Abstract
This chapter describes new methodologies to quantitatively evaluate cell-hydrogel adhesion using advanced optical techniques. The techniques enable one to gain two quantitative measures of cell-hydrogel adhesion, contact area and adhesion strength, which contribute to the rational design of biomaterials in tissue engineering.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Hydrogel is an indispensable material as structural support for cells in nature (i.e., ECM) as well as in tissue engineering. In addition, recent studies revealed that stiffness of hydrogels can influence many cell functions such as motility [1] and differentiation [2]. Such mechano-response was found even at a tissue level. For instance, synchronized beating in culture cardiomyocyte tissue on hydrogel substrate can significantly be modulated with hydrogel elasticity [3]. Local stiffening of liver due to fibrosis results in significant changes in functions within whole organs [4]. These results indicate that hydrogel with adequate stiffness is required to regenerate functional tissues. However, the choice of such hydrogel is still very dependent on trial-and-error although various hydrogels with tunable elasticity have been proposed to mimic natural microenvironments. One of the reasons for the difficulty in the rational design of hydrogel is lack of quantitative measures for cell-hydrogel affinity (adhesion). In fact, although molecular mechanism of cell-hydrogel adhesion was systematically studied, quantitative insights into cell-hydrogel adhesion at a cellular level remain still unknown.
Recently, I have been developing new methodologies to quantitatively evaluate cell-hydrogel adhesion under the collaboration with various scientists [5–8]. Here, two advanced optical techniques were utilized to obtain quantitative measures of cell-hydrogel adhesion, contact area and adhesion strength. In this account, I explain a comprehensive overview of the two optical techniques [7, 8] and their potentials for providing quantitative measures for rational tissue engineering.
2 Quantitative Evaluation of Cell-Hydrogel Contacts by Advanced Interferometric Microscopy
In general, due to small gap between cell membrane and hydrogel at adhesion sites (< 100 nm), visualization of physical contact between cells and hydrogel by optical microscopy is challenging. To date, total internal reflection fluorescence (TIRF) microscopy has been widely used to visualize cell adhesion molecules (e.g., integrin) near substrate. However, height resolution of TIRF (100–200 nm) [9] is not sufficient to identify the physical contact between cells and substrate. In addition, the existence of adhesion molecules does not necessarily guarantee physical contact between cells and hydrogel. It should be also noted that physical contact mediated by other generic interactions such as electrostatic interaction cannot be visualized by the fluorescence imaging. Alternatively, the leading technique for visualizing cell-substrate contacts is RICM , which detects interference of linearly polarized light reflected at cell-liquid (i.e., cell membrane-liquid) and substrate-liquid interfaces [10–14]. If glass substrate is used, RICM is a powerful label-free technique to measure the distance between cell membrane and substrates with a resolution of ~ 2 nm, [14] which is much finer than surface-sensitive fluorescence techniques such as TIRF microscopy. However, in case of hydrogel, contrast of RICM images becomes very poor because intensity and coherence of light are lowered by the following two reasons. Firstly, the refractive index of hydrogels is very close to that of water, which results in low reflectivity at hydrogel-liquid interfaces. Secondly, gels for cell mechano-response study should not be thin monolayer but much thicker (> several μm). As the results, RICM had not been applied to soft, thick hydrogels, which is widely used for cell mechano-response study and tissue engineering.
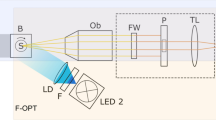
Very recently, I have developed an interferometric optical microscopy for the high contrast visualization of cell-hydrogel contact [8]. As schematically illustrated in Fig. 13.1a, conventional RICM setup utilizes the antiflex method; cross polarizers combined with an objective lens equipped with a quarter-wave plate (Antiflex EC Plan-Neofluar, 63 ×, Numerical aperture = 1.25, Oil Ph3, Zeiss, Gottingen, Germany). A mercury lump with a monochromatic filter (typically λ = 546 nm) has been widely used as a light source. To enhance the contrast of interference images obtained from cell-hydrogel contact, the RICM setup was modified by three points written below (Fig. 13.1b). (1) A confocal unit (FV300, Olympus, Tokyo, Japan) was attached to an inverted microscope (IX70, Olympus, Tokyo, Japan). A confocal aperture in the unit can significantly reduce stray light originating from outside of cell-hydrogel contact zones, e.g., reflection and scattering from glass, gels, cell cytoplasm, and organelle (Fig. 13.1c). (2) High throughput optics mentioned in the following were adopted to increase intensity hence to further improve the signal to noise ratio. Firstly, a polarization beam splitter (PBS, custom-ordered, Olympus Tokyo, Japan) that reflects s-polarized light and transmits p-polarized light over 95 % at the light wavelength was used. The use of the PBS instead of a half mirror and an analyzer can drastically increase light intensity of both illumination and detection in the antiflex setup, while a half mirror splits light at the both sides. Secondly, a polarizer made of aligned silver nanoparticles (colorPol VISIR CWO2, CODIXX AG, Barleben, Germany) was used, because it can enhance the signals by an order of magnitude than the polarizer based on polymer films [15]. (3) As a light source, a diode-pumped solid state laser (λ = 532 nm, 300 mW, SAPPHIRE 532–300-CW-CDPH, Coherent Inc., Santa Clara, USA) or a super luminescent diode (SLD) (λ = 680 nm, 5 mW, coherent length ~ 10 µm, ASLD68–050-B-FA, Amonics, Hong-Kong, China) was used. The use of such an intense monochromatic light source instead of halogen lamps should provide clearer interference patterns. In addition, since coherent length of the solid-state laser and SLD is generally shorter than a gas laser such as a He-Ne laser, [16] the use of light sources with such “moderate” coherence is expected to reduce untargeted interference signals except for cell-hydrogel contact zones [16, 17]. In fact, Sazaki et al. demonstrated the high-contrast visualization of ice and protein crystal surfaces at atomic resolution by using a SLD as a light source for scanning confocal microscopy combined with differential interference contrast microscopy [18].
Schematic illustration of experimental setup of a conventional RICM, b advanced RICM. c A confocal aperture reduces untargeted stray light originating from glass (blue dashed line), cell cytoplasm, and organelle (red dashed line). Reprinted with permission from ref. 8. Copyright (2014) American Chemical Society
Figure 13.2a, 13.1b, 13.2c show RICM images of a polystyrene latex bead with 100 µm in diameter (Φ) on polyacrylamide (PAAm) gel (E = 13.2 ± 0.5 kPa, thickness = 6 ~ 7 μm) taken by a conventional RICM setup and by the advanced RICM system, respectively. The advanced RICM provided much clearer interference patterns than those by a conventional RICM system. It should be also noted that both, laser and SLD, show clear Newton’s ring-like fringes without untargeted interference such as speckle, which is often observed using a He-Ne laser as a light source [16, 17]. From the intensity profile (Fig. 13.2e), the height profile of a bead was reconstructed (Fig. 13.2d) by the following Eq. (13.1) [13, 14] of
Interference images of polystyrene latex bead 100 µm in diameter on PAAm gels (E = 13.2 ± 0.5 kPa) taken by a conventional RICM and advanced RICM with a light source of b laser or c SLD. Scale bars: 10 µm. Size of confocal aperture is 60 µm. d Intensity profiles along the red lines in a–c and e reconstructed and predicted height profiles of a bead. Reprinted with permission from ref. 8. Copyright (2014) American Chemical Society
Here, I is measured intensity. I max and I min are maximum and minimum intensity. λ is wavelength of light, h separation between substrate and sample, and n refractive index of medium (~ 1.333). y represents 2πnsin2(α/2)/λ, where α is a half angle of the cone illumination (55°). The height profile of a bead reconstructed from interference signals is almost equivalent to height profile of a theoretically assumed sphere (Φ = 100 µm) up to 2 µm above the surface. The results clearly demonstrate that the advanced RICM system can provide with clearer interferometric patterns and quantitatively reconstructed height profile of objects on hydrogels.
In the next step, cells adherent on hydrogels were visualized using a conventional RICM setup and the advanced RICM system. Figure 13.3a shows a bright field image of a mouse metastatic melanoma cell (B16–F10) on a PAAm gel (E = 13.2 ± 0.5 kPa) functionalized with fibronectin. Cells were cultured for 3 h in Leibovitz’s L-15 medium without serum and then fixed with a 3.7 w/v % DPBS-formaldehyde solution. With a conventional RICM, the cell came out brighter than its surrounding region, indicating that scattered light from outside of a focal plane (e.g., cell cytoplasm) obscured interference signals (Fig. 13.3b). On the other hand, the advanced RICM yielded a black cell body in sharp contrast against the bright surrounding region (Fig. 13.3e, 13.3d, 13.3e). To assess influence of stray light on the image contrast of our interferometry, RICM images of the cell were taken with different confocal aperture diameters. The image contrast became higher by reducing the aperture diameter from 300 to 60 µm, corresponding to from 2.10 to 0.42 Airy unit, which represents the theoretically derived Airy disk diameter. The advanced RICM could visualize local structures of cell filopodia that are in tight contact with the hydrogel surface. These results clearly indicate that the advanced RICM can clearly visualize adhesion zone of cells adherent to 6 ~ 7 μm thick hydrogels, which is not possible by conventional RICM.
A B16-F10 cell on a PA gel (E = 13.2 ± 0.5 kPa) measured by a bright field microscopy, b conventional RICM, and c–e advanced RICM. Diameter of confocal apertures is c 300 μm, d 150 µm, and e 60 µm. SLD (λ = 680 nm) was used as a light source for the advanced interferometric microscopy. Images at the right of b and e are magnification of cell periphery. Scale bars: 10 μm. The bright dots like the one indicated by a red arrow in c are polystyrene beads embedded in PAAm gels. Reprinted with permission from ref. 8. Copyright (2014) American Chemical Society
In the final step, the advanced RICM was applied to evaluate the impact of substrate elasticity on cell adhesion. B16-F10 cells were cultured for 3 h on PAAm gels with E = 9.8, 13.2, and 62.5 kPa and then fixed. Bright field microscopy images (Fig. 13.4a) implies that B16-F10 exhibited a more pronounced spreading on stiffer gels, and the zones of cell-hydrogel tight contacts are identified as black patches in RICM images (Fig. 13.4b). Such black area can be typically seen for B16-F10 cells on hard glass substrate coated with fibronectin, which binds specifically to its receptor such as integrin. Figure 13.4c shows zone for cell-hydrogel tight contacts estimated by image analysis [8]. It clearly shows patch-like adhesion structures, which were typically observed in RICM images of cells adhered to flat glass substrate [11, 14]. The estimated tight contact area is monotonically increased from 74 to 578 µm2 according to the increase in the stiffness of hydrogel from 9.8 ± 0.4 kPa to 62.5 ± 4.7 kPa, suggesting mechano-response of B16-F10 cells.
B16-F10 cells on PAAm gels with three different stiffness (E = 9.8 ± 0.4 kPa, E = 13.2 ± 0.5 kPa, E = 62.5 ± 4.7 kPa) were measured by a bright field microscopy and b advanced RICM. Laser (λ = 532 nm) was utilized as a light source. c Tight adhesion area. Diameter of a confocal aperture was 60 µm. Scale bars: 10 µm. Reprinted with permission from ref. 8. Copyright (2014) American Chemical Society
In summary, the advanced RICM system drastically enhanced the contrast in interferometric patterns from cell-hydrogel contact zones. Physical contacts between cells and hydrogels could be successfully determined. I foresee that the advanced RICM will provide quantitative insights into mechanical interactions between cells and hydrogels, which contribute to the rational design of biomaterials in tissue engineering.
3 Quantitative Evaluation of Adhesion Strength Between Cells and Hydrogel by Using Laser-Induced Pressure Wave
Gelatin is one of the most widely used biomaterials for tissue engineering. Recently, Kidoaki et al. reported styrenized gelatin gel, whose elastic modulus can be tuned by photo-crosslinking conditions [19]. In a recent account, they reported that elastic pattern of gelatin can be used for inducing mechanotaxis of cells [19, 20]. In addition, synchronized beating in culture cardiomyocyte tissue on photocurable gelatin can significantly be influenced with gelatin elasticity [3]. These clearly indicate that elasticity of gelatin substrate modulates cell-gelatin interaction, which is a key for regulating cell and tissue functions. However, the quantitative insights into the “strength” of interactions between cells and photocurable gelatin still remained unknown.
Recently, adhesion strength of myoblast (C2C12) cells and photocurable gelatin was quantitatively measured by the self-developed cell detachment assay, [7] where the critical pressure for the cell detachment can be determined by pressure waves induced by picosecond laser pulses [5, 6]. The critical pressure required for the detachment of adherent cells P th can be used as a “mechanical” measure to quantitatively assess cell-substrate interaction (adhesion). Figure 13.5 represents bright field images of myoblast (C2C12) cells on gelatin gel before and after the laser-induced cell detachment. Here, the critical detachment pressures of P th = 3.4 MPa or 8.1 MPa can be determined for gels with E = 0.6 and 10 kPa, respectively (Fig. 13.5). Figure 13.6a shows the average pressure as a function of gel elasticity, E, at t = 3 h. The average of P th increased from 4.1 to 34 MPa according to the increase in the stiffness, 0.2 to 140 kPa. The monotonic increase in the adhesion strength of C2C12 cells is consistent with the past studies by micropipetts, [21, 22] but the transition from weak adhesion (P th < 5 MPa) to strong adhesion (P th > 20 MPa) takes place in a narrower E range (1 kPa ≤ E ≤ 20 kPa). To gain more quantitative insights into the dependence of cell adhesion on gelatin mechanics, the plots were fitted with the empirical Hill equation.
Evaluation of adhesion strength of C2C12 cells on gelatin gels. a The average of critical pressure as a function of gel elasticity at t ~ 3 h. The black solid line is the fitting with the Hill equation (Eq. 13.2). The error bar represents standard deviation. b Log-log plots converted from the plot of (A). The black solid line represents the fitting with the power law function (Eq. 13.3). Reprinted with permission from ref. 7. Copyright (2013) American Chemical Society
As indicated by the solid line, the fit with the cooperativity coefficient taken from image analysis of projected area (m = 1.1) [7] can well represent the experimental results. The half-saturation level E 1/2- adhesion, was ~ 7.6 kPa,
Other studies already reported that various cellular processes can phenomenologically be analyzed with power laws, such as the dependence of viscosity of cytoplasm on shear rate, [23] the dependence of projected area of smooth muscle cells on the substrate elasticity, [24] and the dependence of transit time of leukocytes through narrow pores [25]. Figure 13.6b represents the log-log plot of P th vs. E for the regime in which the transition was observed; 0.6 kPa ≤ E ≤ 20 kPa. The solid line corresponds to the power law:
The obtained power law exponent, n ~ 0.53, may be attributed to the establishment of firm adhesion by the formation of stress fibers bound to focal adhesions via talin [26, 27]. Here, contraction force originated from focal adhesions activates the Src kinase, which should lead to an increase in the adhesion strength [28]. In fact, the range of elastic modulus (0.6 kPa ≤ E ≤ 20 kPa) is in good agreement with the regime where the stress fiber formation was formed [7]. The observed power law dependences suggest the transition from weakly adhered to strongly adhered regime can be attributed to the stress fiber formation. The traction force exerted by cells at E ~ 1 kPa may reach the critical threshold for the activation of mechanoreceptors. On the other hand, the plateau regime with E ≥ 20 kPa suggests that the activation of mechanosensors of C2C12 cells is saturated.
In summary, the laser-induced cell detachment assay revealed that the adhesion strength exhibits a transition from weak adhesion to strong adhesion within the distinct elasticity range (E ~ 1–20 kPa). Actually, elastic modulus of environments of myoblast cells in nature is ~ 10 kPa. Engler et al. reported that an optimal elastic modulus of E ~ 12 kPa maximizes myosin striations in muscle [22]. Interestingly, the optimal elastic modulus is very close to that at the middle of adhesion strength transition (E 1/2- adhesion ~ 7.6 kPa). This clearly indicates that adhesion strength can be a quantitative measure to assess optimal cell-substrate interaction for muscle tissue formation. I believe that the approach with the combination of hydrogel and the pressure wave assays is powerful to reveal the optimal interaction between cells and hydrogel for tissue engineering.
4 Conclusion
The advanced optical techniques mentioned in this chapter can provide quantitative measures for cell-hydrogel adhesion. The advanced RICM can visualize cell-hydrogel contact on the basis on distance information reconstructed from interferrometric signals. The laser-induced cell detachment assay can provide with adhesion strength of cell-hydrogel, which is a direct, mechanical measure for the interaction between cells and hydrogel. The systematic application of these techniques will contribute to the understanding of mechanical interactions between cells and hydrogel, which subsequently leads to the rational design of biomaterials in tissue engineering.
References
Pelham RJ, Wang YL (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94(25):13661–13665
Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126(4):677–689. doi:10.1016/j.cell.2006.06.044
Horning M, Kidoaki S, Kawano T, Yoshikawa K (2012) Rigidity matching between cells and the extracellular matrix leads to the stabilization of cardiac conduction. Biophys J 102(3):379–387. doi:10.1016/j.bpj.2011.12.018
Wells RG (2008) The role of matrix stiffness in regulating cell behavior. Hepatology 47(4):1394–1400
Yoshikawa HY, Rossetti FF, Kaufmann S, Kaindl T, Madsen J, Engel U, Lewis AL, Armes SP, Tanaka M (2011) Quantitative evaluation of mechanosensing of cells on dynamically tunable hydrogels. J Am Chem Soc 133(5):1367–1374. doi:10.1021/Ja1060615
Yoshikawa HY, Cui J, Kratz K, Matsuzaki T, Nakabayashi S, Marx A, Engel U, Lendlein A, Tanaka M (2012) Quantitative evaluation of adhesion of osteosarcoma cells to hydrophobic polymer substrate with tunable elasticity. J Phys Chem B 116(28):8024–8030. doi:10.1021/jp212385p
Yoshikawa HY, Kawano T, Matsuda T, Kidoaki S, Tanaka M (2013) Morphology and adhesion strength of myoblast cells on photocurable gelatin under native and non-native micromechanical environments. J Phys Chem B 117(15):4081–4088
Matsuzaki T, Sazaki G, Suganuma M, Watanabe T, Yamazaki T, Tanaka M, Nakabayashi S, Yoshikawa HY (2014) High contrast visualization of cell-hydrogel contact by advanced interferometric optical microscopy. J Phys Chem Lett 5(1):253–257
Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous-solution. Nature 374(6522):555–559
Curtis AS (1964) The mechanism of adhesion of cells to glass. a study by interference reflection microscopy. J Cell Biol 20:199–215
Izzard CS, Lochner LR (1976) Cell-to-substrate contacts in living fibroblasts: an interference reflexion study with an evaluation of the technique. J Cell Sci 21(1):129–159
Gingell D, Todd I (1979) Interference reflection microscopy. A quantitative theory for image interpretation and its application to cell-substratum separation measurement. Biophys J 26(3):507–526. doi:10.1016/S0006-3495(79)85268-6
Rädler J, Sackmann E (1993) Imaging optical thickness and separation distances of phospholipid vesicles at solid surfaces. J Phys II France 3:724–748
Limozin L, Sengupta K (2009) Quantitative reflection interference contrast microscopy (RICM) in soft matter and cell adhesion. Chemphyschem 10(16):2752–2768
Wen R, Lahiri A, Azhagurajan M, Kobayashi S, Itaya K (2010) A new in situ optical microscope with single atomic layer resolution for observation of electrochemical dissolution of AU(111). J Am Chem Soc 132(39):13657–13659
Suzuki Y, Sazaki G, Matsumoto M, Nagasawa M, Nakajima K, Tamura K (2009) First direct observation of elementary steps on the surfaces of glucose Isomerase crystals under high pressure. Cryst Growth Des 9(10):4289–4295
Sazaki G, Matsui T, Tsukamoto K, Usami N, Ujihara T, Fujiwara K, Nakajima K (2004) In situ observation of elementary growth steps on the surface of protein crystals by laser confocal microscopy. J Cryst Growth 262(1–4):536–542
Sazaki G, Zepeda S, Nakatsubo S, Yokoyama E, Furukawa Y (2010) Elementary steps at the surface of ice crystals visualized by advanced optical microscopy. Proc Natl Acad Sci USA 107(46):19702–19707
Kidoaki S, Matsuda T (2008) Microelastic gradient gelatinous gels to induce cellular mechanotaxis. J Biotechnol 133(2):225–230
Kawano T, Kidoaki S (2011) Elasticity boundary conditions required for cell mechanotaxis on microelastically-patterned gels. Biomaterials 32(11):2725–2733
Griffin MA, Engler AJ, Barber TA, Healy KE, Sweeney HL, Discher DE (2004) Patterning, prestress, and peeling dynamics of myocytes. Biophys J 86(2):1209–1222. doi:10.1016/S0006-3495(04)74195-8
Engler AJ, Griffin MA, Sen S, Bonnetnann CG, Sweeney HL, Discher DE (2004) Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol 166(6):877–887
Tsai MA, Waugh RE, Keng PC (1996) Cell cycle-dependence of HL-60 cell deformability. Biophys J 70(4):2023–2029
Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D (2004) Substrate compliance versus ligand density in cell on gel responses. Biophys J 86(1):388a
Nossal R (1998) Cell transit analysis of ligand-induced stiffening of polymorphonuclear leukocytes. Biophys J 75(3):1541–1552
Burridge K, Connell L (1983) A new protein of adhesion plaques and ruffling membranes. J Cell Biol 97(2):359–367
Kaufmann S, Piekenbrock T, Goldmann WH, Barmann M, Isenberg G (1991) Talin binds to actin and promotes filament nucleation. FEBS Lett 284(2):187–191
Vogel V, Sheetz M (2006) Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 7(4):265–275. doi:10.1038/nrm1890
Acknowledgments
I thank Prof. Motomu Tanaka for the tight collaboration for developing the advanced optical techniques. I also thank Prof. Satoru Kidoaki for the support of photocurable gelatin and Prof. Gen Sazaki for the support of the advanced RICM.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Japan
About this chapter
Cite this chapter
Yoshikawa, H. (2015). Quantitative Evaluation of Cell-Hydrogel Adhesion by Advanced Optical Techniques. In: Arai, T., Arai, F., Yamato, M. (eds) Hyper Bio Assembler for 3D Cellular Systems. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55297-0_13
Download citation
DOI: https://doi.org/10.1007/978-4-431-55297-0_13
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55296-3
Online ISBN: 978-4-431-55297-0
eBook Packages: EngineeringEngineering (R0)