Abstract
Both abiotic and biotic stresses adversely affect the plant growth and productivity including crop plants. The development of stress-tolerant plants will be greatly advantageous for modern agriculture in areas that are prone to such stresses. In recent times, several advances have been made towards identifying potential stress-related genes which are capable of increasing the tolerance of plants to both abiotic and biotic stresses. This interaction between biotic and abiotic stresses is controlled by hormone signaling pathways that may induce or antagonize one another, in particular that of abscisic acid. Specificity in multiple stress responses is further controlled by a range of biochemical and molecular mechanisms that act together in a complex regulatory network. Transcription factors, kinase cascades, and reactive oxygen species are key components of this cross talk as are heat shock factors and small RNAs. This review emphasis on elucidating the proteins associated with abiotic and biotic stresses in plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Reactive Oxygen Species
- Abiotic Stress
- Cold Stress
- Late Embryogenesis Abundant
- Late Embryogenesis Abundant Protein
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Plants need suitable physiological condition as well as essential nutrients to grow in the natural environment. As a consequence of their sessile growth habit, plants have adapted dynamic responses to these stresses at the physiological, biochemical, and molecular levels, thus enabling them to survive under different environmental conditions. Most of the crop plants grow in environments that are suboptimal, which prevents the plants from attaining their full genetic potential for growth and reproduction (Bray et al. 2000; Rockstrom and Falkenmark 2000). Abiotic stress factors such as heat, cold, drought, salinity, and nutrient stress have a huge impact on world agriculture, and it has been suggested that they reduce average yields by >50% for most major crop plants (Wang et al. 2003). Further to this, plants must defend themselves from attack by a vast range of pests and pathogens, including fungi, bacteria, viruses, nematodes, and herbivorous insects (Hammond-Kosack and Jones 2000). Each stress elicits a complex cellular and molecular response system implemented by the plant in order to prevent damage and ensure survival, but often at the detriment of growth and yield (Herms and Mattson 1992). Industrial activity combined with a low conscience of the consequences of environmental pollution during a long period created a worldwide problem of soil, air, and water contamination with various pollutants. Heavy metals are among the most widespread soil contaminants. Abiotic stresses usually cause protein dysfunction. The yield and quality of cereals are severely affected by heat stress in many countries (Treglia et al. 1999). Heat stress affects the grain yield and quality. Drought, soil salinity, and heavy metal significantly affect plant growth, development, and productivity, thus posing a severe threat to agriculture throughout the world. Among abiotic stresses, osmotic stress is one of the most severe, caused by drought, high salinity, and cold stresses in nature. Water is the most wide-ranging difficulty among abiotic stresses for production of crop in the world environment. Strategy is to obtain plants with higher performance under water stress conditions by identifying and modifying the molecular mechanisms that take place when the water availability becomes limiting. Plants, as sessile organisms, rely on proteomic plasticity to remodel themselves during periods of developmental change and to respond to biotic and abiotic stresses. In the last decade, methodological improvements have allowed comparative proteomic investigations of plants under stress which have allowed us to analyze biochemical pathways and the complex response of plants to environmental stimuli (Qureshi et al. 2007). More comprehensive approaches that include quantitative and qualitative analyses of gene expression products are necessary at the transcriptome, proteome, and metabolome levels. Abiotic stresses resulted the cellular dehydration, such as freezing and salt and water stress, often lead to similar changes in plant gene expression and metabolism (Cook et al. 2004; Kreps et al. 2002). The phytohormone abscisic acid (ABA) is produced under abiotic stress such as drought and high salinity. ABA is a key mediator in controlling plant response to abiotic stress by regulating stomatal closure and by triggering the activation of many stress-related genes, thereby increasing the tolerance of plants to the stresses. Many abiotic stress-responsive genes have been identified in plants, including rice and Arabidopsis, by using molecular techniques (Fowler and Thomashow 2002; Rabbani et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2006; Nakashima et al. 2009). Stress-induced genes not only function to protect cells from abiotic stress through the production of important enzymes and metabolic proteins (functional proteins) but they also regulate signal transduction and gene expression in the stress response (regulatory proteins). Functional proteins contain hydrophilic proteins including dehydrins and “late embryogenesis abundant” (LEA) proteins and also enzymes that are required for the synthesis of osmoprotectants such as proline and sugars. Regulatory proteins that are activated in response to abiotic stresses, including transcription factors (TFs) such as DREBs (dehydration-responsive element-binding proteins), AREBs (ABA-responsive element-binding proteins), and NAC proteins, have been identified in Arabidopsis and rice (Yamaguchi-Shinozaki and Shinozaki 2006; Nakashima et al. 2009, 2012). Proteomics also makes an essential bridge between the transcriptome and metabolome (Wang et al. 2004; Gray and Heath 2005), complementing genomics research. Only by grouping all this information together is it possible to achieve a comprehensive and exhaustive analysis of the mechanism of plant defense against abiotic and biotic stresses. Upon several stress responses, protein, protein–protein interaction, and posttranslation modification have been also identified (Salekdeh et al. 2002b). Plants have evolved to live in environments where they are often exposed to different stress factors in combination. Being sessile, they have developed specific mechanisms that allow them to detect precise environmental changes and respond to complex stress conditions, minimizing damage while conserving valuable resources for growth and reproduction. Plants activate a specific and unique stress response when subjected to a combination of multiple stresses (Rizhsky et al. 2002). Keeping in the view of the above fact, the present review highlighted the abiotic and biotic stresses in plants in proteomic approach.
Abiotic Stress in Plants
Abiotic stresses limit the productivity and growth potential of plants. Phytohormone like abscisic acid (ABA) is a key mediator in controlling plant response to abiotic stress by triggering the activation of many stress-related genes, thereby increasing the tolerance of plants to the stresses. Stress-induced genes not only function to protect cells from abiotic stress through the production of important enzymes and metabolic proteins (functional proteins) but they also regulate signal transduction and gene expression in the stress response (regulatory proteins). Functional proteins are required for the synthesis of osmoprotectants such as proline and sugars. Regulatory proteins that are activated in response to abiotic stresses, including transcription factors (TFs) such as DREBs (dehydration-responsive element-binding proteins), AREBs (ABA-responsive element-binding proteins), and NAC proteins, have been identified in Arabidopsis and rice (Yamaguchi-Shinozaki and Shinozaki 2006; Nakashima et al. 2009). Nakashima et al. (2012) reported that NAC proteins are plant-specific transcription factors and more than 100 NAC genes have been identified in Arabidopsis and rice to date. Tran et al. (2010) reported that NAC transcriptional factors which constitute one of the largest families of plant specific and help to enhance tolerance against various abiotic stresses. The cDNA encoding a NAC protein was first reported as the responsive to dehydration 26 (RD26) gene in Arabidopsis (Yamaguchi-Shinozaki et al. 1992). Many NAC proteins, including Arabidopsis CUC2, have important functions in plant development. Some NAC genes are upregulated during wounding and bacterial infection (Collinge and Boller 2001; Mysore et al. 2002; Hegedus et al. 2003), whereas others mediate viral resistance (Xie et al. 1999). NAC proteins were thought to be transcriptional activators as the Arabidopsis ATAF1/2 proteins can activate the CaMV 35S promoter in yeast cells. The Arabidopsis AtNAM (NARS2) protein was confirmed by Duval et al. (2002) to function as a transcriptional activator in a yeast system. Kikuchi et al. (2000) reported eight NAC genes in rice (OsNAC1 to OsNAC8) which encode proteins with a single NAC domain. Each OsNAC gene has a unique tissue-specific expression pattern, suggesting this family regulates the development of rice. Ooka et al. (2003) performed a comprehensive analysis of NAC family genes in rice and Arabidopsis. They identified 75 predicted NAC proteins in full-length cDNA datasets of rice and 105 predicted genes in the Arabidopsis genome. Recently, Nuruzzaman et al. (2010) conducted a genome-wide analysis of the NAC transcription factor family in rice and Arabidopsis by investigating 151 nonredundant NAC genes in rice and 117 in Arabidopsis. Morishita et al. (2009) also reported that ANAC078 in the NAC group is responsive to a combination of high light and heat stress. They also demonstrated that ANAC078 regulates flavonoid biosynthesis, leading to the accumulation of anthocyanins under high-light conditions. Hu et al. (2006) reported that the overexpression of the stress-responsive SNAC1 gene increased drought and salt tolerance in rice. SNAC1 enhanced drought resistance in transgenic rice plants at the reproductive stage during growth in fields under severe drought stress without affecting yields (Hu et al. 2006). Nogueira et al. (2005) reported that the SsNAC23 gene, which is homologous to the rice OsNAC5, is associated with cold, herbivory, and water stress in sugarcane. Tran et al. (2009) identified 31 unigenes containing complete open reading frames encoding GmNAC proteins in soybean. Analysis of the C-terminal regulatory domain using a yeast one-hybrid system indicated that among the 31 GmNAC proteins, 28 have transcriptional activation activity. Among them, nine GmNAC genes are induced by dehydration stress with differential induction levels in both shoots and roots. Sperotto et al. (2009) reported that OsNAC5 expression is upregulated by natural (aging) and induced senescence processes. They suggested that OsNAC5 is a novel senescence-associated ABA-dependent NAC transcription factor. The stress-responsive NAC proteins, including OsNAC5, might also function during the process of senescence as well as stress tolerance mediated through ABA. Yang et al. (2011) identified a NAC transcription factor VND INTERACTING2 (VNI2) that mediates signaling cross talk between salt stress response and the process of leaf aging. VNI2 regulates the cold-regulated (COR) and responsive to dehydration (RD) genes by binding directly to their promoters. Overexpression of COR or RD led to prolonged leaf longevity, as observed in the VNI2-overexpressing transgenic plants. Mostafa Kamal et al. (2010a, b) identified GTP-binding proteins, which are involved in signal transduction mechanism in plant systems, and these proteins regulate a flow of kinases that play a vital role in environmental stress signal transduction, and also high temperature seems to upregulate the synthesis of GTP-binding proteins resulting in increased kinase activity (Grover et al. 2001). Some heat-upregulated proteins showed the similarities to elongation factors (EF) and eucaryotic translation initiation factors (eIFs). Heat shock involves changes in the expression patterns of the eIFs, the EF 1-beta, EF 1-alpha, EF Tu, and eIF (4A, 4B, 4E, 4E-1, 4E-2, 5A-1, 5A-2, SUII), in wheat leaves (Gallie et al. 1998). Calcium is a universal molecule in both animals and plants, and the transient increase in Ca2+ level during heat stress is well documented in plants. Heat shock triggers cytosolic Ca2+ bursts, which is transducted by Ca2+ binding proteins(CBP) such as calmodin (CaM), calcineurin (CBL), and annexin and then upregulates the expression of heat shock proteins (HSPs) (Liu et al. 2003). Tubulin proteins are coupled to GTP-binding proteins, which play a role in heat resistance in plant (Segal and Feldman 1996). Mostafa Kamal et al. (2010a, b) also identified serine carboxypeptidase, glucose-1-phosphate, glucose-6-phosphate, and S-adenosyl-methionine synthetase proteins. Different abscisic acid-responsive proteins, LEA protein such as chaperonin, cys peroxiredoxin, ethylene response, and elongation factor TU are responsible for drought stress (Mostafa Kamal et al. 2010a, b). They also observed that cyclin-dependent kinase like zinc finger, transcription factor like MYB, lipid transfer proteins and WRKY are effective for drought stress. Cyclophilin, aquaporin, and chitinase play an important role in cold (upregulated) stress of wheat as reported by Houde et al. (2006). The effects of dehydration, cold temperature treatment, and osmotic and salt stress on the expression of an abscisic acid-responsive protein kinase mRNA (PKABA1) were determined in wheat seedlings (Holappa and Simmons 1995). Mitogen has three protein kinases, MAPK (mitogen-activated protein kinase) and a ribosomal kinase homologue, increased markedly and simultaneously when plants were treated with low temperature (Zhang et al. 2006). GLPs (glucagon-like peptide) function primarily as superoxide dismutase (SOD) to protect plants from the effects of oxidative stress (Khuri et al. 2001). ABA is a central regulator of many plant responses to environmental stresses, particularly osmotic stresses (Chinnusamy et al. 2008; Cramer 2010; Hubbard et al. 2010; Kim et al. 2010). The essential components of ABA signaling include receptors (PYR/PYL/RCAR), protein phosphatases (PP2C), and protein kinases (SnRK2/OST1) (Ma et al. 2009; Park et al. 2009). The PYR/PYL/RCAR proteins were identified as soluble ABA receptors by two independent groups as reported by Park et al. (2009). Leung and Giraudat (1998) identified 2C-type protein phosphatases (PP2C) including ABI1 and ABI2 from the ABA-insensitive Arabidopsis mutants abi1-1 and abi2-1, and they act as global negative regulators of ABA signaling. In yeast, the well-documented central regulators of protein synthesis and energy are SnRK1 (Snf1/MAPK) (mitogen-activated protein kinase), TOR1, and GCN2 (Zaborske et al. 2010; Staschke et al. 2010; Smeekens et al. 2010). These proteins are largely controlled by the phosphorylation of enzymes; all three are protein kinases acting as key hubs in the coordination of metabolism during stressful conditions (Hey et al. 2010). In plants, target of rapamycin (TOR) activity is inhibited by osmotic stress and ABA (Deprost et al. 2007), and GCN2 activity is stimulated by UV light, amino acid starvation, ethylene, and cold stress (Lageix et al. 2008).
Water Stress
Drought is one of the major limiting factors of plant production worldwide. Plant adaptation to drought is the result of many different physiological and molecular mechanisms. Several studies have shown that physiological adaptations to water stress were associated with drought-induced proteins (Bray 1997). Proteomics has proved to be a powerful tool for the identification of proteins and mechanisms involved in drought response and tolerance (Riccardi et al. 1998; Kawasaki et al. 2000; Salekdeh et al. 2002a, b; Hajheidari et al. 2005). Out of 78 responsive proteins, 16 were identified, including proteins involved in the water stress response, the basic metabolic pathway, and lignification. Studies on mild drought stress in rice leaves showed that drought-induced changes in about 42 proteins were reversed completely within 10 days of rewatering. Molecular analyses of wild species such as drought-tolerant grasses would provide a better insight into genes and mechanisms by which plants may adapt to prolonged drought. Tolerant plants are able to maintain tissue water content, or survive a reduction in tissue water content, or recover more completely after rewatering (Cabuslay et al. 1999). The level of proline increased up to 20-fold in response to severe drought conditions as reported earlier (Pedrol et al. 2000; Clifford et al. 1998). Proline is considered as a compatible solute (Samaras et al. 1995) and an osmoprotectant (Serrano and Gaxiola 1994; Okuma et al. 2000). Proline may confer a protective effect by protecting or inducing stress-protective proteins (Khedr et al. 2003). They reported that severe salt stress caused an inhibition of the antioxidative enzymes catalase and peroxidase in Pancratium maritimum L., but the activity of these enzymes was also maintained significantly higher in the presence of proline. Therefore, plant cells require two different mechanisms which will enable the detoxification of excess ROS and fine modulation of ROS for signaling purposes. SOD acts as a first line of defense converting superoxide to the less toxic hydrogen peroxide molecules. The abundance of the cytosolic Cu-Zn SOD of Elymus elongatum increased up to twofold in response to drought. Cytosolic Cu-Zn SOD was shown to be upregulated in rice (Salekdeh et al. 2002b) and sugar beet (Hajheidari et al. 2005). The upregulation of this enzyme in plants grown under progressive stress reveals its important role in response to drought. The detoxification of H2O2 is accomplished with ascorbate peroxidase, glutathione peroxidase, catalase, and 2-Cys peroxiredoxin. Ascorbate peroxidase reduces H2O2 to water, with the concomitant generation of monodehydroascorbate (MDHA). MDHA is a radical with a short lifetime that disproportionates to ascorbate and dehydroascorbate (DHA). DHA is reduced to ascorbate by the action of DHA reductase, using glutathione as the reducing substrate. The most upregulated protein (4.8-fold) was identified as dehydroascorbate reductase suggesting its strong role in the detoxification of H2O2. Hydrogen peroxide (H2O2) is a key regulatory molecule in the response to stresses (Mittler 2002), and its ability to selectively induce a subset of defense genes like glutathione S-transferases and glutathione peroxidases without directly inducing other defense genes. The upregulation of both glutathione S-transferases and glutathione peroxidase may represent such co-expression mechanisms. Wehmeyer et al. (1996) suggested that small heat shock proteins (sHSPs) are among the several factors required for desiccation tolerance. Hajheidari et al. (2005) reported the upregulation of two sHSPs under drought stress in sugar beet grown in the field. These proteins showed sequence homology with abscisic acid- and stress-inducible protein (ASR). The upregulation of this gene has been also reported in response to drought stress in maize (Riccardi et al. 1998) and salt stress in a salt-tolerant rice genotype (Salekdeh et al. 2002a). Iusem et al. (1993) have reported that characterization of cDNA encodes ASR protein whose expression was activated by leaf water deficit and fruit ripening. Silhavy et al. (1995) identified a nuclear-targeting sequence motif in this gene isolated from Solanum chacoense Bitter which is extremely resistant to viruses, insects, and drought.
Phytohormone Stress
A variety of plant hormones, including salicylic acid (SA), jasmonate (JA), ethylene, and abscisic acid, have been implicated in mediating responses to a wide range of biotic and abiotic stresses (Diaz et al. 2002; Thomma et al. 1998; Audenaert et al. 2002). The roles of these hormones are dependent upon the particular host–pathogen interaction (Knoester et al. 1998). On the basis of the interactions of hormones in which resistant responses to biotrophs require SA, whereas responses to necrotrophs require JA and ethylene (Feys and Parker 2000), in some instances, these hormones are involved in determining the level of host basal resistance (Delaney et al. 1994). In case of tomato, ethylene, JA, and SA all independently contribute to its resistance to Botrytis cinerea (Diaz et al. 2002). The host plant actively regulates the Xanthomonas campestris pv. vesicatoria-induced disease response via the sequential action of at least three hormones (JA, ethylene, and SA), which promote expansive cell death of its own tissue (O’Donnell et al. 2003). Further, the effect of phytohormones is also regulated by other factors. For example, the MAPK kinase, EDR1, negatively regulates SA-inducible defenses (Frye et al. 2001), whereas MAPK 4 appears to differentially regulate SA and JA signals (Petersen et al. 2000). These findings also suggest that MAPK modulates cross talk between different plant defense pathways (Hammond-Kosacky and Parkerz 2003).
Osmotic Stress
Among abiotic stresses, osmotic stress is one of the most severe, caused by drought, high salinity and cold stresses in nature. Plants respond to osmotic stress at the morphological, anatomical, cellular, and molecular levels. To cope with osmotic-related stresses, plants have developed various responses such as production of osmolites for osmotic adjustment, synthesis of Na+/H+ antiporters for ion sequestration, and many other mechanisms (Bohnert et al. 1995). AtHKT1, which is a Na+ transporter, mediates osmolality balance between xylem vessels and xylem parenchyma cells (Snarpi et al. 2005). The operation of these responses usually requires three steps: osmotic stress recognition, signal transduction, and production of components for the physiological response (Tamura et al. 2003). In plants, a hybrid-type histidine kinase (AtTHK1) functions as an osmosensor and transmits the stress signal to a downstream mitogen-activated protein kinase (MAPK) cascade (Urao et al. 1999). Lu and Neumann (1999) reported that when rice seedlings were exposed to osmotic stress modulated by polyethylene glycol 6000, growth in emerging first leaves of the intact plant was inhibited. Early inhibition of leaf growth was not related to changes in root size, osmotic potential gradients, or cell wall-yielding characteristics in the leaf expansion zone of stressed seedlings. Deak and Malamy (2005) demonstrated that osmotic stress represses the formation of autonomous lateral roots from lateral root primordia of Arabidopsis, while lateral root initiation was not greatly affected. Abscisic acid (ABA) and a newly identified gene, LRD2, are involved in osmotic repression of lateral root formation. Further examination revealed that both ABA and LRD2 control root system architecture even in the absence of osmotic stress. The molecular mechanisms that mediated responses to environmental cues could also be regulators of intrinsic developmental programs in the root. Zonia and Mnnik (2004) investigated that tobacco pollen tube cell volume changes in response to osmotic perturbation by activation of the phospholipid signaling pathway. Several intermediates in the phospholipid signaling pathway were detected during pollen tube growth. Hypo-osmotic stress induced a rapid increase in phosphatidic acid and a decrease in phosphatidylinositol phosphate. The fact that these signaling molecules are present during normal growth and the mechanism for osmotic response involved components of the biomechanical networks driving pollen tube cell elongation. In osmotically stressed wheat coleoptiles, reduced rates of phenylalanine ammonia-lyase and tyrosine ammonia-lyase activities suppress phenylalanine biosynthesis, resulting in a reduced level of wall-bound ferulic acid. This decrease in wall-bound ferulic acid may lead to reduced levels of diferulic acid, an important contributor to maintaining cell wall extensibility (Wakabayashi et al. 1997). The osmotic potential of soil alters the depth of root systems, the rate of root elongation, and the number of lateral roots (van der Weel et al. 2000); however, there are only a few reports about phenotypic changes in rice under osmotic stress (Lu and Neumann 1999).
Zang and Komatsu (2007) reported that 15 proteins are changed in the basal part of rice leaf sheaths by mannitol treatment. The basal section of leaf sheath may undergo more subtle changes in protein expression than the distal end, where significant morphological changes are evident. They reported that 327 proteins were detected by digital image analysis. Among these, 15 proteins significantly responded to osmotic stress by up- or downregulation: 12 proteins increased in amount and 3 proteins decreased. Ten of the identified proteins are recognized as important components for stress response such as two proteasome degradation system-related proteins, five endoplasmic reticulum (ER)-related proteins, two proteins related to detoxification, and uroporphyrinogen decarboxylase, which is a cell death-related protein (Mock and Grimm 1997). Two proteins matched annotated sequences for putative proteins of unknown function and the three remaining proteins responding to osmotic stress were not similar to other proteins in existing databases.
The glyoxalase system is ubiquitous in nature and consists of two enzymes: glyoxalase I and glyoxalase II, which act coordinately to convert 2-oxoaldehydes into 2-hydroxyacids using reduced glutathione as a cofactor. The glyoxalase pathway involving glyoxalase I and glyoxalase II enzymes is required for glutathione-based detoxification of methylglyoxal. Methylglyoxal is a primary physiological substrate for glyoxalase I. Besides detoxification of methylglyoxal, the glyoxalase system could also play a role in providing tolerance under stress by recycling glutathione that would be “trapped” spontaneously by methylglyoxal to form hemithioacetal, thereby maintaining glutathione homeostasis (Creighyton et al. 1988). Transgenic plants overexpressing glyoxalase I showed significant tolerance to methylglyoxal and high salt (Veena and Sopory 1999; Singla-Pareek et al. 2003).
Heavy Metal Stress
Heavy metals (HMs) are among the most widespread soil contaminants which damage the physiological and metabolic processes. Certain heavy metals such as Cd, Hg, and Pb affect cell systems due to the increasing exposure of living organisms to these metals in the environment (DalCorso et al. 2008). The physiological and molecular basis of plant interactions with the environment has attracted considerable interest in recent years. Being sessile organisms, plants are constantly exposed during their life cycle to adverse environmental conditions that negatively affect growth, development, or productivity. The presence of toxic compounds, such as heavy metals (HMs), is one important factor that can cause damage to plants by altering major plant physiological and metabolic processes (Hossain et al. 2010; Villiers et al. 2011). Different metals and metalloids which are toxic to plants such as copper (Cu), iron (Fe), manganese (Mn), zinc (Zn), nickel (Ni), cobalt (Co), cadmium (Cd), and arsenic (As). Importantly, few HMs and transition metals such as sodium (Na), potassium (K), calcium (Ca), magnesium (Mg), Fe, Cu, Zn, Co, or Ni are essential micronutrients that are critically involved in the functional activities of large numbers of proteins involved in sustaining growth and development. However, at excess concentrations, these metal ions can become detrimental to living organisms, including plants. Although HMs are natural constituents of soils and occur naturally in the environment, contamination of soils by toxic metals and metalloids is of major concern worldwide (Villiers et al. 2011). Depending on their oxidation states, HMs can be highly reactive, resulting in toxicity of plant cells in many ways. At the cellular and molecular level, HM toxicity results in alterations of different plant physiological processes, including inactivation and denaturation of enzymes, proteins, blocking of functional groups of metabolically important molecules, displacement/substitution of essential metal ions from biomolecules and functional cellular units, conformational modifications and disruption of membrane integrity (Sharma and Dubey 2007), which is finally attributed to altered plant metabolism, inhibition of photosynthesis, respiration, and alerted activities of several key enzymes (Sharma and Dietz 2009; Hossain et al. 2010, 2012). In addition, HMs are known to disturb redox homeostasis by stimulating the formation of free radicals and reactive oxygen species (ROS) such as singlet oxygen (1O2), superoxide radicals (O2•−), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH) (Dubey 2011; Anjum et al. CR2402012). Recently, methylglyoxal (MG), a cytotoxic compound, was also found to increase in response various stresses including HMs (Yadav et al. 2005a, b). An increase in MG level in plant cells further intensifies the production of ROS by interfering with different plant physiological and metabolic processes such as inactivation of the antioxidant defense system (Hoque et al. 2010; Hossain et al. 2011a, b) and interfering with vital plant physiological processes such as photosynthesis (Saito et al. 2011). This increase in ROS and MG exposes cells to oxidative stress leading to lipid peroxidation, biological macromolecule deterioration, membrane dismantling, ion leakage, and DNA strand cleavage, and finally death of plants (Navari-Izzo 1998; Romero-Puertas et al. 2002; Barconi et al. 2011).
Heavy metals (HMs) are intracellularly chelated through the synthesis of amino acids, organic acids, GSH, or HM-binding ligands such as metallothioneins (MTs), phytochelatins (PCs), compartmentation fcc within vacuoles, and upregulation of the antioxidant defense and glyoxalase systems to counter the deleterious effects caused by ROS and methylglyoxal (Cobbett 2000; Yadav 2010; Hossain et al. 2012). A large number of recent studies in plants involving sensitive, tolerant, mutant, transgenic, and hyperaccumulator-adopting strategies in the fields of physiology, genomics, proteomics, and metabolomics suggest that GSH by itself and its related metabolizing enzymes, proteins, and peptides play a pivotal role in HM tolerance by controlling different plant physiological processes, including ROS and MG detoxification, heavy metal uptake, translocation, chelation, and detoxification. Yang et al. (2007) reported that a total of 17 Al-responsive proteins were identified, with 12 of those being upregulated and 5 downregulated. Among the upregulated proteins are copper/zinc superoxide dismutase (Cu-Zn SOD), GST, and S-adenosylmethionine synthetase 2, which are the consistently known Al-induced enzymes previously detected at the transcriptional level in other plants. More importantly, a number of other identified proteins including cysteine synthase (CS), 1-aminocyclopropane-1-carboxylate oxidase, G protein b subunit-like protein, abscisic acid- and stress-induced protein, putative Avr9/Cf-9 rapidly elicited protein 141, and a 33 kDa secretory protein are novel Al-induced proteins. Most of these proteins are functionally associated with signaling transduction, antioxidation, and detoxification (Hossain et al. 2012).
Mode of Action of Heavy Metals in Plant Cells
The toxicity of HMs is manifested in many ways when plant cells accumulate them at high levels. HMs are divided into two groups: redox active (Fe, Cu, Cr, Co) and redox inactive (Cd, Zn, Ni, Al, etc.). The redox active HMs are directly involved in the redox reaction in cells and result in the formation of O2 •− and subsequently in H2O2 and OH production via the Haber-Weiss and Fenton reactions (Dietz et al. 1999; Schutzendubel and Polle 2002). Exposure of plants to redox inactive HMs also results in oxidative stress through indirect mechanisms such as interaction with the antioxidant defense system, disruption of the electron transport chain, or induction of lipid peroxidation. The latter can be due to an HM-induced increase in lipoxygenase (LOX) activity. Another important mechanism of HM toxicity is the ability of HMs to bind strongly to oxygen, nitrogen, and sulfur atoms (Nieboer and Richardson 1980). This binding affinity is related to free enthalpy of the formation of the product of the HM and ligand with low solubility of these products. Because of these features, HMs can inactivate enzymes by binding to cysteine residues. Many enzymes need cofactors to work properly for both HM ions (such as Fe2+, Mg2+, Cu2+, Ca2+) and organic molecules (such as heme, biotin, FAD, NAD, or coenzyme A). The displacement of one HM ion by another leads to the inhibition or loss of enzyme activities. Divalent cations such as Co2+, Ni2+, and Zn2+ displace Mg2+ in ribulose-1,5- bisphosphate-carboxylase/oxygenase (RuBisCO) and result in a loss of activity (Wildner and Henkel 1979; Van Assche and Clijsters 1986). Displacement of Ca2+ by Cd2+ in calmodulin, an important protein in cellular signaling, led to the inhibition of calmodulin-dependent phosphodiesterase activity in radish (Rivetta et al. 1997). Additionally, HMs cause membrane damage through various mechanisms, including the oxidation of and cross-linking with protein thiols, inhibition of key membrane protein such as H+-ATPase, or causing changes in the composition and fluidity of membrane lipids (Meharg 1993). Accumulation of MG, a cytotoxic compound, was found to increase in response to HM stress in plants due to impairment of the glyoxalase system that finally elicits oxidative stress by reducing the GSH content (Hossain and Fujita 2010; Hossain et al. 2011a, b, 2012).
Metallothioneins (MTs) are low-molecular-weight (4–8 kDa), Cys-rich, HM-binding, gene-encoded polypeptides that can bind heavy metals via the thiol groups of their Cys residues (Hamer 1986). Although the precise physiological function of MTs has not yet been fully elucidated, proposed roles include participation in maintaining the homeostasis of essential transition HMs, sequestration of toxic HMs, and protection against intracellular oxidative damage (Gasic and Korban 2006). Plant MTs are extremely diverse and have been subdivided into three classes based on the arrangement of the Cys residue (Zhou et al. 2006). The Cys-Cys, Cys-X-Cys, and Cys-X-X-Cys motifs (in which X denotes any amino acid) are characteristic and invariant for MTs. The organization or distribution of cysteine residues confers different MT isoforms and their ability to bind and sequester different HM ions for detoxification and homeostasis. The biosynthesis of MTs is regulated at the transcriptional level and is induced by several factors, including hormones, cytotoxic agents, and HMs, such as Cd, Zn, Hg, Cu, Au, Ag, Co, Ni, and Bi (Kagi 1991). Gene expression studies were performed to quantify mRNA levels in different tissues, at different developmental stages, and under stress conditions such as HM exposure. MT genes appear to be differentially regulated in a tissue-specific manner and in relation to the developmental stage and also in response to a number of stimuli, including heavy metals (Castiglione et al. 2007). Ahn et al. (2012) showed that three Brassica rapa MT genes (BrMT1, BrMT2, and BrMT3) are differentially regulated under various HM stresses.
Cold Stress
Plants are frequently facing different environmental variations. Among these constraints, low temperature is one of the most crucial factors that impair the distribution, growth, and productivity of crops or wild plant species. The mechanisms underlying cold acclimation and tolerance have been intensively studied over the past years, and numerous studies on plant response to cold stress have been reported (Ruelland et al. 2009). Cold weather can result in pollen sterility in plants at the reproductive stage of development. Several studies of plant transcriptomes during cold stress have been performed in rice (Chen et al. 2002a, b; Fowler and Thomashow 2002; Kawamura and Uemura 2003; Amme et al. 2006; Goulas et al. 2006; Cui et al. 2005; Yan et al. 2006; Hashimoto and Komatsu 2007). Hashimoto and Komatsu (2007) reported that some proteins are involved in cold stress, and these are the chilling stress-responsive proteins. In general, the level of mRNA does not always correlate well with the level of protein mainly due to posttranscriptional regulation (Yan et al. 2006). As a global study of the proteins comprising the proteome, proteomics can be playing an increasingly important role in addressing plant response to environmental changes. The proteomes of various plants in response to different environmental factors including drought, salt, heat, and heavy metal have been investigated (Ouerghi et al. 2000; Hajduch et al. 2001; Salekdeh et al. 2002a; Majoul et al. 2003). The differentially expressed proteins have been identified and well-documented stress-responsive proteins and some novel cold-responsive proteins. Plants differ in their cold response and cold-tolerant species may develop efficient strategies to adapt to chilling environment. Bressan et al. (2001) reported that the large-scale transcriptional studies were hindered by the absence of genome sequence information.
ROS-Scavenging Mechanisms in Cold Stress
Cold stress may disturb cellular redox homeostasis and promotes the production of reactive oxygen species (ROS) and reactive aldehydes (as 4-hydroxy-nonenal and methylglyoxal). Plants develop ROS-scavenging mechanisms to cope with the oxidative stress. Because chloroplast is one of the major sources of ROS, four proteins involved in chloroplast redox homeostasis, including 2-cys peroxiredoxin, chloroplast (BAS1), peroxiredoxin-2E, ferritin-1, and alanine-2-oxoglutarate aminotransferase 1 (GGT1) (Gao et al. 2009). BAS1 is involved in the detoxification of alkyl hydroperoxides and its gene expression is regulated under the control of the cellular redox state (Baier and Dietz 1997). Peroxiredoxin-2E reduces hydrogen peroxide and alkyl hydroperoxides with reducing equivalents provided through the thioredoxin or glutaredoxin system and was a component of chloroplast thioredoxin system (Bréhélin et al. 2003). Ferritin-1, an iron binding protein, is to protect plants from oxidative damage induced by manifold stresses (Deák et al. 1999). GGT1, an enzyme involved in photorespiration and photorespiration, has been suggested to be important for maintaining electron flow to prevent photoinhibition under stress conditions (Wingler et al. 2000). Methionine residues of proteins are a major target for oxidation by ROS and peptide methionine sulfoxide reductase plays protective roles in the cellular response to oxidative stress (Kwon et al. 2007). Aldo-keto reductases can detoxify lipid peroxidation products and glycolysis-derived reactive aldehydes that contribute significantly to cellular damages caused by abiotic stresses and improve scavenging capacity of the plant.
Cold stress may impose a great influence on the protein synthesis apparatus in plant. A chloroplast EF-G protein appeared in gel after 2, 5, and 24 days of cold treatment, but there was no spot at the corresponding position in gel of the control group. Because chloroplast EF-G is a housekeeping gene, there must be an EF-G protein spot elsewhere in gel of control group (Gao et al. 2009). It is postulated that posttranslational modification or alternative splicing may occur on EF-G in T. halophila under cold stress to maintain the normal operation of protein synthesis. Although, EF-G was previously reported to play a role in chloroplast biogenesis and development in Arabidopsis. PAB1 was reported to be involved in ubiquitin-dependent protein catabolic process. This may indicate that a high protein turnover rate is needed for plants to eliminate the misfolded polypeptides under cold environment (Amme et al. 2006; Goulas et al. 2006) and also in rice plant (Cui et al. 2005; Yan et al. 2006; Hashimoto and Komatsu 2007). They suggested that the cold stress responses in Thellungiella are similar, in general, to other plant species. Because Thellungiella is a cold-tolerant relative of Arabidopsis, comparison of the cold-induced rosette leaf proteome changes in Thellungiella, and Arabidopsis was performed to advance the understanding of the higher cold tolerance of Thellungiella. The identification of cold-responsive proteins in Thellungiella provides not only new insights into cold stress responses but also a good starting point for further investigation of their functions. However, considering the inability of 2-DE-based proteomic study to resolve membrane proteins and low abundant proteins, integrated transcriptomic, proteomic, and metabolomic approaches should be adopted to gain further insight into plant response to cold stress.
Biotic Stress in Plants
Plants responding to biotic stresses produce several protective compounds and proteins such as pathogenesis-related (PR) proteins, directly disease-related proteins, and other proteins. Biotic is one of the serious stresses affecting plant growth and productivity. A clear perceptive of the molecular mechanisms involved in plants response to biotic stress is of fundamental importance to plant science (Mostafa Kamal et al. 2010a, b). The study of PR proteins is also important for crop production due to the fact that many plant-derived pathogen-related proteins have been identified as members of PR protein families 2, 3, 4, 5, 8, 10, and 14 (Hoffmann-Sommergruber 2002). Among these are pathogenesis-related proteins coded by the host plant that accumulate in response to pathogen infection or other signals related to plant defense responses. Several PR proteins have been characterized at the molecular level and have shown to have antifungal activity in vitro (Datta and Muthukrishnan 1999) and show enzymatic activity such as β-1,3-glucanase and chitinase (PR2 and PR3, respectively), both involved in the degradation of microbial cell wall structural polysaccharides (Legrand et al. 1987) and PR4 and PR9, characterized by ribonuclease and peroxidase activity, respectively (Caporale et al. 2004). Proteomic approaches have been successfully used in discovering the resistance mechanisms in maize against kernel rot caused by Fusarium or Aspergillus (Chen et al. 2004; Chen and Chen 2002; Sonia et al. 2004). Mostafa Kamal (2010a,b) reported that methyl jasmonate (MeJA) and its free acid jasmonic acid (JA) collectively referred to as jasmonates are important cellular regulators involved in diverse developmental process, such as seed germination, root growth, fertility, seed ripening, and senescence. In addition, jasmonate activate plant defense mechanisms in response to insect-driven wounding and various pathogens (Creelman and Mullet 1997). Carmona et al. (1993) reported that different purothionins are active against plant pathogens both in vitro and in vivo. Gamma-1-purothionin showed a higher structural analogy with scorpion toxins and against insect defensins which also present the cystine-stabilized alpha helical (CSH) motif (Bruix et al. 1993) and gamma-2-purothionin inhibits protein translation in cell-free systems resulting in the exhibited plant toxins for pathogen (Colilla et al. 1990). Mostafa Kamal et al. (2010a) indicated that the antifungal activity has been associated with two immunochemically distinct proteins; the proteins are homologous with thaumatin- and pathogenesis-related proteins of the PR5 family. These proteins have intensely sweet properties of thaumatin; multiple unrelated defense functions against insect and fungal pests can now be associated with the family of thaumatin-homologous proteins (Hejgaard et al. 1991). Antimicrobial proteins (MBP-1) inhibit spore germination or hyphal elongation of several plant pathogenic fungi, including two seed pathogens of maize (Fusarium moniliforme Sheld. and Fusarium graminearum (Gibberella zeae (Schw.) Petsch)) and several bacteria, including a bacterial pathogen of maize (Clavibacter michiganense ssp. nebraskense) (Duvick et al. 1992). Wheat calcium-dependent protein kinase (CDPK) genes were found to respond to various biotic and abiotic stimuli, including cold, hydrogen peroxide (H2O2), salt, drought, powdery mildew (Blumeria graminis tritici, BGT), as well as phytohormones abscisic acid (ABA) and gibberellic acid (GA) (Li et al. 2008). Chitinases are important components of plant defense in response to attack by pathogens as F. graminearum (Li et al. 2001). Dilbirligi and Gill (2003) reported that many RGA sequences in wheat are identified as disease resistance gene. Ascorbate peroxidase, peroxidase, and glutathione (GSH)-dependent dehydroascorbate reductase accumulate early in grain fill. SGT1, a component of R-gene-triggered disease resistance, and serpin, a serine protease inhibitor, are also present and may protect the developing grain against various pathogens (Wong et al. 2004). The pathogen resistance proteins such as serpin; chitinase, which hydrolyzes the structural carbohydrate of fungal cell walls; barwin/PR-4 protein, which is induced by fungal pathogens and binds chitin; and xylanase inhibitor protein, which inhibits a fungal enzyme that degrades plant cell walls (Hurkman and Tanaka 2007). Zhang et al. (2008) reported that a total of 375 functional ESTs have been identified by SSH from a cDNA library of resistant soybean varieties associated with Phytophthora root rot. They also observed that system acquired resistance (SAR) induced by Phytophthora in soybean primarily depended on salicylic acid-mediated signaling pathways, which was different from resistance mechanisms in Arabidopsis. Nevertheless, the functions of differentially expressed proteins induced by Phytophthora are almost the same in soybean and Arabidopsis, including material metabolism enzymes and various regulatory factors; this suggests that the molecular basis of system acquired resistance against Phytophthora is much the same in soybean and Arabidopsis (Sun et al. 2008). Plant disease resistance genes usually have conserved motifs such as the nucleotide-binding site (NBS). Based on the NBS domain, two putative resistance genes (KR1 and KR4) were isolated from soybean. Sixteen identified proteins were potentially involved in protein degradation, defense signal transfer, oxidative stress, cell wall reinforcement, and energy and metabolism regulation (Yang, et al. 2010). In addition, differential expression was found by SSH in cDNA libraries from soybean varieties resistant to soybean cyst nematode (Cheng et al. 2007; Lu and Fang 2003).
Interaction with Insect and Pest with Stress
Trichoderma spp. have been known as biocontrol agents for the control of plant diseases (Harman et al. 2004). In many cases, the beneficial fungi may induce systemic resistance that is mediated by alterations in plant gene expression (Alfano et al. 2007; Shoresh et al. 2010). They also reported that the plant in association with Trichoderma strains enhanced plant growth, but the effects, as with other plant growth-promoting microbes (Gamalero et al. 2009), are greater when plants are under suboptimal conditions or biotic, abiotic, or physiological stresses (Yildirim et al. 2006). Several recent reports indicate that the fungi enhances tolerance to abiotic stresses during plant growth, in part due to improved root growth, improvement in water-holding capacity of plants (Harman 2000; Bae et al. 2009), or enhancement in nutrient uptake; whereas, in the absence of stress, plant growth may or may not be enhanced. Although molecular studies indicate greater expression of gene families involved in plant protection against abiotic stresses or oxidative damage in Trichoderma spp.-treated plants, no experimental evidence has been presented correlating enhanced tolerance of plants colonized with biocontrol fungi to these changes in molecular level. These fungi are frequently applied as seed treatments, where they may improve plant stands and induce long-term improvements in plant quality (Harman 2000, 2006).
Effects of Abiotic Stress on Plants: A Systems Biology
Recent advances in biotechnology have dramatically changed our capabilities for gene discovery and functional genomics. For the first time, we can now obtain a holistic “snapshot” of a cell with transcript, protein, and metabolite profiling (Umezawa 2011). Such a “systems biology” approach allows for a deeper understanding of physiologically complex processes and cellular function (Kitano 2002). Understanding the function of genes is a major challenge of the post-genomic era. While many of the functions of individual parts are unknown, their function can sometimes be inferred through association with other known parts, providing a better understanding of the biological system as a whole. High-throughput -omics technologies are facilitating the identification of new genes and gene function. In addition, network reconstructions at the genome scale are key to quantifying and characterizing the genotype to phenotype relationships (Feist and Palsson 2008).
Fundamentally, plants require energy (light), water, carbon, and mineral nutrients for growth. Abiotic stress reduces growth and yield below the optimum levels. Plant responses to abiotic stresses are dynamic and complex system (Skirycz and Inze 2010; Cramer 2010); they are both elastic (reversible) and plastic (irreversible). The plant responses to stress are dependent on the tissue or organ affected by the stress. Transcriptional responses to stress are tissue or cell specific in roots and are quite different depending on the stress involved (Dinneny et al. 2008). In addition, the level and duration of stress (acute vs. chronic) can have a significant effect on the complexity of the response (Tattersall et al. 2007). Water deficit inhibits plant growth by reducing water uptake into the expanding cells and alters enzymatically the rheological properties of the cell wall, for example, by the activity of ROS (reactive oxygen species) on cell wall enzymes (Skirycz and Inze 2010). In addition, water deficit alters the cell wall nonenzymatically, for example, by the interaction of pectate and calcium (Boyer 2009). Furthermore, water conductance to the expanding cells is affected by aquaporin activity and xylem embolism (Parent et al. 2009; Boursiac et al. 2008). With long-term stress, photosynthesis declines due to stomatal limitations for CO2 uptake and increased photoinhibition from difficulties in dissipating excess light energy (Pinheiro and Chaves 2011). One of the earliest metabolic responses to abiotic stresses and the inhibition of growth is the inhibition of protein synthesis (Good and Zaplachinski 1994; Vincent et al. 2007) and an increase in protein folding and processing (Liu and Howell 2010). Energy metabolism is affected as the stress becomes more severe (e.g., sugars, lipids, and photosynthesis) (Cramer et al. 2007; Kilian et al. 2007). Thus, there are gradual and complex changes in metabolism in response to stress.
Regulatory Mechanism
The plant molecular responses to abiotic stresses involve interactions and cross-link with many molecular pathways (Takahashi et al. 2004). Reaction oxygen species (ROS) signaling in response to abiotic stresses and its interactions with hormones has been thoroughly reviewed (Mittler et al. 2010). ROS and RNS (reactive nitrogen species) form a coordinated network that regulates many plant responses to the environment. There are a large number of studies on the oxidative effects of ROS on plant responses to abiotic stress, but only a few studies documenting the nitrosative effects of RNS (Molassiotis and Fotopoulos 2011). Hormones are also important regulators of plant responses to abiotic stress. The two most important hormones are abscisic acid (ABA) and ethylene. ABA is a central regulator of many plant responses to environmental stresses, particularly osmotic stresses (Cramer 2010; Kim et al. 2010; Chinnusamy et al. 2008). There are slower responses to ABA involving transcriptional responses that regulate growth, germination, and protective mechanisms. Recently, the essential components of ABA signaling have been identified, and their mode of action was elaborated (Umezawa et al. 2010). The current model of ABA signaling includes three core components, receptors (PYR/PYL/RCAR), protein phosphatases (PP2C), and protein kinases (SnRK2/OST1). The PYR/PYL/RCAR proteins were identified as soluble ABA receptors by two independent groups (Ma et al. 2009; Park et al. 2009). The 2C-type protein phosphatases (PP2C) including ABI1 and ABI2 were first identified from the ABA-insensitive Arabidopsis mutants abi1-1 and abi2-1, and they act as global negative regulators of ABA signaling (Leung and Giraudat 1998). Yamaguchi-Shinozaki and Shinozaki (2006) reported that the transcriptional regulation of dehydration and salinity stresses have revealed both ABA-dependent and ABA-independent pathways. Cellular dehydration under water-limited conditions induces an increase in endogenous ABA levels that trigger downstream target genes encoding signaling factors, transcription factors, metabolic enzymes, and others. In the post-genomic era, comprehensive analyses using three systematic approaches have increased our understanding of the complex molecular regulatory networks associated with stress adaptation and tolerance. The first one is “transcriptomics” for the analysis of coding and noncoding RNAs and their expression profiles. The second one is “metabolomics” that is a powerful tool to analyze a large number of metabolites. The third one is “proteomics” in which protein and protein modification profiles offer an unprecedented understanding of regulatory networks. Protein complexes involved in signaling have been analyzed by a proteomics approach (Kaufmann et al. 2011). Hirai et al. (2004, 2007) identified MYB transcription factors regulating glucosinolate biosynthesis in Arabidopsis in response to S and N deficiency using an integrated transcriptomics and metabolomics approach. Genes and metabolites in glucosinolate metabolism were found to be coordinately regulated (Hirai et al. 2004). Co-expression analysis was used to identify two MYB transcription factors that positively regulate glucosinolate metabolism (Hirai et al. 2007). Mao et al. (2009) performed a gene co-expression network analysis of 1,094 microarrays of Arabidopsis using a nontargeted approach. They identified 382 modules in this network. The top three modules with the most nodes were photosynthesis, response to oxidative stress, and protein synthesis. Many of the modules also involved responses to environmental stresses. They constructed a cold-induced gene network from a subset of microarrays. Weston et al. (2008) used weighted co-expression analysis to define six modules for Arabidopsis responses to abiotic stress. Two hubs in the common response module were an ankyrin-repeat protein and genes involved in Ca signaling. They created a compendium of genomic signatures and linked them to their co-expression analysis.
Proteomics Approach in Biotic Stress
High temperature and drought, which often occur together during the growing season and likely contribute to poor kernel development, have been reported to increase growth of the fungus and toxin production (Payne 1998). Jones et al. (1981) reported that irrigating corn fields to reduce drought stress also reduced fungal infection and aflatoxin contamination. Irrigation not only relieved drought stress but also reduced soil temperature. Lower soil temperature was found to reduce aflatoxin contamination in peanut (Hill et al. 1983). Increased aflatoxin contamination was observed in drought-treated peanuts with increased soil temperatures (Cole et al. 1985). Dorner et al. (1989) also reported that a higher soil temperature favors Aspergillus flavus growth and aflatoxin production. A study on the effect of drought on peanut resistance to A. flavus by Wotton and Strange (1987) found that fungal colonization was inversely related to water supply as was aflatoxin production. Payne et al. (1986) also concluded that water stress appears to be a major factor affecting aflatoxin contamination. Studies of aflatoxin and fumonisin contamination of corn grown under high or moderate heat stress (Abbas et al. 2002; Chen et al. 2004) demonstrate that heat stress also plays an important role in the susceptibility of corn to both aflatoxin and fumonisin contamination. Tubajika and Damann (2001) compared ear rot and aflatoxin production between nine drought-tolerant and two aflatoxin-resistant corn lines under field and laboratory conditions. They found that drought-tolerant lines all had significantly lower levels of ear rot and aflatoxin contamination compared to the aflatoxin-resistant controls, when grown under drought conditions (Tubajika and Damann 2001). A proteomics approach was recently employed to identify proteins whose level of expression associated with kernel resistance against A. flavus infection and aflatoxin production (Chen et al. 2002a, b). They found over a dozen proteins, either unique or fivefold upregulated in resistant lines, have been identified and sequenced (Chen et al. 2002a, b, 2004). These proteins can be grouped into three categories based on their peptide sequence homology, that is, storage proteins, such as globulin 1, globulin 2, and late embryogenesis abundant proteins (LEA3, LEA14); stress-related proteins, such as an aldose reductase (ALD), a peroxiredoxin antioxidant (PER1), a cold-regulated protein, a water stress-inducible protein, an anionic peroxidase, a glyoxalase I protein (GLX I), and several small heat shock proteins (HSP); and antifungal proteins, which include a trypsin inhibitor and a pathogenesis-related protein. The majority of protein identified was stress-related proteins and highly hydrophilic storage proteins. These data suggest that kernel resistance may require not only the presence of high levels of antifungal proteins but also that of high levels of stress-related proteins and highly hydrophilic storage proteins. Storage proteins have been reported to play an important role in stress tolerance. Chen et al. (2002a, b) reported that the members of the LEA genes family have been associated with plant responses to many different stresses including drought, salt, cold, heat, and wounding (Thomann et al. 1992). Transgenic expression of an LEA protein from barley demonstrated increased tolerance to water and salt stress in rice (Xu et al. 1996). Some stress-related proteins have been reported to not only confer stress tolerance but also enhance disease resistance. The expression of heat shock proteins, especially the small HSPs under stress, has been widely studied (Vierling 1991) and shown to possess molecular chaperone activity (Jacob et al. 1993). Aside from heat stress, HSPs are also induced by other stresses such as cold, drought, or salinity (Sabehat et al. 1998). The role of glyoxalase in stress tolerance is also highlighted in a recent study using transgenic tobacco plants overexpressing a Brassica juncea glyoxalase I (Veena et al. 1999). Park et al. (2001) and Shin et al. (2002) also found that transgenic expression of the tobacco stress-inducible gene 1 (Tsi1) induced expression of several pathogenesis-related genes under normal conditions, resulting in improved tolerance to salt and pathogens. The mechanisms plants use to adapt to abiotic and biotic stresses have been widely studied in a number of plants.
Production of ROS in Plants
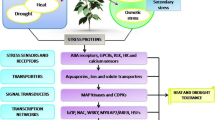
Current research effort has focused on the isolation of stress-responsive genes and their regulation as a means to understand the molecular events underlying the adaptation process. An increasing body of evidence suggests that a subset of plant responses to biotic and abiotic stress is shared, such as the generation of reactive oxygen species (ROS), the activation of mitogen-activated protein kinases (MAPKs), and hormone modulations (Mittler 2002) (Fig. 15.1). Organelles such as chloroplasts, mitochondria, and microbodies are a major source of ROS production in plant cells. Together with an extensive battery of oxidases, the plant cell is well armed for bountiful yet flexible ROS production. In chloroplasts, the primary sources of ROS production are the Mehler reaction and the antenna pigments (Asada and Takahashi 1987). Production of ROS by these sources is enhanced in plants by conditions limiting CO2 fixation, such as drought, salt, and temperature stress, as well as by the combination of these conditions with high-light stress. In C3 plants, limiting CO2 conditions can also activate the photorespiratory pathway (Foyer 2002). As part of this pathway, H2O2 is generated in peroxisomes by the enzymatic activity of glycolate oxidase. Production of H2O2 in microbodies can also occur during lipid catabolism as a side product of fatty acid oxidation. In mitochondria, over-reduction of the electron transport chain is the main source of O2− production under specific stress conditions (Møller 2001). ROS in plant cells include the detoxifying reactions catalyzed by cytochromes in both the cytoplasm and the endoplasmic reticulum. They are thought to play a key role in ROS signaling and contain a multimeric flavocytochrome that forms an electron transport chain capable of reducing O2 to O2−. Chemical inhibitors of mammalian NADPH oxidase (such as diphenyleneiodonium) have been shown to block or impair ROS production during biotic or abiotic stresses in plants (Allan and Fluhr 1997). In addition to NADPH oxidases, pH-dependent cell wall peroxidases, germin-like oxalate oxidases, and amine oxidases have been proposed to generate ROS at the apoplast (Bolwell and Wojtaszek 1997; Walters 2003). Although much attention has been given to NADPH oxidases and their possible role in cell signaling, other ROS-producing mechanisms in the mitochondria, apoplast, and peroxisome are likely to play a role in ROS signaling in response to different stimuli or developmental signals.
Modulation of reactive oxygen species (ROS) signaling by the reactive oxygen gene network of plants (Published by Mittler et al. 2004)
Modulation of ROS Signaling
ROS production is recognized as a common event in plant response to biotic and abiotic stresses (Mittler et al. 2010; Lamb and Dixon 1997). The hypersensitive response (localized plant cell death at the infection site) to halt pathogen invasion during an incompatible host–pathogen interaction has also been reported to involve the production of ROS. Mittler et al. (2004) reported that calcium signaling is predominantly controlled in plants by storage and release; ROS signaling is controlled by production and scavenging. Different developmental or environmental signals feed into the ROS signaling network and perturb ROS homeostasis in a compartment-specific or even cell-specific manner. Perturbed ROS levels are perceived by different proteins, enzymes, or receptors and modulate different developmental, metabolic, and defense pathways. ROS can be generated by various enzymatic activities, of which the best studied are NADPH oxidases, and removed by an array of ROS-scavenging enzymes. The intensity, duration, and localization of the different ROS signals are determined by interplay between the ROS-producing and ROS-scavenging pathways of the cell. This process requires a tight mode of regulation and might involve amplification and/or feedback inhibition loops. In addition to regulating the intensity and duration of the different ROS signals, the ROS-scavenging pathways are also responsible for maintaining a low steady-state baseline of ROS on which the different signals can be registered. The reactive oxygen gene network therefore modulates the steady-state level of ROS in the different cellular compartments for signaling purposes as well as for protection against oxidative damage. It is possible that the use of ROS as versatile signaling molecules originated from their proposed use to sense stress. Most forms of biotic or abiotic stress disrupt the metabolic balance of cells, resulting in enhanced production of ROS. Simple organisms, such as bacteria or yeast, sense the enhanced production of ROS using redox-sensitive transcription factors and other molecular sensors, activate different ROS defense pathways, and regulate their metabolic pathways to lower the production rate of ROS (Costa and Moradas-Ferreira 2001; Georgiou 2002). The signal transduction pathway was demonstrated by Mittler et al. (2004) (Fig. 15.2).
Model of the reactive oxygen species (ROS) signal transduction pathway. ROS can be detected by at least three mechanisms (ROS receptors, redox-sensitive transcription factors, and phosphatases). HSF heat shock factor, PDK phosphoinositide-dependent kinase, TF transcription factor (Published by Mittler et al. 2004)
Stress and Transcription Factors
Studies of transcriptional activation of some stress-responsive genes have also led to the identification of cis-acting elements ABRE (ABA-responsive element) and DRE (dehydration-responsive element)/CRT (C-repeat) that function in ABA-dependent and ABA-independent gene expression in response to stress, respectively (Seki et al. 2003). Transcription factors belonging to the ethylene-responsive element-binding factor family that bind to DRE/CRT were also isolated (Liu et al. 1998). The genes encoding these transcription factors are induced early and transiently in response to cold, and these transcription factors, in turn, activate the expression of target genes. Liu et al. (1998) reported that the transcription factors (DREB2A and DREB2B) are also induced by dehydration and promote the expression of various genes involved in drought stress tolerance. The expression of a new DNA-binding protein DBF1 that specifically interact with the DRE2 cis-element of a corn rab17 gene promoter is induced by ABA, dehydration, and high salinity (Kizis and Pages 2002). Another example of a transcription factor is calcium-dependent protein kinases (CDPKs). CDPKs are implicated as important sensors of Ca2+ flux in plants in response to stress (Ludwig et al. 2004). CDPKs are encoded by multigene families, and expression levels of these genes are spatially and temporally controlled throughout development. In addition, subsets of CDPKs are involved in signal transduction during stress including cold, salt, and drought or pathogen infection. A new transcription factor, BOS1 (Botrytis susceptible 1), was found to be required for both biotic and abiotic stress responses in Arabidopsis as reported by Mengiste et al. (2003). This complex network of interactions allows plants to respond in a highly specific fashion to the exact combination of environmental stresses encountered.
Identification of the Differentially Expressed Proteins
Many cold stress-responsive proteins have been identified (Gao et al. 2009). These proteins includeputative 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, glycine-rich RNA-binding protein 7 (GRP7), ferritin-1, EF-G, phosphoglycerate kinase, ATP synthase CF1 beta chain, UDP-glucose pyrophosphorylase, cysteine proteinase inhibitor, RuBisCO small chain, carbonic anhydrase, and SAL1 phosphatase. New proteins such as isopropylmalate synthase, cytochrome b6–f complex iron–sulfur subunit, and SNF2 domain-containing protein/helicase domain-containing protein/zinc finger (C3HC4 type RING finger) family protein have been identified in response to cold stress. These novel cold-responsive proteins may play important roles in cold tolerance of Thellungiella halophila. The stress related proteins are involved in RNA metabolism and energy pathway, protein synthesis, folding and degradation as well as cell wall and cytoskeleton, metabolisms of nitrogen and sulfur, and signal transduction (Emanuelsson et al. 2000). The cold stress-responsive proteins linked to RNA metabolism, defense response, protein synthesis, and folding and degradation were all upregulated markedly during cold treatment. It suggests that these cellular processes were enhanced by exposure to cold stress. The identified protein involved in photosynthesis and energy pathway displayed diverse change patterns. T. halophila is a valuable model system for investigating the mechanisms involved in plant cold stress tolerance. Proteomic analysis of cold stress response in T. halophila can help to identify key regulators of cold tolerance in plants. Regulation of chloroplast functions under cold stress. The expression regulation of chloroplast proteins is of central importance in cold adaptation (Foyer et al. 1997), and a substantial portion (46%, 23/50) of the identified proteins were predicted to be localized in chloroplast. Eighteen of these have been identified to reside in the chloroplast by previous proteomic studies according to the Plant Proteome Database (PPDB, http://ppdb.tc.cornell.edu/) (Sun et al. 2009). These chloroplast-located cold stress-responsive proteins are associated with various aspects of chloroplast, including Calvin cycle and electron transport (discussed below), chloroplast RNA processing (putative RNA-binding protein cp29), and chloroplast protein synthesis and folding (EF-G and heat-shock protein 70), as well as chloroplast redox homeostasis (BAS1, Peroxiredoxin-2E and Ferritin- 1). At the same time, a chloroplast-localized carbonic anhydrase, which facilitates CO2 move across the chloroplast envelope, was found to decrease in abundance after 5 and 24 days of cold treatment. The changes in Calvin cycle enzymes observed in this study might be associated with decrease in photosynthetic CO2 assimilation, possibly resulting in decreased growth rate in the long term. Enhanced RNA metabolism, including RNA processing, transporting from nucleus to cytoplasm, and mRNA secondary structure stability may be impaired under cold stress, especially for the cold stress-induced defense-related transcripts (Zhu et al. 2007). Several RNA-binding proteins and helicase-like proteins were all found to be upregulated by cold stress in the present study, including GRP7, glycine-rich RNA-binding protein GRP1A (GRP1A), putative RNA-binding protein cp29, and SNF2 domain-containing protein/helicase domain-containing protein/zinc finger (C3HC4-type RING finger) family proteins indicating enhanced RNA metabolism may play an important role in cold tolerance of T. halophila. Among these cold-responsive nucleic acid-binding proteins, GRPs are suggested to play an important role in posttranscriptional regulation of gene expression in plants under various stress conditions (Mousavi and Hotta 2005). GRP7 has been demonstrated to play a role in the export of mRNAs from the nucleus to the cytoplasm under cold stress conditions in Arabidopsis (Kim et al. 2008). RNA-binding protein cp29 is a subunit of the photosystem II and its phosphorylation was reported to relate with the cold tolerance in maize (Mauro et al. 1997). A RNA-binding protein cp29 was proved to be induced by cold stress in Arabidopsis (Amme et al. 2006). The increase in two chloroplast RNA-binding proteins may help to enhance or maintain the chloroplast RNA synthesis under cold conditions. In addition, GRPs and chloroplast RNA-binding proteins are suggested to be part of the plant innate immunity system and increase in their abundance may promote the expression of immunity-related mRNA (Fu et al. 2007).
Conclusion
In conclusion, the proteomic analysis is a very useful tool for providing complex information about differences in the plant proteome during abiotic and biotic stresses. It is almost necessary to clarify the differential function of the individual stress-responsive NAC genes for the control of abiotic stress tolerance and the other biological processes including biotic stress tolerance and growth regulation in order to fully utilize the potential of NAC transcription factors. Large-scale transcriptome analyses coupled with microarray, proteomic, and metabolomic analysis of plants perturbed at the levels of individual or multiple components of the ROS network will be essential for future studies. In the future, a combination of reverse genetics, genomics, and proteomic approaches in various developmental stages and stress conditions will provide us with critical information to elucidate the functional differences of the stress-responsive NAC factors and their relationship in transcriptional control. It is a challenge for plant scientists/environmentalists/biotechnologists in the twenty-first century to develop stable multiple stress tolerance traits in agronomically important crop plants, thus improving yields particularly in areas with adverse environmental conditions and contributing to food security.
References
Abbas HK, Williams WP, Windham GL, Pringle HC, Xie W, Shier WT (2002) Aflatoxin and fumonisin contamination of commercial corn (Zea mays) hybrids in Mississippi. J Agric Food Chem 50:5246–5254
Ahn YO, Kim SH, Lee J, Kim HR, Lee HS, Kwak SS (2012) Three Brassica rapa metallothionein genes are differentially regulated under various stress conditions. Mol Biol Rep 39:2059–2067
Alfano G, Ivey MLL, Cakir C, Bos JIB, Miller SA, Madden LV, Kamoun S, Hoitink HAJ (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97:429–437
Allan AC, Fluhr R (1997) Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9:1559–1572
Amme S, Matros A, Schlesier B, Mock HP (2006) Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE technology. J Exp Bot 57:1537–1546
Anjum NA, Ahmad I, Mohmood I (2012) Modulation of glutathione and its related enzymes in plants responses to toxic metals and metalloids – a review. Environ Exp Bot 75:307–324
Asada K, Takahashi M (1987) Production and scavenging of active oxygen in photosynthesis. In: Kyle DJ et al (eds) Photoinhibition, vol 9, Topics in photosynthesis. Elsevier, Amsterdam/New York, pp 227–287
Audenaert K, De Meyer GB, Hofte MM (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128:491–501
Bae H, Sicher RC, Kim MS, Kim S-H, Strem MD, Melnick RL, Bailey BA (2009) The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J Exp Bot 60:3279–3295
Baier M, Dietz K-J (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 60:282–314
Barconi D, Bernardini G, Santucci A (2011) Linking protein oxidation to environmental pollutants: redox proteome approaches. J Proteomics 74:2324–2337
Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7:1099–1111
Bolwell GP, Wojtaszek P (1997) Mechanisms for the generation of reactive oxygen species in plant defence – broad perspective. Physiol Mol Plant Pathol 51:347–366
Boursiac Y, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Maurel C (2008) Stimulus-induced downregulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J 56(2):207–218
Boyer JS (2009) Evans review: cell wall biosynthesis and the molecular mechanism of plant enlargement. Funct Plant Biol 36(5):383–394
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In: Gruissem W, Buchannan B, Jones R (eds) Biochemistry and molecular biology of plants, vol 23. American Society of Plant Physiologists, Rockville, pp 1158–1203
Bréhélin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y (2003) Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol 132:2045–2057
Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK (2001) Learning from the Arabidopsis experience: the next gene search paradigm. Plant Physiol 127:1354–1360
Bruix M, Jimenez MA, Santoro J, Gonzalez C, Colilla FJ, Mendez E, Rico M (1993) Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins. Biochemistry 32:715–724
Cabuslay G, Ito O, Alejar A (1999) Genotypic differences in physiological responses to water deficit in rice. In: O’Toole J, Ito O, Hardy B (eds) Genetic improvement of rice for water-limited environments. International Rice Research Institute, Los Baños, pp 99–116
Carmona MJ, Molina A, Fernandez JA, Lopez-Fando JJ, Garcia-Olmedo F (1993) Expression of the a-thionin gene from barley in tobacco confers enhanced resistance to bacterial pathogens. Plant J 3:457–462
Caporale C, Di Berardino I, Leonardi L, Bertini L, Cascone A, Buonocore V, Caruso C (2004) Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett 575:71–76
Castiglione S, Franchin C, Fossati T, Lingua G, Torrigiani P, Biondi S (2007) High zinc concentrations reduce rooting capacity and alter metallothionein gene expression in white poplar (Populus alba L. cv. Villafranca). Chemosphere 67:1117–1126
Chen CH, Chen ZX (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen induced Arabidopsis transcription factor. Plant Physiol 129:706–716
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T (2002a) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Chen ZY, Brown RL, Damann KE, Cleveland TE (2002b) Identification of unique or elevated levels of kernel proteins in aflatoxin-resistant maize genotypes through proteome analysis. Phytopathology 92:1084–1094
Chen ZY, Brown RL, Cleveland TE (2004) Evidence for an association in corn between stress tolerance and resistance to Aspergillus flavus infection and aflatoxin contamination. Afr J Biotechnol 3:693–699
Cheng JP, Chen B, Xu YM, Zhou XA, Lu CM, Wei WH (2007) Construction of suppression subtractive cDNA library and analysis of differentially expressed genes during maturing period seed of high and low oil content soybean variety. Chin J Oil Crop Sci 29:365–371
Chinnusamy V, Gong Z, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50(10):1187–1195
Clifford SC, Arndt S, Corlett JE, Joshi S, Sankhla N, Popp M, Jones HG (1998) The role of solute accumulation, osmotic adjustment and changes in cell wall elasticity in drought tolerance in Ziziphus mauritiana (Lamk.). J Exp Bot 49:967–977
Cobbett CS (2000) Phytochelatins and their roles in heavy metal detoxification. Plant Physiol 123:825–832
Cole RJ, Sanders TH, Hill RA, Blankenship PD (1985) Mean geocarposphere temperatures that induce preharvest aflatoxin contamination of peanuts under drought stress. Mycopathologia 91:41–46
Colilla FJ, Rocher A, Mendez E (1990) Gamma-purothionins: amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett 270:191–194
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529
Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci U S A 101:15243–15248
Costa V, Moradas-Ferreira P (2001) Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol Aspects Med 22:217–246
Cramer GR (2010) Abiotic stress & plant responses from the whole vine to the genes. Aust J Grape Wine Res 16:86–93
Cramer GR, Ergul A, Grimplet J, Tillett RL, Tattersall EA, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7(2):111–134
Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Ann Rev Plant Physiol Plant Mol Biol 48:355–381
Creighyton DJ, Magliorini M, Pourmotabbed T, Guha MK (1988) Optimization of efficiency in the glyoxalase pathway. Biochemistry 27:7376–7384
Cui S, Huang F, Wang J, Ma X, Cheng Y, Liu J (2005) A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5:3162–3172
DalCorso G, Farinati S, Maistri S, Furini F (2008) How plants cope with cadmium: staking all on metabolism and gene expression. J Integr Plant Biol 50(10):1268–1280
Datta SK, Muthukrishnan S (1999) Pathogenesis-related proteins in plants. CRC Press, Boca Raton
Deak KI, Malamy J (2005) Osmotic regulation of root system architecture. Plant J 43:17–28
Deák M, Horváth GV, Davletova S, Török K, Sass L, Vass I, Barna B, Király Z, Dudits D (1999) Plants ectopically expressing the iron binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nat Biotechnol 17:192–196
Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E (1994) A central role of salicylic acid in plant disease resistance. Science 266:1247–1250
Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meye C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8(9):864–870
Diaz J, ten Have A, van Kan JA (2002) The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol 129:1341–1351
Dietz KJ, Bair M, Kramer U (1999) Free radical and reactive oxygen species as mediators of heavy metal toxicity in plants. In: Prasad MNV, Hagemeyer J (eds) Heavy metal stress in plants from molecules to ecosystems. Springer, Berlin, pp 73–79
Dilbirligi M, Gill KS (2003) Identification and analysis of expressed resistance gene sequences in wheat. Plant Mol Biol 53:771–787
Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320(5878):942–945
Dorner JW, Cole RJ, Sanders TH, Blankenship PD (1989) Interrelationship of kernel water activity, soil temperature, maturity, and phytoalexin production in preharvest aflatoxin contamination of drought-stressed peanuts. Mycopathologia 105:117–128
Dubey RS (2011) Metal toxicity, oxidative stress and antioxidative defense system in plants. In: Gupta SD (ed) Reactive oxygen species and antioxidants in higher plants. CRC Press, Boca Raton, USA, pp 177–203
Duval M, Hsieh TF, Kim SY, Thomas TL (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily, Plant Mol. Biol 50:237–248
Duvick JP, Rood T, Rao AG, Marshak DR (1992) Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J Biol Chem 267:18814–18820
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Feist AM, Palsson BO (2008) The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol 26(6):659–667
Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16:449–455
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Foyer CH (2002) The contribution of photosynthetic oxygen metabolism to oxidative stress in plants. In: Inzé D, Van Montagu M (eds) Oxidative stress in plants. Taylor and Francis, New York, pp 33–68
Foyer CH, Lelandais M, Kunert KJ (1997) Photo-oxidative stress in plants. Physiol Plant 92:696–717
Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci U S A 98:373–378
Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III eVector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447:284–288
Gallie DR, Le H, Caldwell C, Browning KS (1998) Analysis of translation elongation factors from wheat during development and following heat shock. Biochem Biophys Res Commun 245:295–300
Gamalero G, Erta G, Glick BR (2009) The use of microorganisms to facilitate the growth of plants in saline soils. Springer, Heidelberg
Gao F, Zhou Y, Zhu W, Li X, Fan L, Zhang G (2009) Proteomic analysis of coldstress-responsive proteins in Thellungiella rosette leaves. Planta 230:1033–1046
Gasic K, Korban S (2006) Heavy metal stress. In: Rao KVM, Raghavendra AS, Reddy KJ (eds) Physiology and molecular biology of stress tolerance in plants. Springer, Dordrecht, pp 219–254
Georgiou G (2002) How to flip the (redox) switch. Cell 111:607–610
Good AG, Zaplachinski ST (1994) The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol Plant 90(1):9–14
Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardeström P, Schröder W, Hurry V (2006) The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J 47:720–734
Gray GR, Heath D (2005) A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolome fingerprinting. Plant Physiol 124:236–248
Grover A, Kapoor A, Lakshmi OS, Agarwal S (2001) Understanding molecular alphabets of the plant abiotic stress responses. Plant Mol Biol 80:206–216
Hajduch M, Rakwal R, Agrawal GK, Yonekura M, Pretova A (2001) High resolution two-dimensional electrophoresis separation of proteins from metal-stressed rice (Oryza sativa L.) leaves: drastic reductions/fragmentation of ribulose-1, 5-bisphosphate carboxylase oxygenase and induction of stress-related proteins. Electrophoresis 22:2824–2831
Hajheidari M, Abdollahian-Noghabi M, Askari H, Hedari M, Sadeghian SY, Ober ES, Hosseini Salekdeh GH (2005) Proteome analysis of sugar beet leaves under drought stress. Proteomics 5:950–960
Hamer DH (1986) Metallothionein. Annu Rev Biochem 55:913–951
Hammond-Kosack KE, Jones JDG (2000) Response to plant pathogens. In: Buchannan B, Gruissem W, Jones R (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 1102–1157
Hammond-Kosacky KE, Parkerz JE (2003) Deciphering plant– pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14:177–193
Harman GE (2000) Myths and dogmas of biocontrol: changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis 84:377–393
Harman GE (2006) Overview of mechanisms and uses of Trichoderma spp. Phytopathology 96:190–194
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species—opportunistic, a virulent plant symbionts. Nat Rev Microbiol 2:43–56
Hashimoto M, Komatsu S (2007) Proteomic analysis of rice seedlings during cold stress. Proteomics 2007(7):1293–1302
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcritional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hejgaard J, Jacobsen S, Svendsen I (1991) Two antifungal thaumatin like proteins from barley grain. FEBS Lett 291:27–131
Herms DA, Mattson WJ (1992) The dilemma of plants—to grow or defend. Q Rev Biol 67:283–335
Hey SJ, Byrne E, Halford NG (2010) The interface between metabolic and stress signaling. Ann Bot 105(2):197–203
Hill RA, Blankenship PD, Cole RJ, Sanders TH (1983) Effects of soil moisture and temperature on preharvest invasion of peanuts by the Aspergillus flavus group and subsequent aflatoxin development. Appl Environ Microbiol 45:628–633
Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101:10205–10210
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Hoffmann-Sommergruber K (2002) Pathogenesis-related (PR)-proteins identified as allergens. Biochem Soc Trans 30:930–935
Holappa LD, Simmons MKW (1995) The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol 108:1203–1210
Hoque MA, Uraji M, Akhter Banu MN, Mori IC, Nakamura Y, Murata Y (2010) The effects of methylglyoxal on glutathione S-transferase from Nicotiana tabacum. Biosci Biotechnol Biochem 74(10):2124–2126
Hossain MA, Fujita M (2010) Evidence for a role of exogenous glycinebetaine and proline in antioxidant defense and methylglyoxal detoxification systems in mung bean seedlings under salt stress. Physiol Mol Biol Plant 16:19–29
Hossain MA, Hossain MZ, Fujita M (2009) Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust J Crop Sci 3(2):53–64
Hossain MA, Hasanuzzaman M, Fujita M (2010) Up-regulation of antioxidant and glyoxalase systems by exogenous glycinebetaine and proline in mung bean confertolerance to cadmium stress. Physiol Mol Biol Plant 16(3):259–272
Hossain MA, Hasanuzzaman M, Fujita M (2011a) Coordinate induction of antioxidant defense and glyoxalase system by exogenous proline and glycinebetaine is correlated with salt tolerance in mung bean. Front Agric China 5(1):1–14
Hossain MA, da Silva JAT, Fujita M (2011b) Glyoxalase system and reactive oxygen species detoxification system in plant abiotic stress response and tolerance: an intimate relationship. In: Shanker AK, Venkateswarlu B (eds) Abiotic stress in plants-mechanisms and adaptations. INTECH-Open Access Publisher, Rijeka, pp 235–266
Hossain MA, Hossain MD, Rohman MM, da Silva JAT, Fujita M (2012) Onion major compounds (flavonoids, organosulfurs) and highly expressed glutathione-related enzymes: possible physiological interaction, gene cloning and abiotic stress response. In: Aguirre CB, Jaramillo LM (eds) Onion consumption and health. Nova, New York
Houde M, Belcaid M, Ouellet F, Danyluk J, Monroy AF, Dryanova A, Gulick P, Bergeron A, Laroche A, Links MG, MacCarthy L, Crosby WL, Sarhan F (2006) Wheat EST resources for functional genomics of abiotic stress. BMC Genomics 7:149–166
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Over expressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 103:12987–12992
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes 24(16):1695–1708
Hurkman WJ, Tanaka CK (2007) Extraction of wheat endosperm proteins for proteome analysis. J Chromatogr 849:344–350
Iusem ND, Bartholomew DM, Hitz WD, Scolnik PA (1993) Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiol 102:1353–1354
Jacob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268:1517–1520
Jones RK, Duncan HE, Hamilton PB (1981) Planting date, harvest date, and irrigation effects on infection and aflatoxin production by Aspergillus flavus in field corn. Phytopathology 71:810–816
Kagi JHR (1991) Overview of metallothionein. Methods Enzymol 205:613–626
Kaufmann K, Smaczniak C, De VS, Angenent GC, Karlova R (2011) Proteomics insights into plant signaling and development. Proteomics 11(4):744–755
Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141–154
Kawasaki S, Miyake C, Kohci T, Fujii S, Uchida M, Yokota A (2000) Response of wild watermelon to drought stress: accumulation of an ArgE homologue and citrulline in leaves during water deficits. Plant Cell Physiol 41:864–873
Khedr AHA, Abbas MA, Abdel Wahid AA, Quick WP, Abogadallah GM (2003) Proline induces the expression of salt stress- responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt-stress. J Exp Bot 54:2553–2562
Khuri S, Bakker FT, Dunwell JM (2001) Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol Biol Evol 18:593–605
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 54:1047–1051
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50(2):347–363
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh CH, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine rich RNA-binding protein 7a Vects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466
Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61:561–591
Kitano H (2002) Systems biology: a brief overview. Science 295(5560):1662–1664
Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30:679–689
Knoester M, Van Loon LC, Den V, Heuvel J, Hennig J, Bol JF, Linthorst HJM (1998) Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc Natl Acad Sci U S A 95:1933–1937
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Kwon SJ, Kwon SI, Bae MS, Cho EJ, Park OK (2007) Role of the methionine sulfoxide reductase msrB3 in cold acclimation in Arabidopsis plant and cell physiology. Plant Cell Physiol 48:1713–1723
Lageix S, Lanet E, Pouch-Pelissier MN, Espagnol MC, Robaglia C, Deragon JM, Pelissier T (2008) Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol 8:134
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275
Legrand M, Kauffmann S, Geoffroy P, Fritig B (1987) Biological function of pathogenesis related proteins: four PR proteins of tobacco are chitinases. Proc Natl Acad Sci U S A 84:6750–6754
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Mol Biol 49:199–222
Li W, Faris JD, Muthukrishnan S, Liu D, Chen P, Gill BS (2001) Isolation and characterization of cDNA clones of acidic chitinases and beta-1, 3-Glucanases from wheat spike induced by Fusarium graminearum. Theor Appl Genet 102:353–362
Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY, Jia JZ, Mao L (2008) Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66:429–443
Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22(9):2930–2942
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Liu HT, Li B, Shang ZL, Li XZ, Mu RL, Sun DY, Zhou RG (2003) Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiol 132:1186–1195
Lu B, Fang XJ (2003) Express analysis of cDNA induced by infecting roots of soybean cyst nematode race 4 in soybean ZZD2315. Mol Plant Breed 1:193–200
Lu Z, Neumann PM (1999) Water stress inhibits hydraulic conductance and leaf growth in rice seedling but the transport of water via mercury-sensitive water channels in the root. Plant Physiol 120:143–152
Ludwig AA, Romeis T, Jones JD (2004) CDPK-mediated signaling pathways: specificity and cross-talk. J Exp Bot 55:181–188
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Gril E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068
Majoul T, Bancel E, Triboï E, Ben Hamida J, Branlard G (2003) Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from total endosperm. Proteomics 3:175–183
Mao L, Van HJL, Dash S, Dickerson JA (2009) Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10:346
Mauro S, Dainese P, Lannoye R, Bassi R (1997) Cold-resistant and cold-sensitive maize lines differ in the phosphorylation of the photosystem II subunit, CP29. Plant Physiol 115:171–180
Meharg AA (1993) The role of plasmalemma in metal tolerance in angiosperm. Physiol Plant 88(1):191–198
Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15:2551–2565
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van BF (2010) ROS signaling: the new wave? Trends Plant Sci 16(6):300–309
Mock HP, Grimm B (1997) Reduction of uroporphyrinogen decarboxylase by antisense RNA expression affects activities of other enzymes involved in tetrapyrrole biosynthesis and leads to light dependent necrosis. Plant Physiol 113:1101–1112
Molassiotis A, Fotopoulos V (2011) Oxidative and nitrosative signaling in plants: two branches in the same tree? Plant Signal Behav 6(2):210–214
Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52:561–591
Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol 50:2210–2222
Mostafa Kamal AH, Kim KH, Shin KH, Kim DE, Oh MW, Choi JS, Hirano H, Heo HY, Woo SH (2010a) Proteomics-based dissection of biotic stress responsive proteins in bread wheat (Triticum aestivum L.). Afr J Biotechnol 9:7239–7255
Mostafa Kamal A, Kim AH, Shin KH, Choi JS, Baik NK, Tsujimoto H, Heo HY, Park CS, Wool SH (2010b) Abiotic stress responsive proteins of wheat grain determined using proteomics technique. Aust J Crop Sci 4(3):196–208
Mousavi A, Hotta Y (2005) Glycine-rich proteins: a class of novel proteins. Appl Biochem Biotechnol 120:169–174
Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32:299–315
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Navari-Izzo F (1998) Thylakoid-bound and stromal antioxidative enzymes in wheat treated with excess copper. Physiol Plant 104:630–638
Nieboer E, Richardson DHS (1980) The replacement of the nondescript term heavy Metals’ by a biologically and chemically significant classification of metal ions. Environ Pollut Ser B Chem Phys 1(1):3–26
Nogueira FTS, Schlögl PS, Camargo SR, Fernandez JH, De Rosa VE Jr, Pompermayer P, Arruda P (2005) SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Sci 169:93–106
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
O’Donnell PJ, Schmelz E, Block A, Miersch O, Wasternack C, Jones JB, Klee HJ (2003) Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol 133:1181–1189
Okuma E, Soeda K, Tada M, Murata Y (2000) Exogenous proline mitigates the inhibition of growth of Nicotiana tabacum cultured cells under saline conditions. Soil Sci Plant Nutr 46:257–263
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Ouerghi Z, Rémy R, Ouelhazi L, Ayadi A, Brulfert J (2000) Two dimensional electrophoresis of soluble leaf proteins, isolated from two wheat species (Triticum durum and Triticum aestivum) differing in sensitivity towards NaCl. Electrophoresis 21:248–2491
Parent B, Hachez C, Redondo E, Simonneau T, Chaumont F, Tardieu F (2009) Drought and abscisic acid effects on aquaporin content translate into changes in hydraulic conductivity and leaf growth rate: a trans-scale approach. Plant Physiol 149(4):2000–2012
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Over expression of the tobacco Tsi1 gene encoding an EREBP/AP2- type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13:1035–1046
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071
Payne GA (1998) Process of contamination by aflatoxin producing fungi and their impact on crops. In: Sinha KK, Bhatnagar D (eds) Mycotoxins in agriculture and food safety. Marcel Dekker, New York, pp 279–306
Payne GA, Cassel DK, Adkins CR (1986) Reduction of aflatoxin contamination in corn by irrigation and tillage. Phytopathology 76:679–684
Pedrol N, Ramos P, Reigosa MJ (2000) Phenotypic plasticity and acclimation to water deficits in velvet-grass: a long-term greenhouse experiment. Changes in leaf morphology, photosynthesis and stress-induced metabolites. J Plant Physiol 157:383–393
Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103:1111–1120
Pinheiro C, Chaves MM (2011) Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot 62(3):869–882
Qureshi MI, Qadir S, Zolla L (2007) Proteomics-based dissection of stress-responsive in plants. Plant Physiol 164:1239–1260
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Riccardi F, Gazeau P, de Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize: quantitative variations and identification. Plant Physiol 117:1253–1263
Rivetta A, Negrini N, Cocucci M (1997) Involvement of Ca 2+-calmodulin in Cd 2+ toxicity during the early phases of radish (Raphanus sativus L.) seed germination. Plant Cell Environ 20:600–608
Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130:1143–1151
Rockstrom J, Falkenmark M (2000) Semiarid crop production from a hydrological perspective: gap between potential and actual yields. Crit Rev Plant Sci 19:319–346
Romero-Puertas MC, Palma JM, Gomez M, Del R, Sandalio LM (2002) Cadmium causes the oxidative modification of proteins in pea plants. Plant Cell Environ 25:677–686
Ruelland E, Vaultier MN, Zachowski A, Hurry V (2009) Cold signalling and cold acclimation in plants. Adv Bot Res 49:35–150
Sabehat A, Lurie S, Weiss D (1998) Expression of small heat-shock proteins at low temperatures. A possible role in protecting against chilling injuries. Plant Physiol 117:651–658
Saito R, Yamamoto H, Makino A, Sugimoto T, Miyake C (2011) Methylglyoxal functions as Hill oxidant and stimulates the photoreduction of O2 at photosystem I: a symptom of plant diabetes. Plant Cell Environ 34(9):1454–1464
Salekdeh GH, Siopongco HJ, Wade LJ, Ghareyazie B, Bennett J (2002a) A proteomics approach to analysing drought and salt-responsiveness in rice. Field Crop Res 76:199–219
Salekdeh GH, Siopongco HJ, Wade LJ, Ghareyazie B, Bennett J (2002b) Proteomics analysis of rice leaves during drought stress and recovery. Proteomics 2:1131–1145
Samaras Y, Bressan RA, Csonka LN, Gracia-Rios MG, Paino D’Urzo M, Rhodes D (1995) Proline accumulation during drought and salinity. In: Smirnoff N (ed) Environment and plant metabolism: flexibility and accumulation. Oxford publishers, UK, pp 161–187
Schutzendubel A, Polle A (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53:1351–1365
Segal G, Feldman M (1996) Tubb1, a beta-tubulin cDNA from common wheat. EMBL/GenBank/DDBJ databases
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnology 14:194–199
Serrano R, Gaxiola R (1994) Microbial models and stress tolerance in plants. Crit Rev Plant Sci 13:121–138
Sharma SS, Dietz KJ (2009) The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci 14(1):43–50
Sharma P, Dubey RS (2007) Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep 26(11):2027–2038
Shin R, Park JM, An JM, Paek KH (2002) Ectopic expression of Tsi1 in transgenic hot pepper plants enhances host resistance to viral, bacterial, and oomycete pathogens. Mol Plant Microbe Interact 15:983–989
Shoresh M, Mastouri F, Harman G (2010) Induced systemic resistance and plant responses to fungal biocontrol agents. Annu Rev Phytopathol 48:21–43
Silhavy D, Hutvagner G, Barta E, Banfalvi Z (1995) Isolation and characterization of a water-stress-inducible cDNA clone from Solanum chacoense. Plant Mol Biol 27:587–595
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci U S A 100:14672–14677
Skirycz A, Inze D (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21(2):197–203
Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13(3):274–279
Snarpi SM, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Hrie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44:928–938
Sonia C, Montserrat C, Maria C, Joaquin A, Blanca SS (2004) The defense response of germinating maize embryos against fungal infection: a proteomics approach. Proteomics 4:383–396
Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, Grusak MA, Fett JP (2009) Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230:985–1002
Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC (2010) Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem 285(22):16893–16911
Sun GZ, Zhu ZD, Wu XF, Cui YL, Wang XM (2008) Comparative proteomics of Arabidopsis induced by salicylic acid, wound and Phytophthora sojae. Scientia Agri Sin 4:1030–1039
Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37:969–974
Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) Monitoring the expression profiles of genes induced by hyper-osmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56(1):29–55
Tamura T, Hara K, Yamaguchi Y, Koizumi N, Sano H (2003) Osmotic stress tolerance of transgenic tobacco expressing a gene encoding a membrane-located receptor-like protein from tobacco plants. Plant Physiol 131:454–462
Tattersall EA, Grimplet J, Deluc L, Wheatley MD, Vincent D, Osborne C, Ergul A, Lomen E, Blank RR, Schlauch KA, Cushman JC, Cramer GR (2007) Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct Integr Genomics 7(4):317–333
Thomann EB, Sollinger J, White C, Rivin CJ (1992) Accumulation of group 3 late embryogenesis abundant proteins in Zea mays embryos. Plant Physiol 99:607–614
Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci U S A 95:15107–15111
Tran LS, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyodan B, Nguyen HT (2009) Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics 281:647–664
Tran LSP, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K (2010) Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1(1):32–39
Treglia A, Spano G, Rampino P, Giangrande E, Nocco G, Mita G, Di Fonzo N, Peppotta C (1999) Identification by in vitro translation and Northern blot analysis of heat shock mRNAs isolated from wheat seeds exposed to different temperatures during ripening. Cereal Sci 30:33–38
Tubajika KM, Damann KE (2001) Sources of resistance to aflatoxin production in maize. J Agric Food Chem 49:2652–2656
Umezawa T (2011) Systems biology approaches to abscisic acid signaling. J Plant Res 124(4):539–548
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in abiotic responses: sensing, signaling and transport. Plant Cell Physiol 51:1821–1839
Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11:1743–1754
Van Assche F, Clijsters H (1986) Inhibition of photosynthesis in Phaseolus vulgaris by treatment with toxic concentration of zinc: effect on ribulose-1,5-bisphosphate carboxylase/ oxygenase. J Plant Physiol 125:355–360
van der Weel CM, Spollen WF, Sharo RE, Baskin TI (2000) Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J Exp Bot 51:1555–1562
Veena RVS, Sopory SK (1999) Glyoxalase 1 from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J 17:385–395
Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42:579–620
Villiers F, Ducruix C, Hugouvieux V (2011) Investigating the plant response to cadmium exposure by proteomic and metabolomic approaches. Proteomics 11(9):1650–1663
Vincent D, Ergul A, Bohlman MC, Tattersall EA, Tillett RL, Wheatley MD, Woolsey R, Quilici DR, Joets J, Schlauch K, Schooley DA, Cushman JC, Cramer GR (2007) Proteomic analysis reveals differences between Vitis vinifera L. cv. Chardonnay and cv. Cabernet Sauvignon and their responses to water deficit and salinity. J Exp Bot 58(7):1873–1892
Wakabayashi K, Hoson T, Kamisaka S (1997) Osmotic stress suppresses cell wall stiffening and the increase in cell wall-bound ferulic and diferulic acids in wheat coleoptiles. Plant Physiol 113:967–973
Walters DR (2003) Polyamines and plant disease. Phytochemistry 64:97–107
Wang WX, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E (1996) Synthesis of small heat- shock proteins is part of the developmental program of late seed maturation. Plant Physiol 112:747–757
Weston DJ, Gunter LE, Rogers A, Wullschleger SD (2008) Connecting genes, co- expression modules, and molecular signatures to environmental stress phenotypes in plants. BMC Syst Biol 2:16
Wildner GF, Henkel J (1979) The effect of divalent metal ions on the activity of Mg2+ depleted ribulose-1,5-bisphosphate oxygenase. Planta 146:223–228
Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355:1517–1529
Wong JH, Cai N, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004) Thioredoxin reduction alters the solubility of proteins of wheat starch endosperm: an early event in cereal germination. Plant Cell Physiol 45:407–415
Wotton HR, Strange RN (1987) Increased susceptibility and reduced phytoalexin accumulation in drought-stressed peanut kernels challenged with Aspergillus flavus. Appl Environ Microbiol 53:270–273
Xie Q, Sanz-Burgos AP, Guo H, García JA, Gutiérrez C (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol 39:647–656
Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R (1996) Expression of a late embryogenesis abundant protein gene HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110:249–257
Yadav SK (2010) Heavy metals toxicity in plants: an overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S Afr J Bot 76:167–179
Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK (2005a) Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun 337:61–67
Yadav SK, Singla-Pareek SL, Reddy MK, Sopory SK (2005b) Transgenic tobacco plants over expressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett 579:6265–6271
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K (1992) Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol 33:217–224
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5:484–496
Yang Q, Wang Y, Zhang J, Shi W, Qian C, Peng X (2007) Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7:737–749
Yang H, Huang Y, Zhi H, Yu D (2010) Proteomics-based analysis of novel genes involved in response toward soybean mosaic virus infection. Mol Biol Rep 38:511–521
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168
Yildirim E, Taylor AG, Spittler TD (2006) Ameliorative effects of biological treatments on growth of squash plants under salt stress. Sci Hortic (Amst) 111:1–6
Zaborske JM, Wu X, Wek RC, Pan T (2010) Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem 11:29
Zang X, Komatsu S (2007) A proteomics approach for identifying osmotic-stress-related proteins in rice. Phytochemistry 68:426–437
Zhang T, Liu Y, Yang T, Zhang L, Xu L, An L (2006) Diverse signals converge at MAPK cascades in plants. Plant Physiol Biochem 44:274–283
Zhang C, Meng Q, Gai J, Yu D (2008) Cloning and functional characterization of an O-acetylserine (thiol) lyase-encoding gene in wild soybean (Glycine soja). Mol Biol Rep 35:527–534
Zhou G, Xu Y, Li J, Yang L, Liu JY (2006) Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). J Biochem Mol Biol 39:595–606
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290–295
Zonia L, Mnnik T (2004) Osmotically induced cells swelling versus cell shrinking elicits specific changes in phospholipid signals in tobacco pollen tubes. Plant Physiol 134:813–823
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer India
About this chapter
Cite this chapter
Rout, G.R., Senapati, S.K. (2013). Stress Tolerance in Plants: A Proteomics Approach. In: Rout, G., Das, A. (eds) Molecular Stress Physiology of Plants. Springer, India. https://doi.org/10.1007/978-81-322-0807-5_15
Download citation
DOI: https://doi.org/10.1007/978-81-322-0807-5_15
Published:
Publisher Name: Springer, India
Print ISBN: 978-81-322-0806-8
Online ISBN: 978-81-322-0807-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)