Abstract
The contribution of a single gene to each neurodegenerative disease is diverse in its effect size, depth, and mode of action. In those relatively rare neurodegenerative diseases that show a Mendelian pattern of inheritance, mutations in a major causative gene are solely responsible for the disease phenotype and underlying pathogenesis; whereas, the occurrence of more common neurodegenerative diseases likely requires the combined effect of alterations in many genetic factors and independent environmental factors. The pathological process of neuronal degeneration stems from deficits in neuronal or non-neuronal genes. Therefore, understanding the genetic landscape of neurodegeneration requires delineation of the general or unique effect of individual disease-related genes on the neurodegeneration process. Additionally, the interaction among genes should be determined. This chapter overviews, from distinct aspects, representative examples of genetic factors that have a major impact on the pathophysiology of neurodegenerative diseases.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mendelian inheritance
- Susceptibility
- Alzheimer’s disease
- Amyotrophic lateral sclerosis
- Frontotemporal dementia
- Leukodystrophy
- Astrocyte
- Oligodendrocyte
- Microglia
1 Introduction
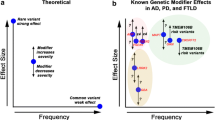
Recent advances in the technology underlying comprehensive genetic and genomic analyses have enabled rapid and high-throughput studies that delineate numerous causative and susceptibility genes for neurodegenerative disorders. It has become apparent, from the many examples reported during the last two decades or so, that genetic factors play a major role in the pathological development of many neurodegenerative diseases. Moreover, we know that the effect size and depth of each genetic factor may vary among different diseases and genes. In a simple model showing the contribution of genetic and environmental factors, Mendelian disorders are placed at the extreme end where an abnormality in a single gene is sufficient to cause a disease (Fig. 6.1). At the other end, some diseases are entirely dependent on environmental factors, such as infections and traumatic accidents. In reality, most neurodegenerative diseases are placed somewhere in the middle of these two extremes. Therefore, an understanding of both genetic and environmental factors is critical to delineating the pathogenesis of neurodegenerative diseases.
The genetic and environmental factors that contribute to the occurrence of neurodegenerative diseases. (Left) Disease defined by a mutation in a single gene (as observed in monogenic Mendelian diseases). (Right) Some diseases are actually the result of environmental factors (such as injuries or nutrition). (Middle) The most common neurodegenerative diseases (in which multiple factors including susceptibility genes and environmental effects contribute to disease occurrence)
What is important yet difficult to understand is that genetic and environmental factors not only contribute to the pathogenesis independently, but also interact with each other to elicit their effect. Investigation and determination of such an interaction between genetic and environmental factors requires analysis of the disease as a systematic disorganization; techniques to uncover such dynamic interactions have not been well developed.
This chapter focuses on the genetic factors that primarily influence the pathology of neurodegenerative diseases at different levels and discusses the effect of environmental factors on the genetic basis of neurodegenerative diseases.
2 Neuronal Autonomous Genetic Factors
2.1 Genetic Factors That Directly Cause Neurodegenerative Disorders
Single genetic factors, mostly alterations in the coding exons of genes, can directly cause neurodegenerative diseases. In rare Mendelian disorders such single-gene defects are responsible for most of the disease mechanisms and phenotypes. This is the most simple and straightforward way of modeling the influence of genetic factors on neurodegeneration. A good example of such modeling is Huntington’s disease (HD). HD is an autosomal-dominant neuropsychiatric disease characterized by progressive movement disorder, most commonly presenting as chorea, progressive cognitive decline leading to dementia, and various psychiatric symptoms that often precede diagnosis (Paulson and Albin 2011). The mutation causing HD is a CAG repeat expansion in the HTT gene (Group THsDCR 1993). This unique mutation is highly specific; no other type of mutation in HTT has been found to cause HD and all affected individuals with HD carry the CAG repeat expansion in HTT. It has been well recognized that the number of repeats is inversely correlated with age of onset (Andrew et al. 1993). Similarly, expanded CAG repeats commonly lead to a lengthy polyglutamine (polyQ) stretch in other autosomal-dominant neurodegenerative disorders including spinocerebellar ataxias, spinobulbar muscular atrophy, and dentatorubral–pallidoluysian atrophy (Zoghbi and Orr 2000). The strong correlation between the number of CAG repeats and age of onset has also been identified in these diseases, suggesting that the disease-associated polymorphic genotype also determines disease onset. Although the presence of a common pathology influencing the age of onset (and presumably disease progression) among these diseases has been expected, molecular mechanisms underlying these polyQ expansions appear to be quite complex, as reviewed elsewhere (La Spada et al. 2011). Nevertheless, CAG expansion involving multiple genes is a unique genetic factor that is specifically associated with neurodegeneration.
Sometimes, a single clinical phenotype can be caused by an alteration in many different genes. This condition is called “genetic heterogeneity.” Charcot–Marie–Tooth (CMT) disease is one of the most common neurogenetic disorders, with a prevalence of 1 in 2500. CMT is characterized by bilateral distal wasting, weakness, and sensory loss that begins in the lower limbs and slowly progresses in a length-dependent manner. CMT is further divided into two forms, CMT1 (demyelinating neuropathy) and CMT2 (axonal neuropathy), based on electrophysiological findings. CMT can be dominant, recessive, or X-linked; more than 60 CMT-associated genes have been identified and the list is still growing (Timmerman et al. 2014). Mutations in each gene likely affect the diverse functions of Schwann cells (associated with CMT1) and neurons (associated with CMT2) in the peripheral nervous system. The most common CMT-causing mutation is the genomic duplication of 17p11.2 that harbors PMP22, which encodes a myelin membrane protein. Another myelin-associated gene, MPZ, is also commonly mutated in patients with CMT1. Mutations in GJB1, encoding connexin32, affect the gap junctions of the myelin membrane. Genes involved in transcription and mRNA processing (EGR2 and CTDP1), cytoskeletal structure (PRX, IFN2, FGD4, and FBLN5), endosomal sorting and cell signaling (LITAF, SH3TC2, MTMR2, MTMR13, SBF1, FIG4, DNM2, and NDRG1), and mitochondria (GDAP1 and HK1) in Schwann cells can be affected in CMT1 (reviewed in Rossor et al. 2013). In CMT2, abnormalities that are even more diverse occur in the subcellular components of the neuronal axon and cell body, including defective nuclear envelope and mRNA processing, endoplasmic reticulum and Golgi apparatus, endosomal sorting and cell signaling, proteasome and protein aggregation, mitochondria, channels, axonal transport, and synaptic transmission (Rossor et al. 2013). These findings suggest that a single neurodegenerative phenotype can occur as a consequence of multi-layered pathological steps affecting tissues, cells, subcellular functional units, and genes, each of which can be responsible for the disease phenotype when mutated.
Single-gene alterations may only explain the common forms of neurodegenerative diseases in a small number of patients, while the vast majority of patients with the same disease may carry no such alterations. Dissecting the molecular pathology of such mutations not only explains the disease in rare cases, but also sheds light on the pathogenesis in most patients, because the rare genetic defect may affect the pathway followed by the common form of the disease. Alzheimer’s disease serves as an example of such a case. Analyses of a small number of early-onset Alzheimer disease families showing an autosomal-dominant pattern of inheritance uncovered highly penetrant mutations in three genes: APP, PSEN1, and PSEN2 (Goate et al. 1991; Sherrington et al. 1995; Rogaev et al. 1995; Levy-Lahad et al. 1995). APP encodes a single transmembrane protein from which amyloid β (Aβ) polypeptides are cleaved by β- and γ-secretases. Aβ polypeptides polymerize to form insoluble Aβ aggregates, the main component of senile plaque, which is the pathological hallmark in patients’ brains. Most APP mutations are heterozygous missense alterations located in or near Aβ-coding exons (Goate et al. 1991). Rare whole-gene duplications (Rovelet-Lecrux et al. 2006) as well as small recessive deletions and missense alterations have also been reported (Di Fede et al. 2009). These APP mutations either change Aβ production, increase the Aβ42 to Aβ40 ratio (which differs in length according to cleavage sites; Aβ42 is more toxic), or enhance fibril formation. Interestingly, one particular APP single-nucleotide polymorphism (SNP), which leads to a change in Ala673Thr, was reported to strongly protect carriers from Alzheimer’s disease (1/OR = 5.29) (Jonsson et al. 2012). This SNP is located close to the cleavage site of BACE1 (β-site APP cleaving enzyme 1) and functionally inhibits BACE1 cleavage to produce Aβ. PSEN1 and PSEN2 are both critical components of γ-secretase, and mutations in either gene result in an increased ratio of Aβ42 to Aβ40 (De Strooper et al. 1998; Scheuner et al. 1996). Findings from these early-onset familial Alzheimer disease cases provided the solid genetic basis for the central role of Aβ in the pathogenesis of the common form of Alzheimer’s disease.
2.2 Genetic Factors That Affect Susceptibility to Neurodegenerative Disorders
In most patients suffering from neurodegenerative diseases, genetic factors play a more complex role in the pathogenesis, onset, phenotype, and prognosis of the disease than in patients suffering from rare Mendelian forms of diseases (discussed in the previous section). Multiple genes appear to orchestrate together with environmental factors to formulate disease status. Recent advances in genome-wide association studies (GWAS) have been successfully delineating the susceptibility genes that contribute to each part of pathogenesis in the complex process of neurodegenerative diseases.
The best known example of such susceptibility genes with the largest effect size is the APOE gene in Alzheimer’s disease (Corder et al. 1993). In fact, the Manhattan Plot (an overview of GWAS results encompassing the entire human genome) demonstrated a single incomparable high peak at the APOE locus, while other associated loci across the entire genome revealed only modest effects. The APOE gene harbors three polymorphic alleles, ε2, ε3, and ε4, of which the ε4 allele is a risk allele for Alzheimer’s disease (Farrer et al. 1997). A meta-analysis in individuals of European descent showed that the risk of Alzheimer’s disease is increased in people carrying one copy of the ε4 allele (ε2/ε4, odds ratio 2.6; ε3/ε4, odds ratio 3.2) or two copies (ε4/ε4, odds ratio 14.9), compared with those carrying an ε3/ε3 genotype (Farrer et al. 1997). This association is also observed in other populations with a weaker (African American ε4/ε4, odds ratio 5.7; Hispanic ε4/ε4, odds ratio 2.2) or a stronger (Japanese ε4/ε4, odds ratio 33.1) effect (Farrer et al. 1997). APOE genotypes strongly affect Aβ deposition as a result of haplotype-specific differences in metabolizing Aβ. Findings from multiple studies in humans and mice suggest that APOE ε4 increases the risk by initiating and accelerating the accumulation, aggregation, and deposition of Aβ in the brain (Liu et al. 2013). APOE ε4 is less efficient at clearing Aβ from the brain. APOE genotypes are also associated with mild cognitive impairment (MCI), which is now considered a pre–Alzheimer disease condition. Individuals with MCI who carry the ε4 genotype are at a higher risk of progression from MCI to Alzheimer-type dementia (Petersen et al. 1995) and suffer a faster cognitive decline than non-carriers (Cosentino et al. 2008). Thus, APOE serves as a major susceptibility gene for Alzheimer’s disease with the ε4 allele increasing the risk by direct enhancement of Aβ pathogenesis.
By contrast, other susceptibility genes in at least nine loci (CLU, CR1, PICALM, BIN1, EPHA1, ABCA7, MS4A cluster, CD33, and CD2AP), identified and confirmed by multiple GWAS, have shown a much smaller effect size (reviewed in Bettens et al. 2013). Pathogenic mechanisms of variations in these nine susceptibility genes have not yet been established; however, these susceptibility genes may be related to specific functions including lipid processing (CLU and ABCA7), functioning of the complement system, inflammation, immune system (CLU, CR1, ABCA7, CD33, and EPHA1), and synaptic cell functions such as endocytosis (PICALM, BIN1, CD33, and CD2AP). However, the mechanisms by which these gene variations affect the pathological process of Alzheimer’s disease remains largely undetermined. Interestingly, common gene variants that have a predominant effect when mutated (i.e., APP, PSEN1, and PSEN2) are not risk factors for the complex forms of Alzheimer’s disease (Harold et al. 2009; Lambert et al. 2009). This contrasts with the findings in Parkinson’s disease, wherein common variants in the genes that are responsible for the Mendelian form of the disease (i.e., MAPT, SNCA, and LRRK2) also serve as risk alleles in the common form of the disease (Simon-Sanchez et al. 2009; Satake et al. 2009).
2.3 Genetics Factors That Contribute to Multiple Neurodegenerative Disorders
Genetic exploration and identification of the genes contributing to neurodegenerative disorders have begun to uncover the fact that clinically distinct neurodegenerative disorders share a common genetic basis to a significant degree. A single gene can contribute to the occurrence of different diseases. There are striking examples of this in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
ALS is a devastating degenerative disorder of motoneurons. It is characterized by the onset of focal weakness, typically in the limbs but sometimes in bulbar muscles. This progresses to paralysis of almost all skeletal muscles, leading to death from respiratory failure typically within 5 years (Sreedharan and Brown 2013). FTD is a distinct form of dementia characterized by progressive degeneration of the frontal or temporal lobe, or both (Loy et al. 2014). FTD is clinically characterized by deterioration in behavior and speech, but memory and visuospatial function are spared to a certain degree. ALS and FTD have been thought of as distinct clinical entities, although rare overlapping cases have been reported. The seminal molecular pathology discovery connecting ALS and FTD was the identification of TDP-43 as the major component of ubiquitinated neuronal cytoplasmic inclusions in both diseases (Neumann et al. 2006), which also led to the identification of mutations in the TARDBP gene (which encodes TDP-43) in autosomal-dominant ALS and FTD families (Sreedharan et al. 2008; Chio et al. 2010). More recently, a massive expansion of a hexanucleotide repeat in an intron of C9ORF72 linked to chromosome 9q21 was found to cause ALS and FTD (Renton et al. 2011; DeJesus-Hernandez et al. 2011). Subsequently, it became apparent that this locus was responsible for a major part of both familial ALS (~40 %) and FTD (~25 %) (Majounie et al. 2012). In addition, at least three other genes are known to link ALS and FTD. FUS encodes an RNA-binding protein that has functional homology with TDP-43. FUS mutations have been found in about 4 % of familial ALS cases without dementia (Kwiatkowski et al. 2009; Vance et al. 2009); however, in rare families, FUS mutations also cause FTD (Blair et al. 2010). Note that FTD exhibiting a FUS-positive histopathological subtype is commonly observed; however, most of these cases do not carry FUS mutations (Snowden et al. 2011). Mutations in the VCP gene, which encodes valosin-containing protein, were initially found to cause inclusion body myopathy, an unusual clinical syndrome characterized by FTD, and Paget’s disease of the bone (Watts et al. 2004); however, they were later found to be responsible for 1–2 % of familial ALS cases too (Johnson et al. 2010). Ubiquitin 2 regulates the proteasome degradation of ubiquitinated proteins. The gene encoding this protein, UBQLN2, is responsible for rare familial cases of ALS and ALS/FTD syndrome (Deng et al. 2011). Since UBQLN2 pathology has been observed in patients with ALS who do not carry mutations in this gene, this protein may play an important role in the final common pathway associated with motor neuron degradation. These cumulative genetic findings have drastically changed our understanding of unexpectedly common molecular pathologies between ALS and FTD.
3 Non-neuronal Genetic Factors Affecting Neurons
In a number of neurodegenerative diseases, the disease genes primarily function in non-neuronal cells including astrocytes, oligodendrocytes, microglia, and vascular cells. Deficits in these non-neuronal genes also affect neurons by causing neurodegeneration. The following examples highlight the contribution of genetic factors affecting cells other than neurons.
3.1 Genetic Deficits in Astroglia That Cause Neurodegeneration
Astrocytes constitute the largest volume of cells in the central nervous system (CNS). Named for its star-like shape, astrocytes are distributed throughout the CNS in both the gray and white matter. Once considered little more than scaffolds for neurons, astrocytes play multiple roles by interacting with many types of cells and structural components in the CNS – including neurons, oligodendrocytes, other astrocytes, blood vessels, pial membranes, and the ependymal layer that lines ventricles – to maintain tissue homeostasis (Lanciotti et al. 2013).
In their interaction with neurons, astrocytes extend many fine processes to contact synapses and establish highly regulated bidirectional communication. By secreting trophic factors – such as brain-derived nerve growth factor (BDNF), glial cell–derived neurotrophic factor (GDNF), nerve growth factor (NGF), insulin-like growth factor (IGF), and thrombospondin – astrocytes contribute to the formation, maintenance, and remodeling of synapses. Astrocytes also modulate neuronal function by secreting cholesterol, apolipoprotein E, glutathione, and hydrogen sulfide. They express receptors to take up neurotransmitters – including glutamate, gamma-aminobutyric acid (GABA), norepinephrine, dopamine, serotonin, acetylcholine, and glycine – to remove them from the synaptic cleft and maintain their low extracellular concentrations. The clearance of glutamate, mainly mediated by a high-affinity excitatory amino acid transporter 1 (EAAT1, also known as “GLT1”), is critical to protecting neurons from excitotoxicity (Rothstein et al. 1996). Dysfunction of EAAT1 and accumulation of excessive extracellular glutamate have been implicated in several neurodegenerative processes and associated diseases, such as epilepsy, stroke, ALS, HD, and Alzheimer’s disease (Coulter and Eid 2012; Rothstein 2009; Faideau et al. 2010; Simpson et al. 2010). Astrocytes also supply neurons with energy substrates. Both glucose and lactate are transported to neurons to maintain the energy source for intense neuronal activity. Astrocytes also control the homeostasis of water and ion fluxes in the synaptic interstitial space through numerous ion channels and transporters.
Astrocytes also provide growth factors – such as platelet-derived growth factor alpha (PDGF-α), fibroblast growth factor 2 (FGF2), neurotrophin-3 (NT-3), IGF-1, and ciliary neurotrophic factor (CNTF) – which are necessary for the survival of oligodendrocytes (Lanciotti et al. 2013). Astrocytes are thought to regulate CNS myelination probably by secreting growth factors; elucidation of the underlying mechanism is important to understanding how astrocytes contribute to CNS repair and remyelination, which could be useful in the treatment of demyelinating diseases.
Astrocytes not only play essential roles to maintain healthy CNS homeostasis, they are also involved in many pathological processes. Specifically, genetic deficits in astrocyte-specific genes can cause neurodegenerative disorders, characterized as cystic leukodystrophies including Alexander disease (AxD), megalencephalic leukodystrophy with subcortical cysts (MLC), and vanishing white matter (VWM). These are all rare genetic diseases that commonly show the characteristic features of demyelinating leukodystrophy with progressive cystic or spongy (vacuolating) degeneration of myelin.
AxD is an autosomal-dominant disease caused by heterozygous mutations in the astrocyte-specific type III intermediate filament GFAP gene (Brenner et al. 2001). Clinical manifestations of AxD vary in severity and onset from more severe infantile and juvenile forms to milder adult onset forms (Messing et al. 2012). Patients with the infantile form show progressive features with seizures, bulbar dysfunction, psychomotor regression, and short lifespan. Pathologically, the brains of patients with severe AxD show loss of oligodendrocytes and myelin; cystic degeneration; neuronal loss most commonly in the hippocampus, striatum, and neocortex; and the presence of Rosenthal fibers in the cytoplasm of astrocytes (Messing et al. 2012). The exact mechanisms for neurodegeneration in AxD remain unknown, but both toxic gain-of-function and impairment of normal astrocyte supportive function have been implicated. Specifically, a marked downregulation of GLT-1 in astrocytes expressing mutant GFAP, and the vulnerability of hippocampal neurons to glutamate-induced cytotoxicity in co-cultures with astrocytes expressing mutant GFAP have been reported (Mignot et al. 2007). These findings may link GFAP mutations to glial glutamate transporter-1 (GLT-1) dysfunction and impairment of the neuron–astrocyte interaction, suggestive of a potential mechanism underlying neuronal loss in AxD. Similarly, dysregulation of the homeostatic functions of astrocytes – including extracellular K+ buffering by Kir4.1 and Na+/K+–ATPase activity – have been implicated in the demyelination of AxD (Hagemann et al. 2005).
3.2 Genetic Deficits in Oligodendrocytes That Cause Neurodegeneration
Mature oligodendrocytes extend cell processes that are modified into thin sheaths of plasma membrane wrapping around a portion of an axon in a spiral fashion (so-called “myelin”). Each oligodendrocyte interacts with as many as 50 different axons by extending multiple processes. Each axon is ensheathed by multiple continuous segments of myelin, separated by small areas of bare axons exposed to interstitial spaces called “nodes of Ranvier.” Myelin functions as an insulator of electronic circuits in neuronal networks, where it enables rapid, efficient, and distant transmission without attenuation. In addition to these unique features of myelin as a structural component for highly efficient axonal conduction, oligodendrocytes communicate with axons through several signaling processes. Axonal signals that support myelin include neuregulins, neurotrophins, and the electrical activity of axons, while oligodendrocytes support axons through the function of some myelin membrane–bound proteins, such as PLP1 and CNP1. Mice in which either PLP1 or CNP1 has been disrupted develop normal myelin, suggesting that these myelin proteins are dispensable for normal myelination. On the other hand, these mice develop broad axonal swelling, indicating that PLP1 and CNP1 support axons independent of myelin (Nave and Trapp 2008). Of these two myelin proteins only PLP1 is directly associated with human disease.
Leukodystrophy comprises a group of rare genetic disorders that primarily target white matter in which myelinating oligodendrocytes and axons predominate. Pelizaeus–Merzbacher disease (PMD) is a hypomyelinating leukodystrophy characterized by deficit in CNS myelination (Inoue 2005). PMD is caused by mutations in PLP1, which encodes a major myelin membrane protein. PMD-causing PLP1 mutations include point mutations that mostly result in amino acid substitutions, genomic duplications that lead to an additional copy of the gene, and null mutations caused by deletions or early-truncating mutations (Inoue 2005). In PMD the structure of the neuronal network is essentially intact. However, slowly progressive axonal degeneration may occur later in the disease probably because of the loss of axonal support by myelin and the resulting metabolic and electronic insufficiency in the axons. Activated microglia have also been found at the site of hypomyelinated axons; thus, microglia may additionally contribute to hypomyelination-associated axonopathy (Ip et al. 2006). In addition to these features (generally observed in all types of mutations), patients with null mutations show an apparent length-dependent progressive axonopathy characterized by axonal swelling (Griffiths et al. 1998; Garbern et al. 2002). This is despite the fact that clinical severity and associated leukodystrophy observed by magnetic resonance imaging (MRI) in patients with null mutations are surprisingly milder than in patients with either point mutations or duplications. It is not completely understood why a milder condition with almost normal myelinating oligodendrocytes with no PLP1 expression is more susceptible to axonal degeneration than severe conditions with bare axons and almost no mature oligodendrocytes caused by other types of mutations. Nevertheless, the expression of myelin-specific genes such as PLP1 appears to play a role in preventing the degeneration of axons.
More than 30 leukodystrophies and their causative genes are known, and most, if not all, involve neurodegeneration (Kohlschutter and Eichler 2011), suggesting that a variety of genetic factors lead to this specific type of neurodegeneration. These diseases commonly show disruption of the myelin sheath in the CNS accompanied by axonal degeneration and inflammatory changes, together called “demyelination.” The genetic bases of leukodystrophies are complex; genes involved in many different cells and functions have been implicated (Kohlschutter and Eichler 2011). X-linked adrenoleukodystrophy is caused by mutations in ABCD1, which encodes a member of the superfamily of ATP-binding cassette transporters (Steinberg et al. 1993). ABCD1 is present on the peroxisomal membrane and is involved in the transport of very long chain fatty acids (VLCFA)–coenzyme A (CoA) synthetase. Defects in ABCD1 result in accumulation of VLCFA, which is toxic to brain cells, especially microglia and oligodendrocytes, and thus leads to axonal degeneration. Mutations in a gene encoding the lysosomal enzyme, arylsulfatase A, cause metachromatic leukodystrophy, which is characterized by abnormal accumulation of cerebroside sulfate (the major component of myelin lipids), which leads to myelin breakdown and neuronal death. Further examples of neurodegenerative leukodystrophies are hypomyelination with atrophy of the basal ganglia and cerebellum (mutations in the β-tubulin4A gene), Canavan disease (mutations in the aspartoacylase gene), and Krabbe disease (mutations in the galactocerebrosidase gene).
3.3 Genetic Deficits in Microglia That Cause Neurodegeneration
Microglial activation and neuroinflammation are major components in neurodegeneration. Numerous genes and pathways are associated with the important role microglia play in neurodegeneration. However, the focus here is on a small number of microglia-specific genes, in which mutations primarily cause neurodegenerative disorders. Nasu–Hakola disease (NHD), also known as “polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy,” is a rare autosomal-recessive neurodegenerative disease characterized by multifocal bone cysts and progressive early-onset dementia. Two genes are known to be responsible for NHD – TYROBP and TREM2 (Paloneva et al. 2000, 2002). Both these genes encode components of cell surface receptors that are expressed not only in blood immune system cells, but also in microglia in the CNS and bone osteoclasts. TYROBP and TREM2 form a receptor-signaling complex that triggers the activation of immune responses in macrophages and dendritic cells (Lanier and Bakker 2000). Although the exact molecular pathogenesis is unknown, loss-of-function mutations in TYROBP and TREM2 are predicted to cause dysfunction of microglia. The brains of patients with NHD show sudanophilic leukodystrophy characterized by loss of myelin and nerve fibers, and gliosis especially in the frontal and temporal lobes (Nasu et al. 1973). Note that these broad neurodegenerative changes involving oligodendrocytes, astrocytes, and neurons primarily result from the abnormal function of microglia. NHD is an extremely rare disease and the vast majority of patients are either Finnish or Japanese (Hakola 1972; Nasu et al. 1973). However, the NHD-associated microglial gene, TREM2, appears to have a broader impact on common neurodegenerative diseases. Heterozygous mutations in TREM2 are associated with an increased risk of several neurodegenerative disorders including Alzheimer’s disease, Parkinson’s disease, ALS, and FTD (Guerreiro et al. 2013; Jonsson et al. 2013; Rayaprolu et al. 2013; Cady et al. 2014; Cuyvers et al. 2014; Borroni et al. 2014). Although the allele frequency of the variant TREM2 is relatively low (~0.3 %), the effect size is remarkably high in Alzheimer’s disease (odds ratio > 3, which is close to that of the APOE4 allele). The exact mechanisms by which TREM2 mutations increase susceptibility to different neurodegenerative diseases is still unknown, but a recent study suggested that reduced function of TREM2 impairs phagocytosis, which may contribute to neurodegeneration (Kleinberger et al. 2014). These findings suggest that normal function of microglia is critical to preventing neurodegeneration in the brain.
4 Conclusions
This chapter overviewed the genetic factors that contribute to the development of neurodegenerative disorders from a wide variety of viewpoints. Neurodegenerative disorders can occur because of a mutation in a single gene or because of multiple contributions from alterations in some genes, or possibly many genes. In addition, environmental factors may also influence the development of neurodegenerative disorders through many different pathophysiological pathways. Such findings have of course been made possible by the rapid and dramatic technological advances in comprehensive genetic and genomic analyses, including genome-wide linkage analysis, GWAS, SNP array analysis, array comparative genomic hybridization (aCGH) analysis, whole-exome sequencing, and whole-genome sequencing. However, despite these advances, we have barely scratched the surface of the genetic contribution to pathogenesis of the most common neurodegenerative disorders. A prominent peak of the pathophysiological landscape of neurodegenerative disorders delineated by the discovery of underlying genetic and genomic bases can now be seen (Fig. 6.2). However, many small peaks representing genes with smaller effect size have yet to be identified. Furthermore, the risk factors stemming from interactions within these genetic factors or between genetic and environmental factors have not been fully elucidated. The fundamental basis of neurodegenerative disorders awaits clarification by future advances in high-throughput deep genomic exploration technology incorporating the multi-factorial interaction with environmental effects.
References
Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA et al (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet 4:398–403
Bettens K, Sleegers K, Van Broeckhoven C (2013) Genetic insights in Alzheimer’s disease. Lancet Neurol 12:92–104
Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA (2010) FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry 81:639–645
Borroni B, Ferrari F, Galimberti D, Nacmias B, Barone C, Bagnoli S, Fenoglio C, Piaceri I, Archetti S, Bonvicini C, Gennarelli M, Turla M, Scarpini E, Sorbi S, Padovani A (2014) Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging 35(934):e7–e10
Brenner M, Johnson AB, Boespflug-Tanguy O, Rodriguez D, Goldman JE, Messing A (2001) Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat Genet 27:117–120
Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, Baloh RH, Ravits J, Simpson E, Appel SH, Pestronk A, Goate AM, Miller TM, Cruchaga C, Harms MB (2014) TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol 71:449–453
Chio A, Calvo A, Moglia C, Restagno G, Ossola I, Brunetti M, Montuschi A, Cistaro A, Ticca A, Traynor BJ, Schymick JC, Mutani R, Marrosu MG, Murru MR, Borghero G (2010) Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p.Ala382Thr TARDBP mutations. Arch Neurol 67:1002–1009
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923
Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, Blacker D, Stern Y (2008) APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology 70:1842–1849
Coulter DA, Eid T (2012) Astrocytic regulation of glutamate homeostasis in epilepsy. Glia 60:1215–1226
Cuyvers E, Bettens K, Philtjens S, Van Langenhove T, Gijselinck I, Van Der Zee J, Engelborghs S, Vandenbulcke M, Van Dongen J, Geerts N, Maes G, Mattheijssens M, Peeters K, Cras P, Vandenberghe R, De Deyn PP, Van Broeckhoven C, Cruts M, Sleegers K, Consortium B (2014) Investigating the role of rare heterozygous TREM2 variants in Alzheimer’s disease and frontotemporal dementia. Neurobiol Aging 35(726):e11–e19
De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391:387–390
Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256
Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477:211–215
Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantu L, Del Favero E, Levy E, Salmona M, Tagliavini F (2009) A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323:1473–1477
Faideau M, Kim J, Cormier K, Gilmore R, Welch M, Auregan G, Dufour N, Guillermier M, Brouillet E, Hantraye P, Deglon N, Ferrante RJ, Bonvento G (2010) In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum Mol Genet 19:3053–3067
Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, Van Duijn CM (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA 278:1349–1356
Garbern JY, Yool DA, Moore GJ, Wilds IB, Faulk MW, Klugmann M, Nave KA, Sistermans EA, Van Der Knaap MS, Bird TD, Shy ME, Kamholz JA, Griffiths IR (2002) Patients lacking the major CNS myelin protein, proteolipid protein 1, develop length-dependent axonal degeneration in the absence of demyelination and inflammation. Brain 125:551–561
Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L et al (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349:704–706
Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, Mcculloch M, Nadon N, Nave KA (1998) Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science 280:1610–1613
Group, T. H. S. D. C. R (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983
Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J, Alzheimer Genetic Analysis, G. (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368:117–127
Hagemann TL, Gaeta SA, Smith MA, Johnson DA, Johnson JA, Messing A (2005) Gene expression analysis in mice with elevated glial fibrillary acidic protein and Rosenthal fibers reveals a stress response followed by glial activation and neuronal dysfunction. Hum Mol Genet 14:2443–2458
Hakola HP (1972) Neuropsychiatric and genetic aspects of a new hereditary disease characterized by progressive dementia and lipomembranous polycystic osteodysplasia. Acta Psychiatr Scand Suppl 232:1–173
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, Mcguinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, Van Den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, Mcquillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’donovan M, Owen MJ, Williams J (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41:1088–1093
Inoue K (2005) PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics 6:1–16
Ip CW, Kroner A, Bendszus M, Leder C, Kobsar I, Fischer S, Wiendl H, Nave KA, Martini R (2006) Immune cells contribute to myelin degeneration and axonopathic changes in mice overexpressing proteolipid protein in oligodendrocytes. J Neurosci 26:8206–8216
Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, Mccluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Consortium I, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ (2010) Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68:857–864
Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jonsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K (2012) A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488:96–99
Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, Van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368:107–116
Kleinberger G, Yamanishi Y, Suarez-Calvet M, Czirr E, Lohmann E, Cuyvers E, Struyfs H, Pettkus N, Wenninger-Weinzierl A, Mazaheri F, Tahirovic S, Lleo A, Alcolea D, Fortea J, Willem M, Lammich S, Molinuevo JL, Sanchez-Valle R, Antonell A, Ramirez A, Heneka MT, Sleegers K, Van Der Zee J, Martin JJ, Engelborghs S, Demirtas-Tatlidede A, Zetterberg H, Van Broeckhoven C, Gurvit H, Wyss-Coray T, Hardy J, Colonna M, Haass C (2014) TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med 6:243ra86
Kohlschutter A, Eichler F (2011) Childhood leukodystrophies: a clinical perspective. Expert Rev Neurother 11:1485–1496
Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, De Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, Mckenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH Jr (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323:1205–1208
La Spada AR, Weydt P, Pineda VV (2011) Huntington’s disease pathogenesis: mechanisms and pathways. In: Lo DC, Hughes RE (eds) Neurobiology of Huntington’s disease: applications to drug discovery. CRC Press, Boca Raton
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer’s disease initiative I, De Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41:1094–1099
Lanciotti A, Brignone MS, Bertini E, Petrucci TC, Aloisi F, Ambrosini E (2013) Astrocytes: emerging stars in leukodystrophy pathogenesis. Transl Neurosci 4:144–164
Lanier LL, Bakker AB (2000) The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol Today 21:611–614
Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K et al (1995) Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269:973–977
Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9:106–118
Loy CT, Schofield PR, Turner AM, Kwok JB (2014) Genetics of dementia. Lancet 383:828–840
Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, Van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome, A. L. S. F. T. D. C., French Research Network On, F. F. A., Consortium I, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, Mccluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 11:323–330
Messing A, Brenner M, Feany MB, Nedergaard M, Goldman JE (2012) Alexander disease. J Neurosci 32:5017–5023
Mignot C, Delarasse C, Escaich S, Della Gaspera B, Noe E, Colucci-Guyon E, Babinet C, Pekny M, Vicart P, Boespflug-Tanguy O, Dautigny A, Rodriguez D, Pham-Dinh D (2007) Dynamics of mutated GFAP aggregates revealed by real-time imaging of an astrocyte model of Alexander disease. Exp Cell Res 313:2766–2779
Nasu T, Tsukahara Y, Terayama K (1973) A lipid metabolic disease-“membranous lipodystrophy”-an autopsy case demonstrating numerous peculiar membrane-structures composed of compound lipid in bone and bone marrow and various adipose tissues. Acta Pathologica Japonica 23:539–558
Nave KA, Trapp BD (2008) Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci 31:535–561
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, Mccluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Paloneva J, Kestila M, Wu J, Salminen A, Bohling T, Ruotsalainen V, Hakola P, Bakker AB, Phillips JH, Pekkarinen P, Lanier LL, Timonen T, Peltonen L (2000) Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 25:357–361
Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71:656–662
Paulson HL, Albin RL (2011) Huntington’s disease: clinical features and routes to therapy. In: Lo DC, Hughes RE (eds) Neurobiology of Huntington’s disease: applications to drug discovery. CRC Press, Boca Raton
Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, Kokmen E, Waring SC, Kurland LT (1995) Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA 273:1274–1278
Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Josephs KA, Knopman DS, White CL, 3rd, Caselli R, Mackenzie IR, Miller BL, Boczarska-Jedynak M, Opala G, Krygowska-Wajs A, Barcikowska M, Younkin SG, Petersen RC, Ertekin-Taner N, Uitti RJ, Meschia JF, Boylan KB, Boeve BF, Graff-Radford NR, Wszolek ZK, Dickson DW, Rademakers R, Ross OA (2013) TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Mol Neurodegener 8:19
Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, Van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Consortium I, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268
Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T et al (1995) Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376:775–778
Rossor AM, Polke JM, Houlden H, Reilly MM (2013) Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat Rev Neurol 9:562–571
Rothstein JD (2009) Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol 65(Suppl 1):S3–S9
Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, Kanai Y, Hediger MA, Wang Y, Schielke JP, Welty DF (1996) Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron 16:675–686
Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38:24–26
Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T (2009) Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet 41:1303–1307
Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S (1996) Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med 2:864–870
Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HA, Haines JL, Perkicak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH (1995) Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375:754–760
Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, Van Der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T (2009) Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 41:1308–1312
Simpson JE, Ince PG, Lace G, Forster G, Shaw PJ, Matthews F, Savva G, Brayne C, Wharton SB, Function, M. R. C. C. & Ageing Neuropathology Study, G (2010) Astrocyte phenotype in relation to Alzheimer-type pathology in the ageing brain. Neurobiol Aging 31:578–590
Snowden JS, Hu Q, Rollinson S, Halliwell N, Robinson A, Davidson YS, Momeni P, Baborie A, Griffiths TD, Jaros E, Perry RH, Richardson A, Pickering-Brown SM, Neary D, Mann DM (2011) The most common type of FTLD-FUS (aFTLD-U) is associated with a distinct clinical form of frontotemporal dementia but is not related to mutations in the FUS gene. Acta Neuropathol 122:99–110
Sreedharan J, Brown RH Jr (2013) Amyotrophic lateral sclerosis: problems and prospects. Ann Neurol 74:309–316
Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, De Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319:1668–1672
Steinberg SJ, Moser AB, Raymond GV (1993) X-linked adrenoleukodystrophy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K (eds) GeneReviews(R). University of Washington, Seattle
Timmerman V, Strickland AV, Zuchner S (2014) Genetics of charcot-marie-tooth (CMT) disease within the frame of the human genome project success. Genes (Basel) 5:13–32
Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, De Belleroche J, Gallo JM, Miller CC, Shaw CE (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323:1208–1211
Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36:377–381
Zoghbi HY, Orr HT (2000) Glutamine repeats and neurodegeneration. Annu Rev Neurosci 23:217–247
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Japan
About this chapter
Cite this chapter
Inoue, K. (2015). Genetic Risk Factors for Neurodegenerative Diseases. In: Wada, K. (eds) Neurodegenerative Disorders as Systemic Diseases. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54541-5_6
Download citation
DOI: https://doi.org/10.1007/978-4-431-54541-5_6
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54540-8
Online ISBN: 978-4-431-54541-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)