CT brain images, in IEEE International Conference on Electronics Computer Technology (2011) pp. 1–4
N. Kumaran, R. Bhavani, Spine MRI image retrieval using texture features. Int. J. Comput. Appl. 46(24), 1–7 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer India
About this paper
Cite this paper
Kumaran, N., Bhavani, R. (2015). PCA-Based Feature Selection for MRI Image Retrieval System Using Texture Features. In: Suresh, L., Dash, S., Panigrahi, B. (eds) Artificial Intelligence and Evolutionary Algorithms in Engineering Systems. Advances in Intelligent Systems and Computing, vol 324. Springer, New Delhi. https://doi.org/10.1007/978-81-322-2126-5_13
Download citation
DOI: https://doi.org/10.1007/978-81-322-2126-5_13
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-2125-8
Online ISBN: 978-81-322-2126-5
eBook Packages: EngineeringEngineering (R0)