Abstract
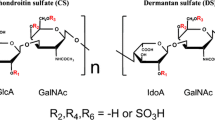
Chondroitin/dermatan sulfate (CS/DS) chains are polysaccharides that are attached to approximately 30 different core proteins, thus giving rise to different CS/DS proteoglycans. The polysaccharide chain is composed of a repeating disaccharide unit [glucuronic (GlcA) or iduronic acid (IdoA) – N-acetyl-galactosamine (GalNAc)]n, where n varies in most cases between 20 and 100 (Fig. 83.1). Dermatan sulfate epimerase 1 and 2 epimerize glucuronic acid to iduronic acid (Maccarana et al. 2006; Pacheco et al. 2009c). This conversion in most cases is not quantitative, meaning that not all available glucuronic acids in a chain are converted. Therefore, a chain has a mixed content of glucuronic and iduronic acid. In the literature a single iduronic acid is sufficient to name the chain DS. To identify the hybrid nature of the GlcA/IdoA chain the term CS/DS is preferred. After epimerization of the chain, the C2 hydroxyl group of GlcA/IdoA and the C4 and/or C6 hydroxyl group of GalNAc can be sulfated, giving rise to seven different disaccharide structures (Kusche-Gullberg and Kjellen 2003). The combination of these disaccharides forms different protein-binding domains, which are generally 2–4 disaccharides long. It is evident that the structural variability of the chain makes CS/DS an informational rich molecule, and it has diverse biological functions (Trowbridge and Gallo 2002; Malmstrom et al. 2012). Iduronic acid in CS/DS is present in vertebrates, e.g., Xenopus (Yamada et al. 2009) and mammals, as well as in some invertebrate phyla of lower organisms such as Echinodermata, e.g., sea urchin and sea cucumber; Tunicata, e.g., ascidian; and Mollusca, e.g., squid (Yamada et al. 2011). It is absent in C. elegans and the fruit fly. DS-epi1 and 2 are expressed in vertebrates.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
- Hepatocyte Growth Factor
- Glucuronic Acid
- Dermatan Sulfate
- Tritiated Water
- Squamous Cell Carcinoma Cell Line
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Chondroitin/dermatan sulfate (CS/DS) chains are polysaccharides that are attached to approximately 30 different core proteins, thus giving rise to different CS/DS proteoglycans. The polysaccharide chain is composed of a repeating disaccharide unit [glucuronic (GlcA) or iduronic acid (IdoA) – N-acetyl-galactosamine (GalNAc)]n, where n varies in most cases between 20 and 100 (Fig. 83.1). Dermatan sulfate epimerase 1 and 2 epimerize glucuronic acid to iduronic acid (Maccarana et al. 2006; Pacheco et al. 2009c). This conversion in most cases is not quantitative, meaning that not all available glucuronic acids in a chain are converted. Therefore, a chain has a mixed content of glucuronic and iduronic acid. In the literature a single iduronic acid is sufficient to name the chain DS. To identify the hybrid nature of the GlcA/IdoA chain the term CS/DS is preferred. After epimerization of the chain, the C2 hydroxyl group of GlcA/IdoA and the C4 and/or C6 hydroxyl group of GalNAc can be sulfated, giving rise to seven different disaccharide structures (Kusche-Gullberg and Kjellen 2003). The combination of these disaccharides forms different protein-binding domains, which are generally 2–4 disaccharides long. It is evident that the structural variability of the chain makes CS/DS an informational rich molecule, and it has diverse biological functions (Trowbridge and Gallo 2002; Malmstrom et al. 2012). Iduronic acid in CS/DS is present in vertebrates, e.g., Xenopus (Yamada et al. 2009) and mammals, as well as in some invertebrate phyla of lower organisms such as Echinodermata, e.g., sea urchin and sea cucumber; Tunicata, e.g., ascidian; and Mollusca, e.g., squid (Yamada et al. 2011). It is absent in C. elegans and the fruit fly. DS-epi1 and 2 are expressed in vertebrates.
Databanks
Dermatan sulfate epimerase 1 and 2
IUBMB nomenclature: the single EC 5.1.3.19 number present in the database is linked only to DS-epi1.
Dermatan sulfate epimerases (DSE, DSEL)
Enzyme | Species | Gene symbol | GeneBank accession no. | NCBI |
|---|---|---|---|---|
protein accession no. | ||||
DS-epi1 | Homo sapiens | DSE | NM_013352 | NP-037484 |
DS-epi2 | Homo sapiens | DSEL(-like) | NM_032160 | NP-115536a |
Name and History
A protein with dermatan sulfate epimerase activity was first purified from bovine spleen (Maccarana et al. 2006). Peptide sequencing led to the cDNA, and it was realized that this gene had been cloned 6 years earlier with the name SART2 (squamous cell carcinoma antigen recognized by T cells 2) (Nakao et al. 2000). The earlier paper isolated SART2 and described it as a protein with unknown function, which is highly expressed in a squamous cell carcinoma cell line recognized by an HLA-A24-restricted tumor-specific cytotoxic T lymphocyte. Since the discovery of the function, the protein was named DS-epi1 and the gene DSE. In the databases DS-epi1 has several synonymous names (chondroitin-glucuronate 5-epimerase, polyglucuronate 5-epimerase, dermatan sulfate 5-epimerase, uronosyl epimerase, chondroitin D-glucuronosyl 5-epimerase), and these originate from different names for the substrate. In the name, DS-epi1, the C5 position of the inverted carboxyl group is not present. However, no epimerases in CS/DS biosynthesis act on any carbon different from C5.
When the gene coding for DS-epi1 was identified (Maccarana et al. 2006), it was immediately apparent that a homologue was present in the genome. This second gene was present in the database as C18orf4 or NCAG1, and the gene had been cloned 3 years earlier as a transcribed gene with unknown function present in a candidate region genetically associated with bipolar disorder (Goossens et al. 2003). C18orf4/NCAG1 was expressed in eukaryotes and shown to have epimerase activity and was therefore renamed DSEL(-like), and it coded for DS-epi2 (Pacheco et al. 2009c). In addition to the epimerase activity, DS-epi2 has a C-terminal domain, which is homologous to CS/DS O-sulfotransferase (see section “Structure”), but the activity has not yet been experimentally proven.
Structure
Human DSE and DSEL are located on chromosomes 6 and 18, respectively. DSE is organized in six exons, with the coding sequence distributed in five exons, whereas DSEL has only two exons with the whole ORF in exon 2 (Fig. 83.2a). DS-epi1 and DS-epi2 have a common N-terminal epimerase domain, with 51 % sequence identity between the two enzymes (Fig. 83.2b). The epimerase domain of the two proteins is identical in secondary structure and 3D predictions (Pacheco et al. 2009a). Critical amino acids present in the catalytic site have been identified, and a mechanism of the reaction has been proposed suggesting an intermediate structure with a broken chain and the presence of a C4-5 double bond in the attacked glucuronic acid (Pacheco et al. 2009a). This intermediate in DS-epimerase reaction is similar to the final product of the reaction catalyzed by chondroitin lyases, a family of enzymes from bacterial origin, which degrade CS/DS at specific positions and which are used as tools in CS/DS structural analysis (Linhardt et al. 2006). Therefore, the DS-epimerases share with several lyases a similar, but not identical, mechanism of action, a moderate sequence similarity, and even a stronger 3D similarity. For these reasons it has been proposed that the DS-epimerase domain has evolved from bacterial lyases. Four N-glycans are essential for the epimerase activity (Pacheco et al. 2009a). Following the epimerase domain in both DS-epi1 and 2, there are two domains with unknown function. These two domains have no 3D similarity with other proteins, and in DS-epi1 and 2 these domains are different. Finally, DS-epi2 has a C-terminal domain recognized in the database as a CS/DS-O-sulfotransferase domain. The DS-epi2 O-sulfotransferase domain shares, between amino acid 841 and 1,222, 16 % identity and 31 % similarity to C6ST1, a 6-O-sulfotransferase active in CS/DS biosynthesis. Intense effort is ongoing in the laboratory of the authors to experimentally establish the O-sulfotransferase activity of DS-epi2. If proven, DS-epi2 would result in an enzyme with dual epimerase and O-sulfotransferase activity.
Enzyme Activity Assay and Substrate Specificity
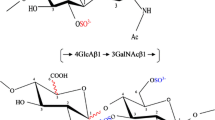
The native substrate for the epimerase is chondroitin, a polymer composed of the repeated disaccharide β1-4-D-GlcA-β−1-3-N-acetyl-d-Galactosamine (Fig. 83.1). The most convenient available method to assay the DS-epimerase activity takes advantage of the E. coli strain K4, which produces, and releases into the medium, a polysaccharide with the linear sequences of chondroitin, to which fructose units are linked to C3 of glucuronic acid. The bacteria are grown in the presence of D-[5-3H]glucose, and the recovered polysaccharide is defructosylated, resulting in 5-3H-labeled chondroitin substrate (Hannesson et al. 1996). Upon incubation with DS-epimerases, the released tritium is recovered in the assay buffer as tritiated water. Labeled water is separated from the labeled substrate either by distillation (Malmstrom and Aberg 1982) or is selectively counted using a biphasic scintillation liquid (Campbell et al. 1983). The assay incubations are at pH 5.5, containing 2 mM Mn2+, which greatly enhances the activity, and must not contain excess salt, because NaCl and KCl inhibit activity approximately 50 % already at 20 mM (Maccarana et al. 2006). A chondroitin polymer or oligosaccharides that are equal or longer than hexasaccharides can be used as substrates (Malmstrom and Aberg 1982; Malmstrom 1984). An alternative assay takes advantage of the fact that the in vitro reaction catalyzed by the DS-epimerases can be driven in both directions, i.e., glucuronic to iduronic or iduronic to glucuronic. Therefore, the reaction can be measured in incubations with unlabeled dermatan, whose structure is identical to the one depicted in Fig. 83.1 on the right side and tritiated water (Malmstrom 1984). The available assays do not distinguish DS-epi1 from DS-epi2. The possibility to differentiate by mass spectrometry deuterium-containing iduronic/glucuronic acid from the hydrogen-containing counterparts might lead to an assay method not using radioactivity, as it has been described for heparan sulfate C5 epimerase (Babu et al. 2011). Further, tandem mass spectrometry and NMR can discriminate between glucuronic and iduronic acid in CS/DS (Miller et al. 2006; Li et al. 2008). These methodologies could be potentially applied to an assay.
Preparation of Enzyme
Epimerase activity is found in all tested tissues except serum, with the spleen containing the highest activity and the brain containing the lowest (Maccarana et al. 2006). DS-epi1 has been purified from bovine spleen and then produced as a recombinant protein in 293HEK, and it is recovered both in the cellular layer and in the medium (Maccarana et al. 2006; Pacheco et al. 2009c). It can be detected as a ∼100 kDa band by Western blot after overexpression (Pacheco et al. 2009c) and as an endogenous protein in tissues and cells with high expression, such as spleen, fibroblasts, and aortic smooth muscle cells (Maccarana et al. 2009). DS-epi2 has been detected as a ∼150 kDa band by Western blot as overexpressed (Pacheco et al. 2009c) or endogenous protein in fibroblasts and skin (Bartolini et al. 2012).
Biological Aspects
DS-epi1 and 2 are both ubiquitously expressed (Nakao et al. 2000; Goossens et al. 2003). Analysis of DS-epi1 KO and DS-epi2 KO mice revealed the relative contribution to the total epimerase activity (Maccarana et al. 2009; Bartolini et al. 2012). DS-epi1 is the predominant enzyme in all tested tissues except in the kidney, where the relative contribution is equal, and in the brain, where DS-epi2 is much more predominant (Bartolini et al. 2012), and this is also in agreement with in situ hybridization evidence (Akatsu et al. 2011). In fibroblasts and smooth muscle cells DS-epi1 is the major contributor of epimerase activity. The amount and relative spatial distribution of the iduronic acids in a CS/DS chain depends on the expression level of the DS-epimerases and of the dermatan 4-O sulfotransferase 1 (D4ST1), which is the CS/DS 4-O-sulfotransferase responsible for the sulfation in vivo of IdoA blocks structures (IdoA-GalNAc-4-O-sulfated)n≥6 (Fig. 83.3) (Miyake et al. 2010). This is in agreement with its specificity in vitro (Mikami et al. 2003), and with the finding that siRNA-mediated D4ST1 downregulation in fibroblasts drastically reduces the IdoA blocks (Pacheco et al. 2009b). In vitro 4-O-sulfation of GalNAc is necessary for IdoA formation when microsomes are incubated with UDP-GlcA and UDP-GalNAc, with a molecular mechanism not fully elucidated. Low expression of these three enzymes produces alternating IdoA-GlcA-IdoA or isolated GlcA-IdoA-GlcA structures (Fig. 83.3), which have distinct biological activities such as being part of the oligosaccharide domains active in neuritogenesis (Li et al. 2010) or interacting to hepatocyte growth factor (Deakin et al. 2009). In a cancer cell line iduronic acid is essential to support migration and invasion with a hepatocyte growth factor-dependent mechanism (Thelin et al. 2012).
It is thought that the structural flexibility of IdoA [25], greater than GlcA, facilitates interactions with proteins. For instance, IdoA and GlcA interact with HCII to regulate coagulation (Maimone and Tollefsen 1990; Tollefsen 2010). They bind to neuronal growth factors such as pleiotrophin and midkine to promote neurite extension (Sugahara and Mikami 2007). IdoA and GlcA interact with several cytokines and adhesion molecules such as RANTES, P-selectin, and INF-γ to influence inflammation and cell recruitment (Handel et al. 2005; Taylor and Gallo 2006). Interaction with fibroblast growth factors affects cellular proliferation and migration (Taylor et al. 2005). Recently dermatan sulfate has been described to play a role in autoimmunity (Rho et al. 2011; Wang et al. 2011) and in differentiation of neural stem cells (Yamauchi et al. 2011).
Knockout Mouse and Transgenic Mice
DS-epi1 KO mice in a mixed C57BL6/129Sv genetic background are vital. However, they are smaller than the controls and have a defect in skin collagen fibril formation (Maccarana et al. 2009). Depending on the genetic background, other multiple defects in the mice have been uncovered (unpublished observations from the laboratory of the authors). DS-epi2 KO mice are apparently normal (Bartolini et al. 2012). Surprisingly, the brain, where DS-epi2 is the major epimerase, does not show obvious abnormalities, and immunostaining for ECM molecules appears normal. Clearly more detailed analyses are needed to study a possible brain phenotype.
Human Disease
There are no mutations in DS-epi1 or DS-epi2 associated with human diseases. However, D4ST1, which functionally collaborates with DS-epi1 to make structures of adjacent IdoA called IdoA blocks (Fig. 83.3), is the cause of a human disease called Ehlers-Danlos syndrome, musculocontractural type, also named, for instance, adducted thumb, clubfoot, and progressive joint and skin laxity syndrome (phenotype MIM number 601776). The disease is characterized by distinctive craniofacial dysmorphism, congenital contractures of thumbs and fingers, clubfeet, severe kyphoscoliosis, muscular hypotonia, hyperextensible thin skin with easy bruisability and atrophic scarring, wrinkled palms, joint hypermobility, and ocular involvement (Malfait et al. 2010). Myopathy has also been described (Voermans et al. 2012). Approximately 20 patients have been diagnosed so far (Dundar et al. 2009; Shimizu et al. 2011). The disease is caused by homozygous or compound heterozygous mutation in D4ST1, which leads to a drastic decrease of IdoA in dermatan sulfate with disappearance of all IdoA blocks (Miyake et al. 2010). This in turn leads to a defect in collagen fibrillization causing weaker tissue. Given the fact that DS-epi1 and D4ST1 are on the same pathway for IdoA block formation, an active search is ongoing to identify idiopathic patients with genetic errors in DS-epi1.
DS-epi1 is aberrantly expressed in most of the cancers and has been isolated as a potential tumor-specific antigen (Nakao et al. 2000). Peptides from DS-epi1 have been used in clinical trials in glioblastoma multiforme (Terasaki et al. 2011), prostate cancer (Noguchi et al. 2003), and hepatocellular carcinoma (Mizukoshi et al. 2011), with elicitation of a partial tumor-specific immune response.
DS-epi2 has been genetically associated with bipolar disorder, a disease affecting approximately 1 % of mankind (Goossens et al. 2003). Interestingly, two single-nucleotide polymorphisms predicted to change amino acid sequence were present in the bipolar disorder group and not in the control group.
Future Perspectives
It is of interest to define the reaction mechanism of the epimerases and the mechanism behind the interplay between DS-epi1 and D4ST1. The presence of iduronic acid changes the biological properties of CS/DS chains. In turn, the activities of approximately 30 CS/DS-PGs depend on the combination of the core proteins and the attached chain. Not surprisingly, the genetic models of ablation of DS-epi1 and 2 are uncovering diverse functions for iduronic acid. Very recently CS/DS was implicated in autoimmunity, inflammation, and stem cell differentiation. Modulation of the amount and distribution of iduronic acid by genetic means or by enzymatic inhibitors could, for instance, direct in vitro differentiation of stem cells before in vivo transplantation. CS/DS has an immediate potential in the differential diagnosis of autoimmune diseases (Rho et al. 2011). Modification of the extracellular matrix by changing CS/DS composition could ameliorate the long-term fibrotic process which is a hallmark of diseases such as asthma and COPD. Other areas that might benefit by pharmacological intervention on iduronic acid are coagulation disorders and spinal cord injury (Yamada and Sugahara 2008). Clearly, additional research is needed to describe the multifaceted interactions between iduronic acid and cancer development. The high epimerase activity in several cancers, combined with the effect of IdoA on migration and the results from vaccination using DS-epi1-derived peptides, motivates further research. Parallel to the studies on biological functions, effort should be put into understanding the biosynthetic regulations that generate such variable CS/DS chains.
Further Reading
-
Trowbridge and Gallo (2002) and Malmstrom et al. (2012): Reviews about dermatan sulfate
-
Pacheco et al. (2009a): Mechanism of action of DS-epi1.
-
Maccarana et al. (2009) and Bartolini et al. (2012): DS-epi1 and 2 KO mice.
-
Tollefsen (2010): Role of dermatan sulfate and heparin cofactor II in arterial injury.
-
Wang et al. (2011) and Rho et al. (2011): Role of dermatan sulfate in autoimmunity.
-
Thelin et al. (2012): Role of IdoA in cancer.
References
Akatsu C, Mizumoto S, Kaneiwa T, Maccarana M, Malmstrom A, Yamada S, Sugahara K (2011) Dermatan sulfate epimerase 2 is the predominant isozyme in the formation of the chondroitin sulfate/dermatan sulfate hybrid structure in postnatal developing mouse brain. Glycobiology 21(5):565–574
Babu P, Victor XV, Nelsen E, Nguyen TK, Raman K, Kuberan B (2011) Hydrogen/deuterium exchange-LC-MS approach to characterize the action of heparan sulfate C5-epimerase. Anal Bioanal Chem 401(1):237–244
Bartolini B, Thelin MA, Rauch U, Feinstein R, Oldberg A, Malmstrom A, Maccarana M (2012) Mouse development is not obviously affected by the absence of dermatan sulfate epimerase 2 in spite of a modified brain dermatan sulfate composition. Glycobiology 22(7):1007–1016
Campbell P, Feingold DS, Jensen JW, Malmstrom A, Roden L (1983) New assay for uronosyl 5-epimerases. Anal Biochem 131(1):146–152
Deakin JA, Blaum BS, Gallagher JT, Uhrin D, Lyon M (2009) The binding properties of minimal oligosaccharides reveal a common heparan sulfate/dermatan sulfate-binding site in hepatocyte growth factor/scatter factor that can accommodate a wide variety of sulfation patterns. J Biol Chem 284(10):6311–6321
Dundar M, Muller T, Zhang Q, Pan J, Steinmann B, Vodopiutz J, Gruber R, Sonoda T, Krabichler B, Utermann G, Baenziger JU, Zhang L, Janecke AR (2009) Loss of dermatan-4-sulfotransferase 1 function results in adducted thumb-clubfoot syndrome. Am J Hum Genet 85(6):873–882
Goossens D, Van Gestel S, Claes S, De Rijk P, Souery D, Massat I, Van den Bossche D, Backhovens H, Mendlewicz J, Van Broeckhoven C, Del-Favero J (2003) A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Mol Psychiatry 8(1):83–89
Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE (2005) Regulation of protein function by glycosaminoglycans – as exemplified by chemokines. Annu Rev Biochem 74:385–410
Hannesson HH, Hagner-McWhirter A, Tiedemann K, Lindahl U, Malmstrom A (1996) Biosynthesis of dermatan sulphate. Defructosylated Escherichia coli K4 capsular polysaccharide as a substrate for the d-glucuronyl C-5 epimerase, and an indication of a two-base reaction mechanism. Biochem J 313(Pt 2):589–596
Kusche-Gullberg M, Kjellen L (2003) Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol 13(5):605–611
Li F, Yamada S, Basappa, Shetty AK, Sugiura M, Sugahara K (2008) Determination of iduronic acid and glucuronic acid in sulfated chondroitin/dermatan hybrid chains by (1)H-nuclear magnetic resonance spectroscopy. Glycoconj J 25(7):603–610
Li F, Nandini CD, Hattori T, Bao X, Murayama D, Nakamura T, Fukushima N, Sugahara K (2010) Structure of pleiotrophin- and hepatocyte growth factor-binding sulfated hexasaccharide determined by biochemical and computational approaches. J Biol Chem 285(36):27673–27685
Linhardt RJ, Avci FY, Toida T, Kim YS, Cygler M (2006) CS lyases: structure, activity, and applications in analysis and the treatment of diseases. Adv Pharmacol 53:187–215
Maccarana M, Olander B, Malmstrom J, Tiedemann K, Aebersold R, Lindahl U, Li JP, Malmstrom A (2006) Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J Biol Chem 281(17):11560–11568
Maccarana M, Kalamajski S, Kongsgaard M, Magnusson SP, Oldberg A, Malmstrom A (2009) Dermatan sulfate epimerase 1-deficient mice have reduced content and changed distribution of iduronic acids in dermatan sulfate and an altered collagen structure in skin. Mol Cell Biol 29(20):5517–5528
Maimone MM, Tollefsen DM (1990) Structure of a dermatan sulfate hexasaccharide that binds to heparin cofactor II with high affinity. J Biol Chem 265(30):18263–18271
Malfait F, Syx D, Vlummens P, Symoens S, Nampoothiri S, Hermanns-Le T, Van Laer L, De Paepe A (2010) Musculocontractural Ehlers-Danlos Syndrome (former EDS type VIB) and adducted thumb clubfoot syndrome (ATCS) represent a single clinical entity caused by mutations in the dermatan-4-sulfotransferase 1 encoding CHST14 gene. Hum Mutat 31(11):1233–1239
Malmstrom A (1984) Biosynthesis of dermatan sulfate. II. Substrate specificity of the C-5 uronosyl epimerase. J Biol Chem 259(1):161–165
Malmstrom A, Aberg L (1982) Biosynthesis of dermatan sulphate. Assay and properties of the uronosyl C-5 epimerase. Biochem J 201(3):489–493
Malmstrom A, Bartolini B, Thelin MA, Pacheco B, Maccarana M (2012) Iduronic acid in chondroitin/dermatan sulfate: biosynthesis and biological function. J Histochem Cytochem 60(12):916–925
Mikami T, Mizumoto S, Kago N, Kitagawa H, Sugahara K (2003) Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulfate biosynthesis. J Biol Chem 278(38):36115–36127
Miller MJ, Costello CE, Malmstrom A, Zaia J (2006) A tandem mass spectrometric approach to determination of chondroitin/dermatan sulfate oligosaccharide glycoforms. Glycobiology 16(6):502–513
Miyake N, Kosho T, Mizumoto S, Furuichi T, Hatamochi A, Nagashima Y, Arai E, Takahashi K, Kawamura R, Wakui K, Takahashi J, Kato H, Yasui H, Ishida T, Ohashi H, Nishimura G, Shiina M, Saitsu H, Tsurusaki Y, Doi H, Fukushima Y, Ikegawa S, Yamada S, Sugahara K, Matsumoto N (2010) Loss-of-function mutations of CHST14 in a new type of Ehlers-Danlos syndrome. Hum Mutat 31(8):966–974
Mizukoshi E, Nakamoto Y, Arai K, Yamashita T, Sakai A, Sakai Y, Kagaya T, Honda M, Kaneko S (2011) Comparative analysis of various tumor-associated antigen-specific t-cell responses in patients with hepatocellular carcinoma. Hepatology 53(4):1206–1216
Nakao M, Shichijo S, Imaizumi T, Inoue Y, Matsunaga K, Yamada A, Kikuchi M, Tsuda N, Ohta K, Takamori S, Yamana H, Fujita H, Itoh K (2000) Identification of a gene coding for a new squamous cell carcinoma antigen recognized by the CTL. J Immunol 164(5):2565–2574
Noguchi M, Kobayashi K, Suetsugu N, Tomiyasu K, Suekane S, Yamada A, Itoh K, Noda S (2003) Induction of cellular and humoral immune responses to tumor cells and peptides in HLA-A24 positive hormone-refractory prostate cancer patients by peptide vaccination. Prostate 57(1):80–92
Pacheco B, Maccarana M, Goodlett DR, Malmstrom A, Malmstrom L (2009a) Identification of the active site of DS-epimerase 1 and requirement of N-glycosylation for enzyme function. J Biol Chem 284(3):1741–1747
Pacheco B, Maccarana M, Malmstrom A (2009b) Dermatan 4-O-sulfotransferase 1 is pivotal in the formation of iduronic acid blocks in dermatan sulfate. Glycobiology 19(11):1197–1203
Pacheco B, Malmstrom A, Maccarana M (2009c) Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate. J Biol Chem 284(15):9788–9795
Rho JH, Zhang W, Murali M, Roehrl MH, Wang JY (2011) Human proteins with affinity for dermatan sulfate have the propensity to become autoantigens. Am J Pathol 178(5):2177–2190
Shimizu K, Okamoto N, Miyake N, Taira K, Sato Y, Matsuda K, Akimaru N, Ohashi H, Wakui K, Fukushima Y, Matsumoto N, Kosho T (2011) Delineation of dermatan 4-O-sulfotransferase 1 deficient Ehlers-Danlos syndrome: observation of two additional patients and comprehensive review of 20 reported patients. Am J Med Genet A 155A(8):1949–1958
Sugahara K, Mikami T (2007) Chondroitin/dermatan sulfate in the central nervous system. Curr Opin Struct Biol 17(5):536–545
Taylor KR, Gallo RL (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J 20(1):9–22
Taylor KR, Rudisill JA, Gallo RL (2005) Structural and sequence motifs in dermatan sulfate for promoting fibroblast growth factor-2 (FGF-2) and FGF-7 activity. J Biol Chem 280(7):5300–5306
Terasaki M, Shibui S, Narita Y, Fujimaki T, Aoki T, Kajiwara K, Sawamura Y, Kurisu K, Mineta T, Yamada A, Itoh K (2011) Phase I trial of a personalized peptide vaccine for patients positive for human leukocyte antigen–A24 with recurrent or progressive glioblastoma multiforme. J Clin Oncol 29(3):337–344
Thelin MA, Svensson KJ, Shi X, Bagher M, Axelsson J, Isinger-Ekstrand A, van Kuppevelt TH, Johansson J, Nilbert M, Zaia J, Belting M, Maccarana M, Malmstrom A (2012) Dermatan sulfate is involved in the tumorigenic properties of esophagus squamous cell carcinoma. Cancer Res 72(8):1943–1952
Tollefsen DM (2010) Vascular dermatan sulfate and heparin cofactor II. Prog Mol Biol Transl Sci 93:351–372
Trowbridge JM, Gallo RL (2002) Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology 12(9):117R–125R
Voermans NC, Kempers M, Lammens M, van Alfen N, Janssen MC, Bonnemann C, van Engelen BG, Hamel BC (2012) Myopathy in a 20-year-old female patient with D4ST-1 deficient Ehlers-Danlos syndrome due to a homozygous CHST14 mutation. Am J Med Genet A 158A(4):850–855
Wang JY, Lee J, Yan M, Rho JH, Roehrl MH (2011) Dermatan sulfate interacts with dead cells and regulates CD5(+) B-cell fate: implications for a key role in autoimmunity. Am J Pathol 178(5):2168–2176
Yamada S, Sugahara K (2008) Potential therapeutic application of chondroitin sulfate/dermatan sulfate. Curr Drug Discov Technol 5(4):289–301
Yamada S, Onishi M, Fujinawa R, Tadokoro Y, Okabayashi K, Asashima M, Sugahara K (2009) Structural and functional changes of sulfated glycosaminoglycans in Xenopus laevis during embryogenesis. Glycobiology 19(5):488–498
Yamada S, Sugahara K, Ozbek S (2011) Evolution of glycosaminoglycans: comparative biochemical study. Commun Integr Biol 4(2):150–158
Yamauchi S, Kurosu A, Hitosugi M, Nagai T, Oohira A, Tokudome S (2011) Differential gene expression of multiple chondroitin sulfate modification enzymes among neural stem cells, neurons and astrocytes. Neurosci Lett 493(3):107–111
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Japan
About this entry
Cite this entry
Maccarana, M., Malmström, A. (2014). Dermatan Sulfate Epimerases (DSE, DSEL). In: Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T. (eds) Handbook of Glycosyltransferases and Related Genes. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54240-7_130
Download citation
DOI: https://doi.org/10.1007/978-4-431-54240-7_130
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54239-1
Online ISBN: 978-4-431-54240-7
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences