Abstract
Caspases are the essential proteases in cellular death pathways and well known as the key players during apoptosis. They are involved in the initiation and execution of the extrinsic and caspase-dependent intrinsic apoptotic pathways as well as in pyroptotic cell deletion. After a short introduction about different cell death modalities, we will have a closer look at these important enzymes that belong to one of the best-characterized protease families.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mitochondrial Outer Membrane Permeabilization
- Death Induce Signaling Complex
- Inflammatory Caspases
- Inflammasome Formation
- Cellular Demise
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
8.1 Introduction
The theory of the cell as the basic unit of life was developed by the German scientists Schleiden and Schwann in 1839. Shortly afterwards, cellular demise was observed by researches in different tissues and cell types. A landmark was in 1885, when Walther Flemming morphologically described the process of dying ovarian follicles containing “chromatin chunks”, which he termed chromatolysis (Flemming 1885). In fact, the condensation of chromatin is nowadays well known as a morphological feature of apoptosis.
In 1972, Kerr et al. (1972) described the morphology of cell deletion in tissues with a reproducible and different mechanism compared to the known process of necrosis. Disappearing cells showed aggregation of nuclear chromatin similar to Flemmings observations. The mechanism could be dissected into two stages: A step of nuclear and cytoplasmic condensation into membrane-bound cellular fragments and a degradation step performed by other cells after phagocytosis. Kerr proposed the term apoptosis for this programmed cell death derived from the Greek word for “falling off”.
Studies on embryonic development of Caenorhabditis elegans (C. elegans) by Ellis and Horvitz (1986) yielded in the discovery of the cell death controlling genes ced-3 and ced-4. A fundamental starting point in death pathway research was in 1993, when this C. elegans gene ced-3 was recognized to encode a protein similar to the mammalian interleukin-1β-converting enzyme (ICE) (Yuan et al. 1993). Several homologues of ICE were identified in mammals forming a proteolytic network as the molecular basis for apoptosis. The increasing amount of identified cysteine-aspartic-dependent proteases with inconsistent and multiple names were unified for simplicity with the name caspase in 1996 (Alnemri et al. 1996).
Caspases were further investigated and revealed a complex network of regulation and activation during apoptosis. They are known as the key-players in apoptosis and besides of this crucial role, caspases are involved in inflammatory pathways and immunity and can regulate proliferation and differentiation.
Besides the caspase-mediated apoptosis, other death pathways like necroptosis, autophagy and pyroptosis were discovered and expanded the research field of death pathways with caspase-dependent and -independent mechanisms.
The numerous different ways of cell removal and destruction underlines the significance of cell deletion. It is crucial for multicellular organisms to maintain the correct numbers of cells in tissues and to shape the architecture of organs. In addition, unspecific cell proliferation and tumor development is related to down-regulation of apoptosis whereas its up-regulation is associated to degenerative diseases like Parkinson and Alzheimer.
8.2 Death Pathways
8.2.1 Nomenclature
Various distinctive death pathways have been discovered till date and were historically classified by cellular morphology. Since 2012, the Nomenclature Committee on Cell Death (NCCD) suggests a classification based not only on morphological observations but also on biochemical data analysis (Galluzzi et al. 2012). It has been revealed that similar morphological features in cell death show biochemical, functional and immunological heterogeneity. This renders the nomenclature based on morphology as not sufficient and the exact identification of a specific pathway as a rather challenging task. The suggested classification on quantitative biochemical analysis is less prone to misinterpretations, since morphological observations require an experienced operator. Thus, the following pathway definitions will consider the NCCD recommendations.
8.2.2 Extrinsic Apoptosis
The extrinsic apoptotic pathway is a caspase-dependent mechanism, which is initiated by extracellular stress signals. Extracellular death ligands bind to death receptors like FAS, tumor necrosis factor (TNF) receptor 1 or TNF-related apoptosis inducing ligand (TRAIL) receptor 1 and 2 leading to receptor trimerization. The activated receptor conformation triggers the assembly of an intracellular death inducing signaling complex (DISC) involving the adaptor protein Fas-associated death domain (FADD) and the initiator pro-caspases-8 or -10.
The DISC recruitment of pro-caspases leads to activation by dimerization and subsequent autocatalytic processing. Then, initiator caspases catalyze the maturation of executioner caspases to start the apoptotic removal of the cell. In particular cells like hepatocytes (type II cells), caspase-8 cleaves the death agonist BB3-interacting domain (Bid) to a truncated form (tBid) instead of activating executioner caspases (Yin et al. 1999). Bid-cleavage leads to mitochondrial outer membrane permeabilization (MOMP) that activates apoptosis in a similar manner as the intrinsic apoptosis driven by caspase-9. Bid-cleavage can thus substitute a direct caspase-8 mediated activation of executioner caspases and can provide an amplification loop of extrinsic apoptosis through the intrinsic apoptotic pathway in selected cells (Yin et al. 1999).
8.2.3 Intrinsic Apoptosis
Intrinsic apoptosis is triggered by intracellular stress signals such as DNA damage, oxidative stress and accumulation of unfolded protein. The NCCD suggests a classification into two different mechanisms due to highly heterogeneous signaling cascades with the mitochondrion as the main control mechanism. Predominance of pro-apoptotic proteins like Bak or Bax can lead to MOMP due to their pore forming activity. Consequently, inter-membrane space (IMS) residential proteins are released into the cytosol where they mediate two apoptotic pathways:
-
Caspase-dependent
The release of cytochrome c upon MOMP triggers the caspase-dependent mechanism of intrinsic apoptosis. Cytochrome c assembles together with the apoptotic protease activating factor 1 (Apaf-1) to the recruitment platform for pro-caspase-9, known as the apoptosome. After the activation by dimerization, caspase-9 facilitates downstream cleavage of executioner caspases leading to cellular demise.
-
Caspase-independent
The release of IMS proteins, such as apoptosis-inducing factor (AIF) and endonuclease g (ENDOG), leads to caspase-independent fragmentation of DNA after relocation into the nucleus. The IMS serine protease high temperature requirement protein A2 (HTRA2) cleaves numerous cytoplasmic substrates including cytoskeleton proteins and contributes to the caspase-independent mechanism of intrinsic apoptosis.
8.2.4 Pyroptosis
Pyroptosis is a regulated death pathway highly connected to caspase-1 activation and inflammatory responses. Brennan and Cookson (2000) described the death of macrophages that were infected by Salmonella typhimurium and named this reproducible mechanism pyroptosis (Cookson and Brennan 2001). Indeed, several other bacteria are able to induce pyroptosis like Shigella flexneri, Bacillus anthracis toxin and Pseudomona aeruginosa.
It became clear that activation of caspase-1 is a crucial step, which is promoted by a multi-protein complex known as the inflammasome. In contrast to the apoptotic pathways, executioner caspases are not involved during pyroptosis.
The caspase-1-activating inflammasome is a large protein complex, which is formed upon infection with bacteria, viruses or parasites, as well as by host-derived danger associated molecular patterns (DAMP). NOD-like receptor (NLR) proteins recognize danger signals and bind via the adaptor protein ASC and its caspase activation and recruitment domain (CARD) to pro-caspase-1. Proximity-induced oligomerization and activation of caspase-1 leads to the maturation of pro-interleukins. The activated cytokines interleukin-1 and -18 are secreted to recruit and activate immune cells for an inflammatory response. The activation of caspase-1 results in cleavage of several other substrates, however the complex mechanism of inflammasome activation and pyroptotic cell death is still not completely understood but has recently been well reviewed by Rathinam et al. (2012).
8.2.5 Regulated Necrosis
Necrosis is commonly known as an accidental cell death with a morphotype of disrupted membranes and release of intracellular content. Hitomi et al. (2008) recently pointed out that there is as well a regulated form of necrosis, which can be triggered under specific energy dependent circumstances. Key mediators of the regulated necrotic pathway are ATP-dependent enzymes like poly(ADP-ribose) polymerase 1 (PARP-1) or receptor-interacting protein kinase 1 (RIP1). Alkylated DNA damage can act as an inducing signal leading to PARP-1 hyperactivation and cytosolic ATP reduction. The depletion of the cellular energy storage results in activation of RIP1 or its homolog RIP3, which triggers the execution of necrosis.
8.2.6 Autophagy
Morphological features like massive cytoplasmic vacuolization were first described in the 1950s. In most cases, autophagy is activated upon cell stress and as a cytoprotective response of dying cells. It is a conserved, kinase-dependent cell degradation pathway in eukaryotes with autophagy related proteins and Beclin-1 as the key players of autophagosome biogenesis. A clear elucidation of autophagosome formation and its regulation is beyond the scope of this chapter but reviewed by Yang and Klionsky (2009) and Rubinsztein et al. (2012). Like apoptosis, the autophagic cell degradation is non-inflammatory due to vacuolization of cytoplasmic content.
8.2.7 Other Modalities
Numerous additional death pathways exist that are in some cases overlapping or bypassing other mechanisms. Thus, a variety of cell death pathways can be observed in cultures induced by the same stimulus. This makes a clear identification of the exact molecular mechanism even more important. In addition to apoptosis as the most important non-inflammatory pathway and pyroptosis as the inflammatory pathway, the following mechanisms are known:
-
Mitotic catastrophe is initiated in an arrested M-phase in the cell cycle due to mitotic machinery aberrations. It finally leads to cell death or senescence.
-
Anoikis is an apoptotic response due to loss of cell-to-matrix interactions
-
Entosis or “cell-cannibalism” describes the complete engulfment of a cell into another cell, which is never released and finally degraded.
-
Parthanatos is initiated by PARP-1 hyperactivation and its resulting ATP-depletion. It is a particular form of regulated necrosis.
-
Netosis is a cell death mechanism restricted to granulocytic cells, sharing biochemical features with autophagy and regulated necrosis.
-
Cornification is important for epidermis formation and restricted to keratinocytes.
8.3 Caspases: The Essential Proteases in Death Pathways
The caspase family members are well known as the key players in apoptosis. They play important roles in initiation and execution of the extrinsic and caspase-dependent intrinsic apoptotic pathways as well as in pyroptotic cell deletion. The first hint of these pro-apoptotic proteins was found in C. elegans in 1986, when Ellis and Horvitz (1986) identified that the genes ced-3 and ced-4 are required for the initiation of apoptosis. Deletion of these two genes leads to the survival of cells, which otherwise usually died during embryonic development. Using the simple model organism C. elegans, more apoptosis-related genes and proteins like ced-9 (Hengartner et al. 1992) and EGL-1 (Conradt and Horvitz 1998) were unraveled decomposing the complex molecular system of apoptosis into its relevant proteins. CED-4 dimeric proteins are blocked in an inactive form by CED-9 in living cells. Apoptotic stimuli induce the production of EGL-1, which binds to CED-9 and thus liberates the CED-4 dimers. The released dimers build the apoptosome by tetramerization of the asymmetric dimers and recruitment of proCED-3, where CED-3 is activated by proteolysis or conformational changes and then released to trigger apoptosis.
Apoptotic research on mammalian cells and human proteins was boosted after the discovery of ICE as a unique cysteine protease (Thornberry et al. 1992) and its homology to the nematodic CED-3 (Yuan et al. 1993). The unified name caspase was finally introduced after the identification of several ICE homologues and their confusing nomenclature (Alnemri et al. 1996). In fact, the mammalian genomes encode numerous ced-like genes that may have arisen by gene duplication during evolution. Compared to nematodes, this emphasizes a much more complex apoptotic system with more redundancy and regulatory effectors.
In 1994, the first high-resolution crystal structure of human ICE was reported at a resolution of 2.6 Å and gave first insights into the catalytic mechanism and its regulation (Wilson et al. 1994). The tetrameric enzyme consists of two identical heterodimers made by a small (p10) and a large (p20) subunit. Two substrate-binding pockets are shaped between the p10 and p20 of the individual heterodimer. The active sites are formed by a catalytic diad (Cys-285 and His-237) and cleave substrates specifically after aspartate residues. Both, the conserved catalytic diad residues Cys-285 and His-237 as well as the requirement of a substrate with an aspartic acid at P1 are common for all caspase family members.
Based on the structure and selective mutagenesis, an allosteric regulation of ICE activity was proposed (Wilson et al. 1994). Mutations in the dimer interface of the heterodimeric subunits, far away from the active site cleft, resulted in an inactive ICE suggesting the tetrameric enzyme as a prerequisite for activity.
During the last two decades, 12 different human caspases have been identified and biochemically characterized. Seven of them could be crystallized or studied by NMR spectroscopy till date. Even though this progress led to one of the best-characterized protease families among proteolytic enzymes, open questions are still remaining.
8.3.1 Classification, Nomenclature and Family Members
The Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) classifies the family of caspases as cysteine endopeptidases with the enzyme category (EC) number 3.4.22 where today caspase-1 to caspase-11 are listed in the IUBMB database with individual EC numbers. The MEROPS database assigned these enzymes to the clan CD with the family C14 [http://merops.sanger.ac.uk/ (Rawlings et al. 2012)].
The term caspase was introduced in 1996 with “c” abbreviating the catalytic residue cysteine and “aspase” referring to the required aspartic acid at position P1 of the peptidase (Alnemri et al. 1996). Other similar terminologies for caspase can be found in the literature like “cysteine-dependent aspartate-directed proteases” (Eckhart et al. 2008).
The number of each family member is assigned to the date of publication and reflects the history of caspase research. Caspases are expressed as zymogens, indicated by the prefix “pro” (e.g. pro-caspase-1). The polypeptide chain consists of either a N-terminal pro-domain or a short pro-peptide and a catalytic domain that can be proteolytically separated from the former.
The architecture of the catalytic domain is made by a large and a small subunit, connected by a short linker. The individual subunits are particularly named by their exact molecular weight (e.g. caspase-1-p10 for the small subunit of caspase-1). Complete caspase activation is achieved by proteolytic removal of the N-terminal pro-domain or short pro-peptide as well as the inter-subunit linker. In addition, two catalytic domains dimerize and form a heterotetrameric structure also known as the caspase-fold.
A total of 16 different mammalian caspases could be identified by comparative genomics and evolutionary analysis (Eckhart et al. 2008). The individual family members can be classified according to their function, their length and structure of the N-terminal pro-domain, their optimal cleavage motives or their phylogenetic analysis of the amino acid sequence. The existing human homologues are generally grouped according to sequence identity and functionality.
8.3.1.1 Human Caspases
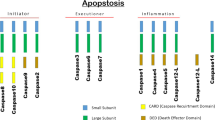
Homology based analysis of the human caspases divides the family into three subgroups that can be assigned as well to their function (Fig. 8.1).
The caspase family. The human caspase family members are ordered by their function into inflammatory and apoptotic initiator and executioner caspases. Caspase-14 is mainly expressed in keratinocytes and assigned to cornification processes. A human majority expresses a truncated form of caspase-12 due to a single nucleotide polymorphism, which leads to a stop codon at position 125 (indicated by asterisk). Numbers indicate terminal residues of domains. UniProtKB accession numbers and alternative names are listed for the human members. More mammalian caspase family members have been identified in other species. Caspase-11 and caspase-13 are orthologues of human caspase-4 and thus not listed
Caspase-1, -4, -5 and -12 are inflammatory caspases according to their roles in cytokine activation. For instance, caspase-1 gets activated by the inflammasome formation and triggers pyroptosis, an inflammatory death pathway (Rathinam et al. 2012). The role of caspase-5, a close homologue to caspase-1, could be assigned to cytokine maturation and inflammasome formation (reviewed by Martinon and Tschopp 2004) whereas caspase-4 was recently shown to be involved in inflammasome activation (Sollberger et al. 2012). A special case is the full-length caspase-12, known for increased susceptibility to severe sepsis. It is expressed only in a small population of African descent, while the human majority expresses a truncated, pro-domain-only form due to a single nucleotide polymorphism (Saleh et al. 2004).
Caspase-2, -8, -9 and -10 are known as apoptotic-inducing peptidases, which are activated in response to either extra- or intracellular apoptotic stimuli. The characteristic N-terminal CARD or two death effector domains (DED) are a prerequisite for initiator caspase activation. These domains control the oligomerization at activation platforms like the apoptosome and provoke autocatalytic activation by induced-proximity (Salvesen and Dixit 1999). In this manner, a pro-apoptotic signal can be translated into a proteolytic response.
Activated initiator caspases are responsible for the proteolytic maturation of the executioner caspases-3, -6 and -7. Executioner caspases are lacking the structured N-terminal domains and contain only a short N-terminal peptide sequence, which is thought to be a regulatory signal (Meergans et al. 2000; Denault and Salvesen 2003). In cells, these enzymes exist as heterotetrameric pro-forms and are activated by proteolysis of the N-terminal peptide and the inter-subunit linker. Once activated, they provoke the majority of proteolytic events leading to the known morphology of apoptotic cells.
Caspase-14 is a special member and mainly expressed in keratinocytes (reviewed by Denecker et al. 2008). Its function is assigned to cornification processes and mice experiments recently showed a relationship between caspase-14-deficiency and UVB-induced photodamage (Hoste et al. 2013).
8.3.1.2 Caspases in Mammals
Besides the well-characterized human members, numerous other homologues have been discovered in different mammalian species increasing the family members from caspase-1 to caspase-18 (Eckhart et al. 2008). Notably, murine caspase-11 and bovine caspase-13 are orthologues of human caspase-4 (Koenig et al. 2001; Eckhart et al. 2008) and thus named inconsistently with respect to the nomenclature guidelines of Alnemri et al. (1996).
A remarkable finding was published in 2011, when Kayagaki et al. (2011) discovered that the generally used caspase-1 knockout mouse (strain 129) is particularly a double knockout of caspase-1 and -11. A new caspase-1-only-deficient mouse strain depicts a crucial role of caspase-11 in the non-canonical activation of inflammation. Translated to humans, this suggests a similar role for caspase-4 and -5 and thus a specific function in incidents such as sepsis. The findings suggest a careful interpretation of the role of caspase-1 versus -11, which so far only has been determined experimentally using the mouse strain 129.
The evolutionary relationship of mammalian caspases has recently been studied. This revealed that the caspase family has gone through extensive changes with numerous gene deletions and duplications (Eckhart et al. 2008) (Fig. 8.1).
8.3.1.3 Non-mammalian Caspases and Metacaspases
Identification of the cell death related gene ced-3 in C. elegans was a landmark in the discovery of caspases (Ellis and Horvitz 1986; Yuan et al. 1993). CED-3 gets activated upon binding to the nematodic apoptosome formed by a CED-4 octamer and then executes the apoptotic pathway in C. elegans. A crystal structure of the formed complex between CED-3 and CED-4 revealed a funnel shaped octamer formed by CED-4, on which CED-3 is supposed to become activated, although no electron density could be observed for CED-3 in the structure (Qi et al. 2010).
Caspase homologues exist as well in insects. The initiator caspase Dronc is a caspase-9 ortholog in Drosophila and involved in apoptosis and cell proliferation (Steller 2008). Additional initiator (Dredd, Strica) and executioner (Drice, Dcp-1, Decay, Damm) caspases have been discovered in Drosophila (reviewed by Kumar and Doumanis 2000; Steller 2008). Recently, another Dredd homologue was characterized from an insect vector of human disease: Aedis Dredd (AeDredd) from the yellow fever mosquito Aedis aegypti (Cooper et al. 2007). This finding could be a first step to unravel the role of apoptosis in innate immune response of insects and in the particular case of mosquitoes to understand their exceptional vectorial capacity.
While caspases can be found in metazoan, distant homologues could be identified as well in protozoa, fungi and plants. These distantly related cysteine-dependent peptidases cleave after arginine or lysine and are called metacaspases (Tsiatsiani et al. 2011).
8.3.2 The Caspase Fold
The tetrameric caspase structure can be described as a globular fold made by two equivalent and symmetry-related catalytic domains with a α/β topology.
Each subdomain derives from a single polypeptide chain, encoding a N-terminal pro-domain or -peptide, and a large (17–20 kDa) and a small subunit (10–12 kDa). The N-terminal domains are separated from the caspase structure by an inter-domain linker, which can be cleaved during activation processes. Due to this flexible linker, the crystallization of a full-length caspase with its pro-domain was not successful and thus their hypothetical interactions remain unclear. However, several pro-domain homologues have been separately crystallized or investigated by NMR spectroscopy. These dead domains revealed a globular fold of six antiparallel α-helices with α1–α5 forming a conserved Greek key motive (Vaughn et al. 1999; reviewed by Kersse et al. 2011).
A tightly packed caspase catalytic domain roughly forms a cuboid with the approximate dimension of 25 Å × 35 Å × 42 Å. It is composed of a β-sheet, which is sandwiched between two layers of overall five α-helices. The twisted β-sheet involves five parallel strands (β1–β5) and one antiparallel strand (β6). The latter aligns with the β6 strand of the second catalytic domain in antiparallel manner. This results in a continuous β-sheet with 12 strands and together with ten α-helices in a twofold symmetry related structure (Fig. 8.2a).
The caspase fold. (a) Cartoon representation of caspase-3 crystal structure (2DKO) representing a dimer of two catalytic subunits (colored in green/orange and gray). Twelve central β-strands are sandwiched between ten α-helices. Active site residues His-237 and Cys-285 are colored in blue and located after strand β3 and β4, respectively. The five parallel β-strands of a catalytic domain points towards the substrate-binding cleft. A 90° rotation reveals a square-like arrangement of the α-helices. (b) Substrate-binding groove of caspase-3 (2DKO) covalently bound to substrate-analogue inhibitor DEVD-chloromethylketone. Binding cleft forming loops are colored in green (loop-1), orange (loop-2), blue (loop-3) and red (loop-4). (c) Topology map of caspases colored according to crystal structure shown in (a). Active site residues His-237 and Cys-285 are indicated. Loop-2 is also known as 240-loop. (d) Catalytic mechanism according to Fuentes-Prior and Salvesen (2004)
8.3.3 The Active Site, Substrate Recognition and Cleavage Mechanism
The numbering of a specific amino acid in different caspase family members can diverge due to residue insertions or deletions. In general, a facilitated numbering based on caspase-1 sequence is used as proposed by Fuentes-Prior and Salvesen (2004).
8.3.3.1 Active Site Architecture and Substrate Recognition
The active site and binding groove of the catalytic domain is constructed by loops of its large and small subunits (Fig. 8.2b). The catalytic Cys-285 resides at the end of the central β-strand 4 whereas His-237 is located at the end of β-strand 3 (Fig. 8.2c). In a cartoon representation, the five parallel β-strands point towards the substrate-binding groove, with the tip of β-strand 3 and 4 indicating the position of the active site residues.
A strictly conserved Arg-179 positioned in front of helix α1 in loop-1 is crucial for the P1 specificity. It shapes the aspartate specific binding pocket S1 together with the conserved residues Gln-283 and Arg-341. In addition, loop-3 residue Arg-341 is involved in the formation of the binding subsite S3 mediating main-chain-main-chain hydrogen bonds and interacting with the carboxylate group of a preferred glutamate at P3 (Fuentes-Prior and Salvesen 2004).
The S2 and S4 pocket residues are less conserved. This leads to more substrate variability at P2 and P4 that can be observed directly by comparing small, peptide-based substrates (Roschitzki-Voser et al. 2012).
In inflammatory and initiator caspases, Val-338 of loop-3 shapes a large hydrophobic S2 pocket that tolerates bulky side chains like His or Thr as P2 residue. In contrast, in executioner caspases Val-338 is substituted by Tyr leading to a preference for small aliphatic substrate residues such as Val or Ala. A similar effect is observed at the S4 subsite, where a bulky Trp-348 in apoptotic caspases shapes a small S4 pocket with an increased affinity for branched aliphatic residues or aspartate at position P4 (Fuentes-Prior and Salvesen 2004). A substitution in caspase-1 (Trp-348 by Val) or caspase-4 and -5 (Trp-348 by Ile) reshapes the S4 subsite that favors large and hydrophobic residues.
A S5 pocket could only be characterized in caspase-2 with its preferential accommodation of a small hydrophobic substrate residue (Fuentes-Prior and Salvesen 2004). Furthermore, residues after the scissile bond do not require special properties to be accommodated in the less restrictive primed subsite. An exception is the S1′ pocket that mildly discriminates for charged or bulky residues (Timmer and Salvesen 2007).
In summary, a consensus caspase substrate sequence can be defined as (D or W)-E-X-D ↓ ϕ with P4-P3-P2-P1 ↓ P1′ where Asp at position P4 is preferred by caspases of the apoptotic subfamily, X can be substituted by any amino acid and ϕ stands for a small uncharged residue (Timmer and Salvesen 2007). Today, a large number of caspase substrates are commercially available with a proposed high specificity for a single caspase family member. When looking at the consensus substrate sequence, it is evident that such specificity is difficult to achieve and in fact could not be observed experimentally (McStay et al. 2008). This effect is not only observed for specific caspase substrates but also for peptide inhibitors, which are designed based on the consensus sequence and thus are not feasible for a highly specific caspase inhibition.
8.3.3.2 Catalytic Mechanism
Fuentes-Prior and Salvesen (2004) proposed a first catalytic mechanism in 2004. The authors suggested a mechanism with close similarity to the proteolytic reaction of serine peptidases (Fig. 8.2d). Their mechanism is based on crystal structures with active site covalently bound inhibitors mimicking a tetrahedral intermediate.
In a hypothetical enzyme-substrate complex, the P1 carbonyl oxygen is oriented by hydrogen bonds towards the oxyanion hole, which is formed by backbone amine groups of the conserved residues Gly-238 and Cys-285. A nucleophilic attack of the Cys-285 sulfur atom on the carbonyl carbon forms a tetrahedral intermediate with covalently bound substrate. This state is stabilized by the imidaloze moiety of His-237, which protonates the α-amino group of the P1′ residue and thus triggers the release of the C-terminal peptide product from the S1′ subsite. The formed acyl intermediate is subsequently hydrolytically processed. The deprotonated π-nitrogen of His-237 abstracts a proton from a water molecule, which itself attacks the thioester carbonyl. This forms a second tetrahedral intermediate followed by a cleavage of the covalent bond formed between the carbonyl carbon and the sulfur of Cys-285. The non-covalently bound N-terminal peptide product is released and the regenerated enzyme starts a new catalytic cycle.
8.3.4 Activation in Death Pathways
Caspases are expressed as zymogens that undergo proteolytic activation in a highly regulated manner. Monomeric initiator and inflammatory caspases are activated at macromolecular platforms formed by adapter proteins. According to the induced-proximity model, the recruitment to these platforms results in a dimerization of two catalytic subunits followed by activation and autocatalytic processing (Salvesen and Dixit 1999). In contrast, the executioner caspases exist as intracellular dimers of two catalytic subunits and are activated upon cleavage by inducer or as well active executioner caspases without the requirement of additional adapter proteins.
8.3.4.1 Caspase-Dependent Intrinsic Apoptosis: The Apoptosome
Formation of the macromolecular complex known as the apoptosome is the pivotal point of caspase activation in intrinsic apoptosis (Fig. 8.3a). MOMP-mediated release of cytochrome c from the mitochondrial IMS into the cytosol triggers the assembly of Apaf-1 into a wheel-like heptamer (Yuan et al. 2010).
Caspase activation platforms. Inflammatory and apoptotic initator caspases are activated at macromolecular complexes, which induces dimerization and autoproteolytic cleavage. (a) Apoptosome formation: released cytochrome c from the mitochondrion binds to Apaf-1 and induces a conformational change that reorients its NOD and CARD. Seven Apaf-1 molecules then form a wheel-like heptamer, which have been analyzed by electron microscopy (3IZA). The structure revealed a CARD-CARD interaction between the Apaf-1 and caspase-9 that form a disc above the central hub. (b) Death inducing signaling complex: activation of caspase-8 or -10 occurs at the DISC, which is initiated by trimerization of death receptors. Trimeric receptors recruit pro-caspase-8 or -10 via adapter proteins (FADD, TRADD). The assembly of FAS/FADD DDs have been structurally characterized (3OQ9) and showed a pentameric arrangement of 5–7 Fas DD and 5 FADD DDs. First, this structure shows a ring-like arrangement of the DDs similar to the Apaf-1 arrangement. Second, it indicates a requirement of more than one trimeric Fas receptor to initiate the DISC formation. (c) Proposed mechanism of inflammasome formation: NLRP as well as NLRC proteins exist in a conformation, where its NACHT domain is blocked. Pathogen and danger associated molecular patterns can be recognized by the LRR, which triggers a conformational change leading to a freed NACHT domain and subsequent oligomerization. Adapter proteins like ASC are then recruiting caspase-1 or -5 for activation. (d) PIDDosome formation: sequential cleavage of PIDD releases its DD, which recruits pro-caspase-2 via the adapter protein RAIDD. The crystal structure of PIDD-RAIDD DD assembly (2OF5) revealed a ring like structure with five RAIDD DD (dark green) stacked on top of five PIDD DD (blue). Two additional RAIDD DD (light green) are located on a third level. (a–d) The available complex structures of these oligomeric activation platforms point out a stoichiometric asymmetry: in principal, one of the pro-caspase monomers will not dimerize due to the odd-numbered adapter proteins
The adapter protein Apaf-1 consists of a N-terminal CARD, a nucleotide binding and oligomerization domain (NOD) and a C-terminal regulatory region with two domains (WD1 and WD2). Its fundamental role in apoptosome formation relies on the Apaf-1 oligomerization with a molecular mechanism proposed based on the crystal structure of a full-length murine Apaf-1 protein (Reubold et al. 2011). Binding of cytochrome c to the regulatory region of monomeric Apaf-1 releases the attached NOD, which rotates upon binding of ATP to expose its oligomerization areas and thereby relocates the now accessible CARD. This conformational change leads to the assembly into a circular structure of seven Apaf-1 subunits with central NOD and propeller-like regulatory regions (Reubold et al. 2011). The N-terminal CARDs form a recruitment disk on top of the protomer, accessible for the targeted pro-caspase-9.
Recruitment of pro-caspase-9 to the apoptosome via homotypic interactions between the Apaf-1 and the pro-caspase-9 CARDs results in dimerization of two catalytic domains and activation of the enzyme by cleavage (Pop et al. 2006). Notably, inter-subunit linker cleavage is not necessary for activity (Stennicke et al. 1999) but may provide stability to the active dimer (Fuentes-Prior and Salvesen 2004). Thus, the Apaf-1 based apoptosome can be defined as a cofactor of caspase-9 (Pop et al. 2006). As a last step, limited proteolysis of downstream executioner caspases by caspase-9 promotes the execution of apoptosis.
8.3.4.2 Extrinsic Apoptosis: Death Inducing Signaling Complex (DISC)
Numerous extracellular death stimuli provoke the intracellular formation of a macromolecular recruitment platform for caspase-8 or -10, known as the death inducing signaling complex (DISC) (Kischkel et al. 1995). Initially, a death ligand binds to its receptor and induces oligomerization (Fig. 8.3b). A prominent and well-studied example is the receptor Fas (also known as APO-1 or CD95), which binds to Fas ligand and thereby forms a trimeric complex (Kischkel et al. 1995). TRAIL receptors (death receptor 4 and 5) are other examples that form a homotrimer upon binding to their ligand (Chaudhary et al. 1997; Pan et al. 1997; Walczak et al. 1997). Both receptor types are members of the death receptor family with its superfamily of TNF receptors and trigger the intracellular DISC formation.
Death receptor oligomerization results in a conformational change that exposes the intracellular death domains (DD) of the receptor as structurally shown for Fas (Scott et al. 2009). Then, homotypic interactions of the accessible DDs with the DDs of the adaptor protein FADD build the recruitment platform for the apical caspases.
FADD is a cytosolic protein with a N-terminal DED and a C-terminal DD (Chinnaiyan et al. 1995). The crystal structure of the protein-complex between DDs from Fas and FADD revealed a circular assembly of 5–7 Fas-DDs and 5 FADD-DDs, which optimally orients the FADD-DEDs for caspase recruitment (Wang et al. 2010). Additionally, both recent structural studies on DISC formation described the observation of higher oligomeric clusters and denote at least two trimeric Fas receptors as a prerequisite for signal transduction (Scott et al. 2009; Wang et al. 2010).
The complete DISC is formed upon the recruitment of pro-caspase-8 or -10 via homotypic interactions of their N-terminal DED to FADD-DEDs. This leads to an increased local pro-caspase concentration, a dimerization of two catalytic domains by induced proximity and enzymatic activity of the caspase dimer (Salvesen and Dixit 1999; Pop et al. 2007).
Caspase-8 is autocatalytically processed in the inter-subunit linker to stabilize the active dimeric conformation (Pop et al. 2007) and cleavage of the pro-domain linker allows the release of the peptidase into the cytosole (Martin et al. 1998).
Structural investigations on the unprocessed catalytic domain of caspase-8 by NMR spectroscopy visualized a highly flexible inter-subunit linker and an activation induced rearrangement of loop-1 and -3 in comparison to the X-ray structure of an active dimer (Keller et al. 2009). Included mutational studies stressed the combination of proteolytic cleavage and dimerization as a prerequisite for full activity.
Further studies on caspase-8 using a reconstituted DISC revealed a substrate specificity switch between pro- and active caspase-8 (Hughes et al. 2009). Initial dimerization of pro-caspase-8 induces activity for a limited substrate range, including autoproteolysis of pro-caspase-8 or cleavage of c-FLIP, a caspase-related regulatory protein involved in death survival signaling (Scaffidi et al. 1999; Micheau et al. 2002). Autocatalytic processing then forms the fully active enzyme with the complete substrate repertoire, including the executioner caspases and Bid that are responsible for the downstream signaling in apoptosis.
The activation mechanism for caspase-10 is similar to caspase-8 initiation (Wachmann et al. 2010). Induced proximity results in the formation of an active dimer and the further autocatalytic processing accelerates the cleavage of downstream proteins. However, the authors could show that Bid cleavage does not require processed caspase-10 in contrast to tBid production by caspase-8. This finding suggested a pro-apoptotic role of this enzyme in the uncleaved form and further research will be needed to clarify this function.
8.3.4.3 Pyroptosis: The Inflammasome
The activation of inflammatory caspases remained elusive until 2002 with the discovery of a molecular activation platform termed the inflammasome (Martinon et al. 2002). This intracellular protein complex triggers the activation of caspase-1 and -5 and is formed by NLRP1 and an adapter protein ASC (Fig. 8.3c). Subsequently, researchers identified more NLR family members that are involved in inflammasome formation, like NLRP3 (Agostini et al. 2004), NLRC4 (Mariathasan et al. 2004) and others (reviewed by Rathinam et al. 2012).
NLR proteins act as the danger-sensing molecule in the inflammasome and contain three important domains: A leucine-reach repeat (LRR) with an assumed function of danger signal recognition similar to Toll like receptors (Martinon et al. 2002), a nucleotide-binding and oligomerization domain NACHT and a death mediating domain, which is either a PYD in NLRP1 and NLRP3 or a CARD in NLRC4. Although the initiation of inflammasomes remains unclear, Martinon and Tschopp (2005) proposed a possible mechanism: Binding of a danger-associated ligand to LRR induces a conformational change that exposes the NACHT domain and thereby triggers oligomerization and recruitment of adapter proteins and inflammatory caspases.
The activation of pro-caspase-1 by NLRP1- or NLRP3-inflammasomes involves the adapter protein ASC, which contains a N-terminal CARD and a C-terminal PYD. The solution structure of full-length ASC visualized the two isolated domains connected via a flexible linker in a back-to-back orientation (de Alba 2009). This may prevent steric hindrance between the two domains and raises the capture radius for pro-caspases and NRL molecules. In addition, NLRP1 contains a C-terminal CARD that is able to directly recruit pro-caspase-1 although the presence of ASC increases caspase-1 activation (Faustin et al. 2007). A similar observation has been reported for the NLRC4 inflammasome, which can directly recruit pro-caspase-1 via its N-terminal CARD (Miao et al. 2010) but showed an ASC-dependent increase of activation (Mariathasan et al. 2004; Miao et al. 2010).
Insights into the activation mechanism of inflammatory caspases have been obtained with the crystal structure of pro-caspase-1 (Elliott et al. 2009). The inter-subunit linker of one chain occupies the central cavity of the zymogen dimer and locates its residue Asp-297 in close proximity to the active site of the other chain. After a possible interdimeric cleavage at Asp-297, the peptide chain is released and the central cavity can then harbor the other inter-subunit linker to trigger a second interdimeric processing. Additional autoproteolysis at a second cleavage site (Asp-316) then stabilizes the active dimer by conversion of an α-helix from one chain into an intradimer β-sheet (Elliott et al. 2009). These findings support the induced proximity model (Salvesen and Dixit 1999) for inflammatory caspases and emphasizes the crucial role of macromolecular oligomerization platforms for dimerization.
8.3.4.4 Caspase-2 Activation: The PIDDosome
The activation platform for caspase-2 was termed PIDDosome after the identification of the involved proteins by Tinel and Tschopp (2004). This large protein complex is formed by oligomerization of PIDD (p53-induced protein with a death domain) and the adapter protein RAIDD (Fig. 8.3d).
PIDD consists of a N-terminal LRR domain, two central ZU-5 domains and a C-terminal DD. Autoproteolysis between or after the two ZU-5 domains has been observed as a switch for pro-survival or pro-death signaling (Tinel et al. 2007). The adapter protein RAIDD, a N-terminal CARD and a C-terminal DD, is responsible for caspase-2 recruitment via CARD-CARD interactions. Besides its involvement in the PIDDosome, RAIDD has been observed in specific binding of RIP1, a serine/threonine protein kinase involved in death pathway (Duan and Dixit 1997).
The crystal structure of a complex of the DDs that are involved in PIDDosome formation discovered a stoichiometry of seven RAIDD DDs and five PIDD DD molecules (Park et al. 2007). Five RAIDD DDs are stacked in a ring-like structure on top of an asymmetric, pentameric ring of PIDD DDs. Two additional RAIDD DD molecules are located in a third level on top of the RAIDD DD ring-like structure. In principal, this complete assembly is able to recruit seven pro-caspase-2 molecules for activation although only three active enzymes can be formed.
8.3.4.5 Activation of Executioner Caspases
Executioner caspases exist as stable dimers in solution in contrast to the zymogens of the inflammatory and initiator related family members. Activation occurs upon partial proteolysis by initiator or executioner caspases and results in a rearrangement of the inter-subunit linker (Riedl et al. 2001a).
The pro-caspase-7 crystal structures revealed an asymmetric blocking of the central cavity by the inter-subunit linker (Chai et al. 2001b; Riedl et al. 2001a). Linker cleavage at Asp-297 promotes its release from the central cavity. It then stabilizes the active conformation via bundle formation, which is promoted between the new C-terminus of the large subunit of one catalytic domain and the newly formed N-terminus of the small subunit of the other adjacent catalytic domain. This bundle is further interacting with loop-4 and enables the formation of the active site as shown in peptide-bound caspase-7 structures (Riedl et al. 2001a; Fuentes-Prior and Salvesen 2004).
The important role of the central cavity in caspase activation is not only depicted by the zymogen structures of caspase-1 and -7 (Chai et al. 2001b; Riedl et al. 2001a; Elliott et al. 2009) but also by the identification of allosteric inhibitors of caspase-3 and -7 (Hardy et al. 2004). These inhibitors bind in the central cavity and trap the protease in a zymogen-like conformation.
8.3.5 Caspase Substrates
The commercially available short peptide substrates have been derived from a combinatorial approach by scanning of large peptide libraries (Thornberry et al. 1997) unraveling the caspase substrate consensus. These small molecule substrates are coupled to a fluorophore reporter and have generally been used for caspase characterization in-vitro (Roschitzki-Voser et al. 2012).
Diverse methods to identify in-vivo cleavage sites have been developed in the field of degradomics, like scanning of the transcriptome using bioinformatics (Boyd et al. 2005) as well as cell-based degradation methods, recently reviewed by Agard and Wells (2009). A growing list of identified in-vivo caspase substrates is available in the MEROPS database [http://merops.sanger.ac.uk/ (Rawlings et al. 2012)] with almost 900 physiological relevant entries. Undoubtedly, some of these entries are cleaved only in specific cell types or not explicitly in cell death pathways while others are well known for being processed during apoptosis. Proteolysis of those proteins typically leads to a loss-of-function or a gain-of-function. While the executioner caspases are prominent examples for the latter, several other important substrates have been described.
8.3.5.1 Rho-Associated Kinase I (ROCK I)
The family of Rho GTPases is known as an actin cytoskeleton regulator with the Rho-associated kinases (ROCK) as their effectors (Hall 1998). These serine/threonine kinase isoforms are activated upon interaction with GTP-bound Rho proteins.
During apoptosis, caspases cleave ROCK 1 at a DETD ↓ G motive near the C-terminus and thereby remove a putative autophosphorylation/auto-inhibitory domain (Coleman et al. 2001). The result is an intrinsically more active kinase, which promotes membrane blebbing in apoptotic cells by phosphorylation of downstream targets. It was further shown that a truncated ROCK 1 is independent of Rho GTPases and triggers membrane blebbing without activated caspases.
8.3.5.2 Caspase-Activated DNAse (CAD)
A morphological feature of apoptosis is the degradation of chromosomal DNA observable as a characteristic DNA ladder. A crucial nuclease for this process has been identified as the caspase-activated DNAse (CAD, also known as DNA fragmentation factor DFF40) (Nagata et al. 2003). In cells, this enzyme forms an inactive dimeric complex with the inhibitor of CAD (ICAD, also known as DFF45).
The cleavage sites DETD ↓ S and DAVD ↓ T are located in two flexible inter-domain linkers of CAD and get cleaved by caspase-3. This triggers the release from ICAD and a formation of active CAD dimers. Notably, CAD is not only responsible for apoptotic degradation of nucleosomal DNA, it also promotes cell differentiation induced by DNA strand breaks (Larsen et al. 2010). Thus, CAD is an excellent example for the caspase-3 functional dualism in cell death execution and cell proliferation.
8.3.5.3 Poly(ADP-Ribose) Polymerase (PARP)
First notices of cleavage of poly(ADP-ribose) polymerase (PARP) have been described in chicken cells in 1994 (Lazebnik et al. 1994). PARP is involved in repair mechanisms of single and double strand DNA breaks. It forms branched poly(ADP-ribose) molecules, attaches them to acceptor proteins, including itself, leading to the recruitment of repair proteins to the DNA breakage site (Javle and Curtin 2011).
The executioner caspase-3 and -7 cleave the 116 kDa PARP after a canonical sequence DEVD ↓ G into an 85 kDa fragment having no enzymatic activity. This loss-of-function prevents the repair of DNA fragments induced by CAD. Remarkably, hyperactivation of PARP can lead to a low ATP level inside cells that is a known trigger of regulated necrosis. In this prospect, the loss-of-function supports not only the apoptotic pathway but also prevents the activation of necrotic cell death and inflammatory responses.
Furthermore, PARP cleavage is a good example for exosite interactions between a protease and its target protein. It could be shown that PARP, which has been modified with long branched poly(ADP-ribose) molecules, is cleaved with higher efficiency by caspase-7 than by caspase-3 (Germain et al. 1999) although the latter cleaves short peptide substrates more efficiently. The enhanced affinity to caspase-7 was assigned to exosite interactions between the caspase-7-p20 and the branched poly(ADP-ribose) molecule.
8.3.5.4 Human Telomerase Reverse Transcriptase (hTERT)
The human telomerase hTERT maintains the length of DNA telomeres by its reverse transcriptase activity and ensures chromosomal stability. The reverse transcriptase as the catalytic domain of the protein was recently identified as a unique caspase-6 and -7 substrate that is cleaved during apoptosis (Soares et al. 2011). Four cleavage sites have been identified with TVTD ↓ A and VNMD ↓ Y cleaved by caspase-6 and TSLE ↓ G and PKPD ↓ G cleaved by caspase-7. These cleavage sites are conserved in old world monkeys and apes although the function of the persistent fragments during apoptosis remains unclear. hTERT is a recent example for a non-canonical substrate recognition of caspases.
8.3.5.5 Pro-interleukins and Interleukins
The starting point of caspase research was the identification of the ICE, today known as caspase-1 (Thornberry et al. 1992). Indeed, interleukin precursors and their active forms are caspase substrates that either provoke inflammatory responses or prevent inflammation during apoptosis.
For example, bacterial infections of cells can lead to caspase-mediated processing of pro-interleukin-1β (YVHD ↓ A) and -18 (LESD ↓ Y) with followed secretion of these activated cytokines. Caspases-1 and -4 are able to process the pro-forms (Thornberry et al. 1992; Kamens et al. 1995; Akita et al. 1997; Gu et al. 1997) and have been reported to be involved in the inflammasome-mediated response (Sahoo et al. 2011; Sollberger et al. 2012).
Remarkably, pro-interleukins are not only activated by inflammatory caspases but also by apoptotic members as reported for pro-interleukin-16 activation (PKPD ↓ G) by caspase-3 (Zhang et al. 1998). Pro-interleukin-16 is expressed in more than 90 % of all T cells (Chupp et al. 1998), is located in the cytoplasm and the nucleus, while a nuclear presence inhibits progression (Center et al. 2004). Thus, proteolytic modification of such a cell cycle regulator by caspase-3 illustrates the important role of these proteases in other molecular mechanisms.
In contrast to other cytokines, interleukin-33 is expressed in its active form and is not secreted after inflammatory stimuli (Lüthi et al. 2009). It is more likely to be released after membrane disruption during necrosis, while apoptotic cell death leads to interleukin-33 deactivation mediated by caspase-3 and -7 after cleavage at DGVD ↓ G (Lüthi et al. 2009). In principal, the proteolysis of interleukin-33 by apoptotic executioner caspases acts as an intracellular proinflammatory extinguisher in a tissue protective manner.
The enormous variety of caspase substrates illustrates the important roles of these enzymes in cell death, proliferation and inflammatory pathways. Although, a consensus sequence has been defined (Timmer and Salvesen 2007), natural substrates can as well be cleaved at non-canonical sites as demonstrated in rare cases (Krippner-Heidenreich et al. 2001; Soares et al. 2011).
8.3.6 Morphological Changes upon Activation
Activation of cellular death pathways results in major morphological changes that can be observed under the light microscope. Biochemical techniques have been used to characterize death pathways (Galluzzi et al. 2012). Particular morphological features (reviewed by Ziegler and Groscurth 2004) are well characterized and especially the apoptotic morphological hallmarks can be explained by caspase-mediated activation of effector proteins.
Nuclear condensation and DNA fragmentation during apoptosis relies on executioner caspases. On one hand, the cleavage of nuclear lamins by caspase-3 and -6 induces a loss of structural integrity and a nuclear shrinkage, which promotes a collapse into smaller nuclear particles in a late apoptotic state (Rao et al. 1996) (Fig. 8.4). On the other hand, CAD is activated by caspase-3 and produces DNA fragments of 180- to 200-bp length, visible by gel electrophoresis (Nagata et al. 2003; Larsen et al. 2010). In addition, cleavage of the DNA repair enzyme PARP by caspase-3 and -7 (Germain et al. 1999) prevents the repair response thus further provoking apoptosis.
Morphology of apoptosis. Jurkat cells were growing in media containing etoposide (100 μM, lower panels) or without etoposide (upper panels). Paraformaldehyde fixation was performed after 6 h. Etoposide treated cells show condensed and fragmented nuclei while untreated cells possess round shaped nuclei (left panels, DNA staining with Hoechst33342). Transmission images show round shaped healthy cells, whereas etoposide treatment leads to apoptotic cells displaying membrane blebbing (right panels)
ROCK 1 cleavage by executioner caspases leads to an intrinsically more active kinase that phosphorylates downstream targets in the actin-myosin system (Coleman et al. 2001). Actin-myosin related cell contractility initiates the formation of membrane protrusions commonly known as membrane blebs (Fig. 8.4). Further progression of cell shrinkage separates these blebs into apoptotic bodies, packed with organelles and nuclear fragments, which are subsequently engulfed by surrounding cells via phagocytosis (Ziegler and Groscurth 2004).
Other phosphorylation targets in the Rho-ROCK signaling pathway are flippases, which maintain the membrane asymmetry of phosphatidylserine (PS) that is located within the inner membrane leaflet of a normal cell (Krijnen et al. 2010). In an early stage of apoptosis, PS is presented at the outer membrane as an “eat-me” signal for phagocytes (Grimsley and Ravichandran 2003). First studies on oxidatively stressed erythrocytes indicated caspase-related PS externalization (Mandal et al. 2002) while research on cardiomyocytes identified a Rho-ROCK signaling to be crucial for PS exposure (Krijnen et al. 2010).
8.3.7 Regulation and Specific Inhibition
A tight regulation of caspase activation is essential to avoid accidental induction of cellular demise. Thus, a complex network involving numerous proteins has been evolved, which in particular cases guarantees the survival or decease of a cell (Fig. 8.5). Most of those proteins belong to the Bcl-2 family or to the inhibitor of apoptosis protein (IAP) family. In addition, released mitochondrial proteins upon MOMP can interact with members of these two families and facilitate the progression of apoptosis.
Regulatory network. Activation of caspase-8 or -10 at the DISC can be repressed by cFLIPs. Once activated, these caspases cleave downstream executioner caspases that trigger the apoptotic pathway. In addition, caspase-8 or -10 can produce tBid that stimulates the intrinsic caspase-dependent apoptotic pathway. This pathway is also triggered by intracellular death stimuli like DNA damage and activates pro-apoptotic Bcl-2 proteins, known as the subfamily BH3-only proteins. These proteins are regulated by anti-apoptotic Bcl-2 family members that provide a first threshold for death stimuli. The BH3-only proteins Bax and Bak possess a pore forming activity to induce MOMP. Released mitochondrial proteins then trigger the progression of the intrinsic apoptotic pathway: cytochrome c binds to Apaf-1 and induces apoptosome formation and caspase-9 activation. Released Smac/DIABLO is binding and inhibiting IAPs and HtrA2/Omi not only antagonizes IAPs but also cleave cellular substrates. Additional important regulators of apoptosis are the IAPs: they specifically inhibit caspases and are able to counteract with Smac/DIABLO in anti-apoptotic manner
Members of the Bcl-2 family are known to have cell regulatory functions and often possess either pro- or anti-apoptotic activity, depending on the environmental conditions (reviewed by Hardwick et al. 2012). Bcl-2 proteins (e.g. Bcl-2, Bcl-B, Bax, Bak,…) consist of 3–4 Bcl-2 homology (BH) domains enumerated from BH1 to BH4. Members of this family (e.g. Bim, Bid, Puma,…) contain only one BH3 domain, are thus called BH3-only proteins and known as promoters of apoptosis. Certain family members contain an additional C-terminal transmembrane domain (e.g. Bcl-2, Bax, Bim,…) that localizes them at membranes. Bcl-2 proteins exist in the cytosol, at outer mitochondrial membranes and supposedly inside mitochondria, however this submitochondrial localization remains controversial (Hardwick et al. 2012).
The IAP family members are multi domain proteins that directly suppress apoptosis by caspase inhibition (Deveraux et al. 1997) or indirectly via interaction with pro-apoptotic signaling complexes (Fulda and Vucic 2012). In addition, several members contain a C-terminal RING domain that functions as an E3 ubiquitin ligase with increasing significance in cell survival (Vaux and Silke 2005; Vucic et al. 2011).
Intracellular apoptotic stimuli can activate BH3-only proteins, which promotes Bax and Bak oligomerization and pore formation at the outer mitochondrial membrane (Kuwana et al. 2002). This leads to the release of cytochrome c and other mitochondrial proteins that function in a pro-apoptotic manner. The second mitochondria-derived activator of caspases (Smac, also termed DIABLO) is known to bind and inhibit IAP (Verhagen et al. 2000) but can itself be antagonized by IAPs (Vucic et al. 2005). Another released protein is HtrA2 (also known as Omi), a serine protease that is involved in apoptosis and suppression of IAP (Verhagen et al. 2002). In addition, caspase-8 can cleave the BH3-only protein Bid, forming a truncated form tBid and thus integrates the extracellular apoptotic stimulus by inducing MOMP.
Furthermore, phosphorylation of caspases and upstream adapter proteins has been reported as regulatory mechanisms in cell death pathways (Kitazumi and Tsukahara 2011). For instance, caspase-9 can be phosphorylated at the catalytic domain residue Ser-196 and in the CARD at Thr-125, which both prevents caspase-9 activation (Cardone et al. 1998; Allan et al. 2003). Phosphorylation of caspase-8 at Ser-364 and caspase-3 at Ser-150 has been reported as survival signals during neutrophil apoptosis (Alvarado-Kristensson et al. 2004). Other phosphorylation sites on caspase-8 have been discovered (Tyr-310, Tyr-397, Tyr-465) and revealed a dynamic post-translational regulation of apoptosis in neutrophils (Jia et al. 2008). These findings strongly emphasize the important control functions of kinases and phosphatases in death pathways.
8.3.7.1 X-Linked Inhibitor of Apoptosis Protein (XIAP)
A well-studied IAP family member is the X-linked IAP (XIAP) that is able to directly inhibit caspases-3, -7 and -9 (Deveraux et al. 1997, 1998). It is characterized by three baculoviral IAP repeat (BIR1–3) modules, an ubiquitin associated (UBA) domain and a C-terminal RING domain. Structural investigations on XIAP binding to caspase-3, -7 and -9 unraveled two different binding and inhibition mechanisms, which are mainly related to binding of linker regions near the BIR2 and BIR3 domains, respectively (Chai et al. 2001a; Riedl et al. 2001b; Shiozaki et al. 2003). In addition, XIAP can simultaneously bind and inhibit both, initiator caspase-9 and executioner caspase-3 (Bratton et al. 2002) at a tightly packed “holo-apoptosome” that not only recruits pro-caspase-9 but also binds active caspase-3 (Yuan et al. 2011).
Crystal structures of caspase-3 and -7 have been reported with a truncated XIAP containing a short N-terminal linker peptide and the BIR2 domain (Fig. 8.6a). Remarkably, both homologue caspases showed the same principle of inhibition: The XIAP residues involved in binding interactions are located in the peptide linker, which lies inside the substrate-binding cleft of the enzyme in opposite direction when compared to the observed natural binding of substrate (Riedl et al. 2001b). Furthermore, the S4 pocket is occupied by Asp-148 of XIAP whereas upstream residues sterically block the access to the S1 and S1′ subsites and thus competitively inhibit the enzyme.
Specific inhibition. (a) XIAP inhibition of caspase-3 and -7. Crystal structures of XIAP-BIR2 domain in complex with either caspase-3 (1I3O) or caspase-7 (1I51) revealed a competitive inhibition mechanism by direct binding into the substrate-binding groove. The BIR2 domain was crystallized in both complexes, although no electron density for the domain could be observed in the complex with caspase-7. (b) The crystal structure of XIAP-BIR3 domain in complex with caspase-9 (1NW9) unraveled a different inhibition mechanism: BIR3 interacts with residues in the dimer interface and thus prevents a dimerization of caspase-9. (c) Complex structure of caspase-2 with DARPin F8 (2P2C) showed a binding of the DARPin to loop-4. This binding stabilizes the loop in a distinct conformation compared to substrate bound caspase-2 and thus inhibits the enzyme allosteric. (d) DARPin D3.4 binds to caspase-3 (2XZD) at the substrate-binding cleft and inhibits the enzyme in a linear competitive mechanism
The complex of caspase-9 with the XIAP-BIR3 domain revealed a completely different inhibition mechanism: The BIR3 domain binds at the caspase-9 dimer interface of the catalytic domain and consequently traps caspase-9 in an inactive, monomeric form thus preventing active site formation (Shiozaki et al. 2003) (Fig. 8.6b). This interaction is further provoked by the N-terminal peptide of the small subunit of caspase-9, which interacts with BIR3 at the so-called Smac pocket (Shiozaki et al. 2003).
As other IAPs, XIAP can regulate the apoptotic pathway via its C-terminal RING domain. Ubiquitination of caspase-3 mediates proteosomal degradation (Suzuki et al. 2001) while RING domain truncations of XIAP lead to elevated caspase-3 activity (Schile et al. 2008). In contrast, ubiquitination of AIF is nondegradative but also attenuates its death-inducing activity (Lewis et al. 2011).
8.3.7.2 c-FLIP: A Structural Homologue of Caspase-8
The family of FLICE-inhibitory proteins (FLIPs) has been discovered in viral genomes (termed vFLIPs) and supports viral infections by down regulation of cellular death responses (Thome et al. 1997). In humans, numerous splice variants of cellular FLIPs (c-FLIPs) have been reported whereas three isoforms are expressed (Krueger et al. 2001; Golks et al. 2005; Safa 2012). The two known short forms, c-FLIPS and c-FLIPR, consist of two DEDs that can be recruited to the DISC where they block binding of caspase-8 and thus prevent a death-receptor mediated apoptosis (Krueger et al. 2001; Golks et al. 2005).
A longer isoform c-FLIPL is structurally similar to caspase-8 and comprises two DED and a C-terminal caspase-like domain lacking enzymatic activity (Yu et al. 2009). Its regulation of the extrinsic-apoptotic pathway is more adaptive and concentration dependent compared to its short isoforms. On one hand, FLIPL is recruited to the DISC where it can form heterodimers with catalytic domains of caspase-8 at physiological conditions and thus mediates its activation (Chang et al. 2002). In contrast, a high expression level of FLIPL induces partial autocatalytic processing of caspase-8 (Krueger et al. 2001), which then remains at the DISC-platform, cleaves local substrates like RIP1 and prevents apoptosis (Micheau et al. 2002; Kavuri et al. 2011). Furthermore, c-FLIPL is also known as a limiting factor of kinase-dependent necrosis (reviewed by Green et al. 2011; Oberst et al. 2011).
The highly variable signal transduction from death receptors to caspase-8 and c-FLIPL strongly emphasizes the critical interplay of these proteins in regulation of pro- or anti-apoptotic responses.
8.3.7.3 Non-natural Inhibition: Designed Ankyrin Repeat Proteins (DARPins)
Gene knockouts or artificial inhibition of a specific protein are general methods used in cell biology to target its distinct role in diverse cellular processes. These methods have also been applied to unravel the known functions of the caspase family members, which have been broadly studied in cells derived from gene knockout mice. However, the production of a knockout mouse is a time-consuming process, not always successful or occasionally inaccurate, as shown for the caspase-1 deficient strain 129 that also harbors a deficient caspase-11 (Kayagaki et al. 2011).
A protein knock-out by specific inhibition is a second approach to study the role of a specific protein in cellular processes. In the case of caspases this is rather difficult to achieve due to the high structural homology of these enzymes. The commercially available substrate-based inhibitors target the conserved active site and are thus not very specific (McStay et al. 2008). In contrast, selected binding proteins like anti- and nanobodies or designed ankyrin repeat proteins (DARPins) bind via an increased protein-protein interaction surface that may enhance the specificity. Furthermore, the binding may induce functional modification of the target protein, as shown in several examples like a Fas receptor agonistic antibody (Chodorge et al. 2012) or an ABC transporter modulating DARPin (Seeger et al. 2012).
Highly specific inhibition of caspase-2 and -3 has been achieved with two different DARPins (Schweizer et al. 2007; Schroeder et al. 2013). These binding proteins were designed based on the ankyrin protein scaffold and usually contain two or three internal repeats that are sandwiched between a N- and C-terminal capping repeat (Binz et al. 2004). Each internal repeat obtains six randomized positions that are potentially involved in interactions, forming a diverse library with at least 1010 different molecules that can be selected by phage- or ribosome display (Hanes and Plückthun 1997; Steiner et al. 2008). Selected DARPins combine the advantages of high affinity and stability with high purity and expression yields in a cysteine less framework.
The caspase-2 specific DARPin (AR_F8) binds and inhibits the enzyme with a tight binding mixed-type inhibition mechanism (Schweizer et al. 2007). AR_F8 binding to caspase-2 occurs primarily at the binding groove forming loop-4 and stabilizes a distinct conformation of this loop compared to substrate analogue bound caspase-2 (Fig. 8.6c). Additional structural rearrangements have been observed N-terminal at the small subunit of the adjacent catalytic domain that finally results in a displacement of the catalytic residue Cys-285. Altogether, the structural and kinetic analysis of this DARPin clearly revealed an allosteric inhibition of caspase-2 with high specificity.
Another selected DARPin (D3.4) has been reported to specifically target caspase-3 (Schroeder et al. 2013). This binder inhibits the protease in a tight binding and purely competitive mechanism (Fig. 8.6d). Structural data uncovered a complex formation of D3.4 and caspase-3 at the active site cleft leading to obstructed binding pockets. D3.4 positions its residue Asp-45 inside the S4 pocket mimicking a natural peptide binding. Further interactions of the second internal repeat lock the caspase-3 residue Tyr-204 in a closed state that occupies the subsite S2 as seen in the XIAP caspase-3 complex structure (Riedl et al. 2001b). Although the structural mechanism of inhibition displays similarities to XIAP, the active site occupying peptide is not in a reversal orientation. Furthermore, this binder is a decent example for active site directed inhibition that is combined with an enlarged target-inhibitor interaction surface to achieve high specificity.
8.4 Caspase-Related Diseases
The important roles that caspases mediate in cell death and differentiation pathways are highly regulated and often include redundant mechanisms. However, their importance in cellular processes also increases the susceptibility for severe diseases in case of misregulation. For instance, alteration of the apoptotic pathway is related to cancer, neurological and cardiovascular disorders as well as autoimmune diseases (reviewed by Favaloro et al. 2012).
Cancer development and progression is highly connected to overexpression of regulatory Bcl-2 proteins, down-regulation of apoptosome forming Apaf-1 and modulation of death receptor pathways by reduced or increased expression of death receptors or death ligands, respectively (Favaloro et al. 2012). Direct mutations of the caspase genes can lead to a reduction or loss of enzymatic activity, while other studies reported dominant negative caspase variants that prevent activation of the wild type form (reviewed by Olsson and Zhivotovsky 2011). This results in a decrease of apoptotic capacity that may favor cancer manifestation. However, there is little evidence for tumorigenesis induced by an individual caspase-mutant that indeed emphasizes a redundant network with tumor suppressor functionalities (Olsson and Zhivotovsky 2011).
Caspases have been reported as major conductors in the development of Alzheimer’s disease (reviewed by Rohn 2010). Two of the major pathological markers in Alzheimer’s disease are the formation of neurofibrillary tangles (NFTs), consisting of hyperphosphorylated microtubule-associated protein tau and the extracellular deposition of amyloid-β (Aβ) in plaques. Aβ is created after sequential proteolysis of the amyloid precursor protein (APP) by β- and γ-secretases. However, caspase involvement of APP cleavage has also been observed (Zhang et al. 2011).
In addition, it has been proposed that the presence of Aβ stimulates caspase activation that leads to tau cleavage, predominantly mediated by caspase-3 and -6, and a following tau hyperphosphorylation (Rohn 2010). Moreover, this hypothesis of Aβ-linked tau cleavage has been verified using a transgenic mouse model that overexpresses the antiapoptotic protein Bcl-2 (Rohn et al. 2008). Thus, the role of caspases in Alzheimer’s disease lays assumably not only terminal in the execution of neuronal cell death but also proximal in the initiation of the disease mechanism.
Caspase involvement has also been shown in Huntington’s disease, a neuronal disorder that is caused by mutation of the huntingtin (htt) protein (reviewed by Ehrnhoefer et al. 2011). Disease related htt mutations lead to an abnormal elongation of a polyglutamine stretch in the N-terminal region, which is supposed to prevent aggregation in wildtype proteins by turnover regulation (Ehrnhoefer et al. 2011). Both, either wildtype or mutant proteins can be processed by proteases, including the caspase family.
N-terminal cleavage fragments of the htt mutant have been reported to increase the apoptotic susceptibility in dependence of fragment and polyglutamine extension length (Hackam et al. 1998). Removal of the five predicted caspase cleavage site in htt could reduce its toxicity and furthermore unraveled a specific caspase-6 cleavage site that is crucial in the development of Huntington’s disease symptoms (Graham et al. 2006). Active caspase-6 has been proposed as an early disease marker due to its activation before the manifestation of the first motor abnormalities (Graham et al. 2010).
Activation of caspase-6 and downstream cleavage of this enzyme has been shown to be a fundamental step in the development of neurodegenerative diseases and highlights its potential as a novel therapeutic target (reviewed by Graham et al. 2011).
Alterations in the inflammasome formation mechanisms can lead to hyperactivation of caspase-1 and extensive production of IL-1β and IL-18, which can be related to autoimmune diseases (reviewed by Lamkanfi et al. 2011). For instance, enhanced expression of caspase-1 and IL-18 has been found in patients suffering from multiple sclerosis (Huang et al. 2004).
8.5 Concluding Remarks
Death pathways are fundamental mechanisms of multicellular organisms to ensure the correct development, tissue maintenance and homeostasis. The diverse processes are tightly regulated to prevent accidental cellular demise but also to facilitate a fast removal of cells upon cell death activation. Its redundant system of overlapping, distinctive pathways secures the multicellular integrity upon various cytotoxic stimuli in an exceptional manner.
The particular death pathways of apoptosis and pyroptosis are primarily driven by caspases. These enzymes are classified according to their activity related to inflammation and apoptosis. Active caspases cleave specific substrates, which results in either activation of demise effectors or deactivation of survival proteins that both accelerate cell destruction. Thus, a precise regulation is indispensable and occurs at numerous steps. Although this highly specific and minimal error-prone regulatory system has been evolved with redundancy, imbalances result in severe diseases including cancer and neurodegenerative disorders.
In conclusion, modern research unraveled a broad contribution of caspases not only in cell death mechanism but also in inflammatory responses and tissue protection by anti-inflammatory activity during apoptosis. Thus, researchers began to reconsider the general classification of inflammatory and apoptotic caspases. Martin et al. (2012) recently suggested a new classification of positive and negative regulators of inflammation: Besides the pro-inflammatory caspases-1, -4 and -5, the nowadays known apoptotic caspases are categorized as anti-inflammatory proteases. Overall, it further underlines the remarkable role of this protease family in numerous distinctive cellular processes besides death pathways.
Abbreviations
- Aβ:
-
Amyloid-β
- AIF:
-
Apoptosis-inducing factor
- Apaf-1:
-
Apoptotic protease activating factor 1
- APP:
-
Amyloid precursor protein
- BH:
-
Bcl-2 homology
- Bid:
-
BB3-interacting domain
- BIR:
-
Baculoviral IAP repeat
- C. elegans :
-
Caenorhabditis elegans
- CAD:
-
Caspase-activated DNAse
- CARD:
-
Caspase activation and recruitment domain
- DAMP:
-
danger associated molecular pattern
- DARPin:
-
Designed ankyrin repeat protein
- DD:
-
Death domain
- DED:
-
Death effector domain
- DFF40:
-
DNA fragmentation factor 40 also termed CAD
- DIABLO:
-
Direct IAP binding protein with low pI, also termed Smac
- DISC:
-
Death-inducing signaling complex
- ENDOG:
-
Endonuclease g
- FADD:
-
Fas-associated death domain
- FLICE:
-
FADD-like interleukin-1 beta-converting enzyme, today known as caspase-8
- FLIP:
-
FLICE-inhibitory protein
- HTRA2:
-
High temperature requirement protein 2
- htt:
-
Huntingtin
- IAP:
-
Inhibitor of apoptosis protein
- ICAD:
-
Inhibitor of CAD, also known as DFF45
- ICE:
-
Interleukin-1β-converting enzyme
- IMS:
-
Inter-membrane space
- LRR:
-
Leucine-rich repeat
- MOMP:
-
Mitochondrial outer membrane permeabilization
- NC-IUBMB:
-
Nomenclature Committee of the International Union of Biochemistry and Molecular Biology
- NCCD:
-
Nomenclature Committee on Cell Death
- NFT:
-
Neurofibrillary tangle
- NLR:
-
NOD-like receptor
- NOD:
-
Nucleotide binding and oligomerization domain
- PARP:
-
Poly(ADP-ribose) polymerase
- PIDD:
-
p53-induced protein with a death domain
- PS:
-
Phosphatidylserine
- RIP:
-
Receptor-interacting protein
- ROCK:
-
Rho-associated kinase
- Smac:
-
Second mitochondria-derived activator of caspase, also termed DIABLO
- tBid:
-
Truncated Bid
- TNF:
-
Tumor necrosis factor
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- UBA:
-
Ubiquitin associated
- XIAP:
-
X-linked inhibitor of apoptosis protein
References
Agard NJ, Wells JA (2009) Methods for the proteomic identification of protease substrates. Curr Opin Chem Biol 13(5–6):503–509
Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J (2004) NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20(3):319–325
Akita K, Ohtsuki T, Nukada Y, Tanimoto T, Namba M, Okura T, Takakura-Yamamoto R, Torigoe K, Gu Y, Su MS, Fujii M, Satoh-Itoh M, Yamamoto K, Kohno K, Ikeda M, Kurimoto M (1997) Involvement of caspase-1 and caspase-3 in the production and processing of mature human interleukin 18 in monocytic THP.1 cells. J Biol Chem 272(42):26595–26603
Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR (2003) Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5(7):647–654
Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J (1996) Human ICE/CED-3 protease nomenclature. Cell 87(2):171
Alvarado-Kristensson M, Melander F, Leandersson K, Rönnstrand L, Wernstedt C, Andersson T (2004) p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J Exp Med 199(4):449–458
Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grütter MG, Plückthun A (2004) High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol 22(5):575–582
Boyd SE, Pike RN, Rudy GB, Whisstock JC, Garcia de la Banda M (2005) PoPS: a computational tool for modeling and predicting protease specificity. J Bioinform Comput Biol 3(3):551–585
Bratton SB, Lewis J, Butterworth M, Duckett CS, Cohen GM (2002) XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ 9(9):881–892
Brennan MA, Cookson BT (2000) Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol 38(1):31–40
Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science 282(5392):1318–1321
Center DM, Cruikshank WW, Zhang Y (2004) Nuclear pro-IL-16 regulation of T cell proliferation: p27(KIP1)-dependent G0/G1 arrest mediated by inhibition of Skp2 transcription. J Immunol 172(3):1654–1660
Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y, Dataa P (2001a) Structural basis of caspase-7 inhibition by XIAP. Cell 104(5):769–780
Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y (2001b) Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell 107(3):399–407
Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X (2002) c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J 21(14):3704–3714
Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity 7(6):821–830
Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81(4):505–512
Chodorge M, Züger S, Stirnimann C, Briand C, Jermutus L, Grütter MG, Minter RR (2012) A series of Fas receptor agonist antibodies that demonstrate an inverse correlation between affinity and potency. Cell Death Differ 19(7):1187–1195
Chupp GL, Wright EA, Wu D, Vallen-Mashikian M, Cruikshank WW, Center DM, Kornfeld H, Berman JS (1998) Tissue and T cell distribution of precursor and mature IL-16. J Immunol 161(6):3114–3119
Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF (2001) Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol 3(4):339–345
Conradt B, Horvitz HR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93(4):519–529
Cookson BT, Brennan MA (2001) Pro-inflammatory programmed cell death. Trends Microbiol 9(3):113–114
Cooper DM, Pio F, Thi EP, Theilmann D, Lowenberger C (2007) Characterization of Aedes Dredd: a novel initiator caspase from the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol 37(6):559–569
de Alba E (2009) Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J Biol Chem 284(47):32932–32941
Denault J-B, Salvesen GS (2003) Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem 278(36):34042–34050
Denecker G, Ovaere P, Vandenabeele P, Declercq W (2008) Caspase-14 reveals its secrets. J Cell Biol 180(3):451–458
Deveraux QL, Takahashi R, Salvesen GS, Reed JC (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388(6639):300–304
Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC (1998) IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 17(8):2215–2223
Duan H, Dixit VM (1997) RAIDD is a new ‘death’ adaptor molecule. Nature 385(6611):86–89
Eckhart L, Ballaun C, Hermann M, VandeBerg JL, Sipos W, Uthman A, Fischer H, Tschachler E (2008) Identification of novel mammalian caspases reveals an important role of gene loss in shaping the human caspase repertoire. Mol Biol Evol 25(5):831–841
Ehrnhoefer DE, Sutton L, Hayden MR (2011) Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist 17(5):475–492
Elliott JM, Rouge L, Wiesmann C, Scheer JM (2009) Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation. J Biol Chem 284(10):6546–6553
Ellis HM, Horvitz HR (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44(6):817–829
Faustin B, Lartigue L, Bruey J-M, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 25(5):713–724
Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V (2012) Role of apoptosis in disease. Aging 4(5):330–349
Flemming W (1885) Ueber die Bildung von Richtungsfiguren in Säugethiereiern beim Untergang Graaf’scher Follikel. Arch Anat Entw Gesch (Veit & Comp). 221–244
Fuentes-Prior P, Salvesen GS (2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 384(Pt 2):201–232
Fulda S, Vucic D (2012) Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov 11(2):109–124
Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, Mehlen P, Nuñez G, Peter ME, Piacentini M, Rubinsztein DC, Shi Y, Simon H-U, Vandenabeele P, White E, Yuan J, Zhivotovsky B, Melino G, Kroemer G (2012) Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 19(1):107–120
Germain M, Affar EB, D’Amours D, Dixit VM, Salvesen GS, Poirier GG (1999) Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J Biol Chem 274(40):28379–28384
Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN (2005) c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 280(15):14507–14513
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125(6):1179–1191
Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA, Metzler M, Bissada N, Wang L, Faull RLM, Gray M, Yang XW, Raymond LA, Hayden MR (2010) Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci 30(45):15019–15029
Graham RK, Ehrnhoefer DE, Hayden MR (2011) Caspase-6 and neurodegeneration. Trends Neurosci 34(12):646–656
Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS (2011) RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol Cell 44(1):9–16
Grimsley C, Ravichandran KS (2003) Cues for apoptotic cell engulfment: eat-me, don’t eat-me and come-get-me signals. Trends Cell Biol 13(12):648–656
Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, Hayashi N, Higashino K, Okamura H, Nakanishi K, Kurimoto M, Tanimoto T, Flavell RA, Sato V, Harding MW, Livingston DJ, Su MS (1997) Activation of interferon-gamma inducing factor mediated by interleukin-1beta converting enzyme. Science 275(5297):206–209
Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR (1998) The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol 141(5):1097–1105
Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279(5350):509–514
Hanes J, Plückthun A (1997) In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A 94(10):4937–4942
Hardwick JM, Chen Y-b, Jonas EA (2012) Multipolar functions of BCL-2 proteins link energetics to apoptosis. Trends Cell Biol 22(6):318–328
Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA (2004) Discovery of an allosteric site in the caspases. Proc Natl Acad Sci U S A 101(34):12461–12466
Hengartner MO, Ellis RE, Horvitz HR (1992) Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356(6369):494–499
Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135(7):1311–1323
Hoste E, Denecker G, Gilbert B, Van Nieuwerburgh F, van der Fits L, Asselbergh B, De Rycke R, Hachem J-P, Deforce D, Prens EP, Vandenabeele P, Declercq W (2013) Caspase-14-deficient mice are more prone to the development of parakeratosis. J Invest Dermatol 133(3):742–750
Huang W-X, Huang P, Hillert J (2004) Increased expression of caspase-1 and interleukin-18 in peripheral blood mononuclear cells in patients with multiple sclerosis. Mult Scler 10(5):482–487
Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, Macfarlane M (2009) Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell 35(3):265–279
Javle M, Curtin NJ (2011) The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer 105(8):1114–1122
Jia SH, Parodo J, Kapus A, Rotstein OD, Marshall JC (2008) Dynamic regulation of neutrophil survival through tyrosine phosphorylation or dephosphorylation of caspase-8. J Biol Chem 283(9):5402–5413
Kamens J, Paskind M, Hugunin M, Talanian RV, Allen H, Banach D, Bump N, Hackett M, Johnston CG, Li P (1995) Identification and characterization of ICH-2, a novel member of the interleukin-1 beta-converting enzyme family of cysteine proteases. J Biol Chem 270(25):15250–15256
Kavuri SM, Geserick P, Berg D, Dimitrova DP, Feoktistova M, Siegmund D, Gollnick H, Neumann M, Wajant H, Leverkus M (2011) Cellular FLICE-inhibitory protein (cFLIP) isoforms block CD95- and TRAIL death receptor-induced gene induction irrespective of processing of caspase-8 or cFLIP in the death-inducing signaling complex. J Biol Chem 286(19):16631–16646
Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479(7371):117–121
Keller N, Mares J, Zerbe O, Grütter MG (2009) Structural and biochemical studies on procaspase-8: new insights on initiator caspase activation. Structure 17(3):438–448
Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26(4):239–257
Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P (2011) The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci 36(10):541–552
Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 14(22):5579–5588
Kitazumi I, Tsukahara M (2011) Regulation of DNA fragmentation: the role of caspases and phosphorylation. FEBS J 278(3):427–441
Koenig U, Eckhart L, Tschachler E (2001) Evidence that caspase-13 is not a human but a bovine gene. Biochem Biophys Res Commun 285(5):1150–1154
Krijnen PAJ, Sipkens JA, Molling JW, Rauwerda JA, Stehouwer CDA, Muller A, Paulus WJ, van Nieuw Amerongen GP, Hack CE, Verhoeven AJ, van Hinsbergh VWM, Niessen HWM (2010) Inhibition of Rho-ROCK signaling induces apoptotic and non-apoptotic PS exposure in cardiomyocytes via inhibition of flippase. J Mol Cell Cardiol 49(5):781–790
Krippner-Heidenreich A, Talanian RV, Sekul R, Kraft R, Thole H, Ottleben H, Lüscher B (2001) Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem J 358(Pt 3):705–715
Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S (2001) Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem 276(23):20633–20640
Kumar S, Doumanis J (2000) The fly caspases. Cell Death Differ 7(11):1039–1044
Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111(3):331–342
Lamkanfi M, Walle LV, Kanneganti T-D (2011) Deregulated inflammasome signaling in disease. Immunol Rev 243(1):163–173
Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA (2010) Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A 107(9):4230–4235
Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC (1994) Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371(6495):346–347
Lewis EM, Wilkinson AS, Davis NY, Horita DA, Wilkinson JC (2011) Nondegradative ubiquitination of apoptosis inducing factor (AIF) by X-linked inhibitor of apoptosis at a residue critical for AIF-mediated chromatin degradation. Biochemistry 50(51):11084–11096
Lüthi AU, Cullen SP, Mcneela EA, Duriez PJ, Afonina IS, Sheridan C, Brumatti G, Taylor RC, Kersse K, Vandenabeele P, Lavelle EC, Martin SJ (2009) Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity 31(1):84–98
Mandal D, Moitra PK, Saha S, Basu J (2002) Caspase 3 regulates phosphatidylserine externalization and phagocytosis of oxidatively stressed erythrocytes. FEBS Lett 513(2–3):184–188
Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430(6996):213–218
Martin DA, Siegel RM, Zheng L, Lenardo MJ (1998) Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem 273(8):4345–4349
Martin SJ, Henry CM, Cullen SP (2012) A perspective on mammalian caspases as positive and negative regulators of inflammation. Mol Cell 46(4):387–397
Martinon F, Tschopp J (2004) Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell 117(5):561–574
Martinon F, Tschopp J (2005) NLRs join TLRs as innate sensors of pathogens. Trends Immunol 26(8):447–454
Martinon F, Burns K, Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10(2):417–426
McStay GP, Salvesen GS, Green DR (2008) Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ 15(2):322–331
Meergans T, Hildebrandt AK, Horak D, Haenisch C, Wendel A (2000) The short prodomain influences caspase-3 activation in HeLa cells. Biochem J 349(Pt 1):135–140
Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A (2010) Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107(7):3076–3080
Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grütter MG (2002) The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 277(47):45162–45171
Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H (2003) Degradation of chromosomal DNA during apoptosis. Cell Death Differ 10(1):108–116
Oberst A, Dillon CP, Weinlich R, Mccormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471(7338):363–367
Olsson M, Zhivotovsky B (2011) Caspases and cancer. Cell Death Differ 18(9):1441–1449
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM (1997) The receptor for the cytotoxic ligand TRAIL. Science 276(5309):111–113
Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H (2007) Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128(3):533–546
Pop C, Timmer J, Sperandio S, Salvesen GS (2006) The apoptosome activates caspase-9 by dimerization. Mol Cell 22(2):269–275
Pop C, Fitzgerald P, Green DR, Salvesen GS (2007) Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 46(14):4398–4407
Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, Luo G, Li H, Wang J, Yan N, Shi Y (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141(3):446–457
Rao L, Perez D, White E (1996) Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol 135(6 Pt 1):1441–1455
Rathinam VAK, Vanaja SK, Fitzgerald KA (2012) Regulation of inflammasome signaling. Nat Immunol 13(4):333–342
Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40:D343–D350
Reubold TF, Wohlgemuth S, Eschenburg S (2011) Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure 19(8):1074–1083
Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, Salvesen GS, Bode W (2001a) Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci U S A 98(26):14790–14795
Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS (2001b) Structural basis for the inhibition of caspase-3 by XIAP. Cell 104(5):791–800
Rohn TT (2010) The role of caspases in Alzheimer’s disease; potential novel therapeutic opportunities. Apoptosis 15(11):1403–1409
Rohn TT, Vyas V, Hernandez-Estrada T, Nichol KE, Christie L-A, Head E (2008) Lack of pathology in a triple transgenic mouse model of Alzheimer’s disease after overexpression of the anti-apoptotic protein Bcl-2. J Neurosci 28(12):3051–3059
Roschitzki-Voser H, Schroeder T, Lenherr ED, Frölich F, Schweizer A, Donepudi M, Ganesan R, Mittl PRE, Baici A, Grütter MG (2012) Human caspases in vitro: expression, purification and kinetic characterization. Protein Expr Purif 84(2):236–246
Rubinsztein DC, Shpilka T, Elazar Z (2012) Mechanisms of autophagosome biogenesis. Curr Biol 22(1):R29–R34
Safa AR (2012) c-FLIP, a master anti-apoptotic regulator. Exp Oncol 34(3):176–184
Sahoo M, Ceballos-Olvera I, Del Barrio L, Re F (2011) Role of the inflammasome, IL-1β, and IL-18 in bacterial infections. Scientific World J 11:2037–2050
Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS, Buchman TG, Zehnbauer BA, Hayden MR, Farrer LA, Roy S, Nicholson DW (2004) Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature 429(6987):75–79
Salvesen GS, Dixit VM (1999) Caspase activation: the induced-proximity model. Proc Natl Acad Sci U S A 96(20):10964–10967
Scaffidi C, Schmitz I, Krammer PH, Peter ME (1999) The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 274(3):1541–1548
Schile AJ, García-Fernández M, Steller H (2008) Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 22(16):2256–2266
Schroeder T, Barandun J, Flütsch A, Briand C, Mittl PRE, Grütter MG (2013) Specific inhibition of caspase-3 by a competitive DARPin: molecular mimicry between native and designed inhibitors. Structure 21(2):277–289
Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Plückthun A, Grütter MG (2007) Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechanism. Structure 15(5):625–636
Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ (2009) The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 457(7232):1019–1022
Seeger MA, Mittal A, Velamakanni S, Hohl M, Schauer S, Salaa I, Grütter MG, Van Veen HW (2012) Tuning the drug efflux activity of an ABC transporter in vivo by in vitro selected DARPin binders. PLoS One 7(6):e37845
Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y (2003) Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11(2):519–527
Soares J, Lowe MM, Jarstfer MB (2011) The catalytic subunit of human telomerase is a unique caspase-6 and caspase-7 substrate. Biochemistry 50(42):9046–9055
Sollberger G, Strittmatter GE, Kistowska M, French LE, Beer H-D (2012) Caspase-4 is required for activation of inflammasomes. J Immunol 188(4):1992–2000
Steiner D, Forrer P, Plückthun A (2008) Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol 382(5):1211–1227
Steller H (2008) Regulation of apoptosis in Drosophila. Cell Death Differ 15(7):1132–1138
Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS (1999) Caspase-9 can be activated without proteolytic processing. J Biol Chem 274(13):8359–8362
Suzuki Y, Nakabayashi Y, Takahashi R (2001) Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A 98(15):8662–8667
Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schröter M, Scaffidi C, Krammer PH, Peter ME, Tschopp J (1997) Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386(6624):517–521
Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J (1992) A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356(6372):768–774
Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272(29):17907–17911
Timmer JC, Salvesen GS (2007) Caspase substrates. Cell Death Differ 14(1):66–72
Tinel A, Tschopp J (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304(5672):843–846
Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J (2007) Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J 26(1):197–208
Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV (2011) Metacaspases. Cell Death Differ 18(8):1279–1288
Vaughn DE, Rodriguez J, Lazebnik Y, Joshua-Tor L (1999) Crystal structure of Apaf-1 caspase recruitment domain: an alpha-helical Greek key fold for apoptotic signaling. J Mol Biol 293(3):439–447
Vaux DL, Silke J (2005) IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6(4):287–297
Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102(1):43–53
Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, Moritz RL, Simpson RJ, Vaux DL (2002) HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J Biol Chem 277(1):445–454
Vucic D, Franklin MC, Wallweber HJA, Das K, Eckelman BP, Shin H, Elliott LO, Kadkhodayan S, Deshayes K, Salvesen GS, Fairbrother WJ (2005) Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP. Biochem J 385(Pt 1):11–20
Vucic D, Dixit VM, Wertz IE (2011) Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol 12(7):439–452
Wachmann K, Pop C, Van Raam BJ, Drag M, Mace PD, Snipas SJ, Zmasek C, Schwarzenbacher R, Salvesen GS, Riedl SJ (2010) Activation and specificity of human caspase-10. Biochemistry 49(38):8307–8315
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, Rauch CT (1997) TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 16(17):5386–5397
Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, Robinson CV, Siegel RM, Walz T, Wu H (2010) The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol 17(11):1324–1329
Wilson KP, Black JA, Thomson JA, Kim EE, Griffith JP, Navia MA, Murcko MA, Chambers SP, Aldape RA, Raybuck SA (1994) Structure and mechanism of interleukin-1 beta converting enzyme. Nature 370(6487):270–275
Yang Z, Klionsky DJ (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335:1–32
Yin XM, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, Roth KA, Korsmeyer SJ (1999) Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400(6747):886–891
Yu JW, Jeffrey PD, Shi Y (2009) Mechanism of procaspase-8 activation by c-FLIPL. Proc Natl Acad Sci U S A 106(20):8169–8174
Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75(4):641–652
Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW (2010) Structure of an apoptosome-procaspase-9 CARD complex. Structure 18(5):571–583
Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW (2011) The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure 19(8):1084–1096
Zhang Y-w, Thompson R, Zhang H, Xu H (2011) APP processing in Alzheimer’s disease. Mol Brain 4:3
Zhang Y, Center DM, Wu DM, Cruikshank WW, Yuan J, Andrews DW, Kornfeld H (1998) Processing and activation of pro-interleukin-16 by caspase-3. J Biol Chem 273(2):1144–1149
Ziegler U, Groscurth P (2004) Morphological features of cell death. News Physiol Sci 19:124–128
Acknowledgement
This work was supported by the Swiss National Science Foundation grant 310030-122342 to Markus G. Grütter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Wien
About this chapter
Cite this chapter
Flütsch, A., Grütter, M.G. (2013). Proteases in Death Pathways. In: Brix, K., Stöcker, W. (eds) Proteases: Structure and Function. Springer, Vienna. https://doi.org/10.1007/978-3-7091-0885-7_8
Download citation
DOI: https://doi.org/10.1007/978-3-7091-0885-7_8
Published:
Publisher Name: Springer, Vienna
Print ISBN: 978-3-7091-0884-0
Online ISBN: 978-3-7091-0885-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)