Abstract
Aptamer and SELEX technology have been proposed over 20 years. Despite the huge success in developing aptamers against all kinds of targets, the effort to cover the natural shortages of nucleic acids, including lack of binding functional diversity and low information densities, has never been stopped. Strategies proposed to improve the aptamer properties include post-selective modifications and introducing unnatural nucleic acids in SELEX process. As a perfect binding ligand can hardly be designed and modified on purpose due to the poor understanding of the intricate biological system, the best way to generate improved aptamers would be through modified SELEX experiment, in which unnatural nucleotides are incorporated into library and perform the in vitro evolution. Those unnatural nucleotides include the modifications on almost every components of nucleic acids, (deoxy) ribose, phosphate linkage, and nucleobases. To increase the chemical diversity and the information density of nucleic acids, researchers developed methods to append functional groups as well as to create replicable expanded genetic systems. Some of these unnatural nucleic acids have now been utilized in SELEX, and a panel of improved aptamers has been delivered. In this book chapter, we mainly discuss the emerged unnatural bases which have been used in SELEX or at least have the potential to be used in SELEX, and how these unnatural nucleic acid help generate improved aptamers.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Unnatural nucleic acids
- Modified SELEX
- Artificial expanded genetic information system (AEGIS)
- Chemical diversity
- Information density
- Synthetic biology
3.1 General
It has been a quarter century since scientists proposed that binding ligands could be generated toward any targets of interest in the forms of nucleic acids [1–3]. The method proposed, termed systematic evolution of ligands by exponential enrichment (SELEX) [1] has been inspiring innumerable efforts committing to deliver aptamers against all kinds of target. Theoretically, with a simple recipe, the DNA or RNA aptamers could be delivered quickly, reliably, and inexpensively, toward any target researchers have in hand. It mimics the natural Darwinism (survival the fittest) that the best fitted nucleic acids will survive and be enriched during the evolution process in the exposure of the environment of targets.
Naturally, the emergence of this method aroused the comparison between nucleic acid aptamers and antibodies, one of the nature’s solutions for the generation of biomolecule binders and by far the most useful binding tools in biomedicine. Generally speaking, nucleic acid aptamers share some advantages of antibodies, including high specificity and binding affinity, generated by evolution process, enzymatic pathways available for amplification and degradation, as well as highly biocompatibility. Beyond that, nucleic acid aptamers possess some unique merits. For instance, it could be reproduced simply by chemical synthesis method, equipped with desired chemical modification, with little batch-to-batch variance. They are more likely a product that combining the power of ‘chemistry’ and power of ‘biology’ [4]. Indeed, considering the disadvantages of antibodies, including tedious generation process and intrinsic property of instability and immunogenicity, the nucleic acid aptamers and SELEX method were believed to be very competitive.
Despite the high enthusiasm at very beginning, the hope of aptamer rivaling antibodies [5] has never been fully realized. Aptamers have been generated toward a variety of targets ranging from small molecules [6], biomacromolecules [7, 8], to various cancer cells [9–12]. However, very few of these aptamers are now entering the clinical trial phases to be considered as drug leads, nor to be used as detection methods for in vivo application [13]. This fact has always been cited by the sceptics to question the practicability of aptamers.
Honestly, those queries are understandable. The capacity in combinatorial assembly has determined how far natural nucleic acids can go as a therapeutic or diagnostic component. Nucleic acids, after all, are built from merely four different nucleotide building blocks, which render it have therefore, compared to antibodies, much weaker power in terms of binding diversity [14–16]. To be specific, those ligand-receptor co-structure studies revealed that nucleic acid aptamers mainly employ hydrogen bonds, π–π stacking, and electrostatic interactions in the binding process, however, carries little of other key functional groups commonly found in protein, including hydrophobic groups (Leu and Ile), positively charged groups (Lys and Arg), negatively charged groups (Asp and Glu), polarizable binding groups (indole in Trp and thioether in Met), catalytic groups (His), and metal coordinating groups (Cys and His). From this point of view, nucleic acids were indeed far inferior to proteins as matrices for functional performance. Besides, the pharmacokinetic profile of aptamers is not satisfactory either. Particularly, it is embodied in short plasma half-life due to the nature of small molecule and enzyme vulnerability due to the self-maintaining mechanism of our bodies. Undoubtedly, all above chemical characteristics determined that application of nucleic acid aptamers would be largely limited.
Nevertheless, it would not be wise to throw away the apple because of the core. On the contrary, our scientists are smart enough to overcome those drawbacks by exploiting the chemical knowledge we have. The efforts of dealing with imperfection of aptamers started shortly after the discovery of aptamer, when people realizing the necessity and urgency to do so.
The first strategy is straightforward and feasible, which is usually referred to as ‘post-selective’ modification. This strategy focuses mainly on improving properties of the existing nucleic acid aptamer. For example, modifications on the ribose–phosphate backbone by introducing phosphorothioates [17] or locked nucleic acids (LNA) [18, 19], both of which are not accepted as substrates by the majority of nucleases, will help maintain stability when applied in vivo. Some other functional groups, those could help aptamers to carry ‘cargoes’ [20], to cross-linking with its target [21], or to gain improved pharmacokinetic properties [22], have been introduced into post-selective modification method in order to make ‘perfect’ aptamers. Nowadays, most of these modified phosphoramidites carrying amine, thiol, or carboxyl groups are commercially available, along with a variety of appendents which could be directly labeled on nucleic acid molecules.
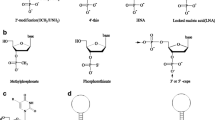
The most famous example of successfully utilizing post-selective modification strategies to improve aptamer properties is Pegaptanib (Macugen®, Fig. 3.1) [23]. It is the first and so far the only aptamer therapeutic approved by FDA for the treatment of wet form of age-related macular degeneration (AMD). It was delivered through SELEX against vascular endothelial growth factor (VEGF)-165 in the year of 1994 [24] and it was found that this aptamer can block the actions of its target VEGF. However, it took another 4 years to modify this aptamer to make it resistant to enzyme degradation and renal clearance [25]. Resulting from these modifications, the in vivo stability of nucleic acid aptamer has been largely improved, despite a certain degree of reduction in binding affinity for VEGF. The success of Pegaptanib sheds light on the whole aptamer research area and highlighted again the urgency and necessity of chemical modification on natural nucleic acids in aptamer research.
Chemical structure and predicted secondary structure of Pegaptanib. a Chemical structure of Pegaptanib. b Predicted secondary structure of Pegaptanib. High degree of modification including 2′-fluoro-pyrimidines (in blue) and 2′-methoxy-purines (in red) replaced nucleotides, capped 3′ end, additional two 20 kDa PEG groups via 5′-end coupling
However, those post-selective strategies could not cope with the essential drawback that the building blocks of nucleic acids cannot provide enough chemical diversity, therefore cannot guarantee to generate aptamers with desired affinity when confronting any target. The factors attributed by experimental work as well as theory to low performance of SELEX include the low level of functional groups in nucleic acid molecules and the low information density due to merely four building blocks [26]. Those issues have to be dealt with in the evolution process but not after. Accordingly, another strategy to modify the nucleic acid library in the SELEX experiment has been proposed to cover the shortages, which could be further divided into two methods.
On one hand, various functional groups were introduced onto four standard nucleotides in an attempt to obtain those functional nucleic acid molecules [27–31]. On the other hand, people attempted to increase the number of independently replicating nucleotides in the nucleic acid library, with the purpose of increasing information density carried by nucleic acid and the resultant folding possibility [32–34].
All these efforts of introducing new functionality or novel nucleotides into SELEX process followed similar principles, that is, the biotechnologies developed for SELEX. Three key technologies are involved which include the following: (1) chemical synthesis of phosphoramidites for solid-phase synthesis of nucleic acid and triphosphates for enzymatic amplification, (2) engineering and identifying polymerases to efficiently amplify nucleic acids library using standard four nucleotides plus unnatural nucleotide triphosphates, and (3) sequencing techniques to identify the survivor sequences containing unnatural nucleotides. It is well known that these technologies were originally developed only to natural nucleic acids. To apply those unnatural nucleic acids in SELEX procedure, these technologies need to be adjusted or even compromised to accept unnatural nucleic acids, with satisfactory efficiency and feasibility. From this angle, whether ideally designed unnatural nucleotides can be utilized in SELEX or not largely depends on whether corresponding key technologies could be well developed or not.
Synthesis of phosphoramidites and triphosphates is the first yet fundamental step. It is different from post-selective strategy where only modified phosphoramidites (most of them are commercially available) are needed. To implement those modified nucleic acid building blocks into SELEX process, not only unnatural nucleic acid phosphoramidites, but also the triphosphates, as a key component of recipe for enzymatic amplification, need to be prepared by organic synthetic method.
Maybe sounds a piece of cake for those masters in synthetic chemistry; however, the enzymes for amplification that has been evolved for billion years to accept only natural NTP are never easy to be satisfied. For those who is willing to add functional groups onto natural DNA, the inevasible enzymatic polymerization reactions for amplification involved in SELEX imposes the restriction that the modified NTP has to be perfectly compatible with the usually over 200 rounds of amplification during SELEX process. By mentioning compatible here, it means these functional groups added should not interfere with the recognition and binding sites of polymerase and should also match with the complementary nucleotides.
To prevent the classical base-pairing interface from disturbing, the modification are usually restricted on those sites which will not be involved in the base paring and polymerase binding process [4, 35] (Fig. 3.2). On nucleobases, the non-Watson-Crick sites, more specifically C5 of pyrimidines are usually the positions where modification proceeded on, which is in the major groove in duplex and will not interfere with approaching by polymerase [35, 36]. Besides, some modifications on ribose–phosphate backbone are also feasible, as long as it will not distort the whole DNA/RNA structure to a large degree. For example, ribose 2′-replacement with fluoro, amino, or methoxy group are commonly used generate enzyme resistent nucleic acid library, similarly in the example of Pegaptanib. Another famous example is displacing natural d-ribose with artificial l-ribose to generate the enantiomer of natural DNA (termed Spiegelmer) and thus prevent its degradation by nucleases. These examples will be detailed discussed in the later sections of this chapter.
For those who are dedicating to expand the armory of nucleotides, this concern is even bigger. Artificial designed nucleobases need to be both stable in duplex nucleic acid and replicable by natural existent polymerases. After the idea proposed by Benner in the late 1980s [37, 38], many efforts have been devoted into this area in light of the great potential of encoding additional information by expanded genetic alphabets in both in vitro and in vivo applications, and considerable achievements have been made since then. By adding additional base pair, the drawbacks of nucleic acids mentioned above, that is, lack of functionality and low information density, could be potentially solved at the same time. By far, two main types of approaches to develop such type of unnatural bases have been reported. The first, developed by Benner and coworkers [39], exploited unnatural hydrogen bond patterns between two complementary nucleobases, while maintaining the Watson–Crick geometry. Another one, proposed by Kool and colleagues [40, 41], that interbase hydrogen bonding is not imperative for nucleotide replication has broaden the way of thinking that focused more on van der Waals and hydrophobic interactions. The followers include Hirao [32, 33] and Romesberg [42, 43] and their coworkers have contributed tremendously in this field. The detailed story will also be discussed in later sections.
Since this is a book focusing on cell-SELEX-related technologies, in this chapter, we will mainly pay attention to introduce those unnatural nucleic acids involved strategies which have been used or at least have the potential to be used for SELEX. We will start with strategies that conducting modifications on ribose–phosphate backbone and their application in SELEX will be discussed, including the 2′-modified nucleotides [44], oxygen replacement on phosphate and Spiegelmers developed by Klussmann group [45]. Next, the strategies of adding functional groups to natural nucleobases, including hydrophobic groups, positive charged groups, and amino acid side chain mimicking groups, will be introduced. Following that we will discuss currently emerged replicable artificial nucleobase pairs and one application in SELEX done by Hirao and coworkers [33]. At the end, we will introduce so far the only full SELEX utilizing artificially expanded genetic information system (AEGIS) done by Benner, Tan, and their colleagues.
3.2 Evolution of Aptamers with Artificial Nucleic Acids
One key component of the recipe for success of SELEX technique is that this process highly mimics the natural evolution process. Those binders enriched in the selection process are exactly those survivors who can adjust oneself and adapt to the target environment. Similar to the reason why human being must maintain species diversity on earth, addition of chemical diversities into nucleic acid library will make this process even more lifelike.
Thanks to the solid fundamental established by organic chemists. Nowadays, human being is processing a huge toolbox with which we design and synthesize almost any chemical structure in need. Of course the ligands are not among these accessible chemical structures, not because of the lack of chemistry knowledge, but simply because the interaction of ligands with their receptors are too complicated for human being to interpret. However, the matured organic chemistry knowledge provides us with all kinds of strategies to modify natural biological molecules. With all this chemical modifications, we can now fulfill our goal of obtaining binders on demand, or even to the bigger picture we could ever imagined.
3.2.1 Modification on Nucleic Acid Backbone to Deliver Improved Aptamers
When people realized that aptamers made of natural nucleic acids have all sorts of drawbacks that severely hindered its wide application, the first solution emerged in mind would be appending functional groups on nucleic acid molecules. These ideas directly root in traditional chemical thought pattern that the function of chemicals could be improved by further chemical modification. If one has a glimpse history of drug discovery, you will find these patterns were applied to almost every type of chemical drugs people ever developed.
It indeed works. To cope with a specific problem emerged, there is always a genius method developed accordingly. That is exactly where the charm of chemistry lies on.
3.2.1.1 Modification on 2′ Site of Ribose
The earliest efforts to apply modified nuclei acids into SELEX were focusing on 2′ site on ribose. The purpose was to increase the nuclease resistance property of aptamers. The functional groups added onto 2′ site of ribose include amino [46], fluoro [47], and methoxyl group [44, 48] (Fig. 3.2). Those techniques were originally proved in post-selective modification, such as in the example of Pegaptanib. However, in order to acquire aptamers more rapidly and conveniently, meanwhile to avoid the potential affinity decreasing resulting from post-selective modification, researchers started to apply those modifications in the selection process. Y639F mutant T7 RNA polymerase was usually used for these modified RNA transcription [47]. 2′-amino pyrimidines, both cytidine (C) and uridine (U), have been used in RNA libraries for SELEX. A variety of targets have now been selected toward using these 2′-modified nucleic acid. These examples include basic fibroblast growth factor (bFGF) [49], L-selectin [50], human keratinocyte growth factor (hKGF) [51], human neutrophil elastase (HNE) [46], human thyroid stimulating hormone (hTSH) [52], immunoglobulin E (IgE) [53], and interferon-γ (IFN-γ) [54]. Usually the stability could be enhanced that lifetime could be prolonged by 1,000–80,000 times [49, 51].
2′-fluoro replacement method generated a couple of aptamers which has been entered clinical trial. Besides Pegaptanib, there are also anti-Factor IXa aptamer RB006 that binds to members of the coagulation cascade and act as anticoagulants [55] and anti-C5 aptamer which inhibits the complement cascade [56]. Again, 2′-fluoro U and 2′-fluoro C were exploited in library to select aptamers against KGF [51], IFN-γ [54], human thrombin [57], and cluster of differentiation 4 (CD4) [58]. However, someone may concern about its side effect as it was suggested that the administration and degradation of 2′-F-pyrimidine could lead to incorporation into cellular DNA [59, 60].
2′-methoxyl group might be a good alternative. The advantage of this method is that the synthesis is less expensive, and more importantly, the methoxyl group is naturally occurred in as a common moiety in posttranscriptional modification. Methylation of the 2′-hydroxyl groups is a selective protection system used by nature and thus introduced here to reduce the safety concerns. SELEXs using library containing this modification were conducted recently on VEGF-165 [44] and Interleukin 23 (IL-23) [48]. The affinities were high, with a Kd value within nanomolar range.
3.2.1.2 Replacing Oxygen on Phosphate
Another site which could be replaced to functionalize the nucleic acid library is on 5′-α-P-site on phosphate. The functional groups used to replace phosphate linkage this site include boranophosphate [61] and phosphorothioate [62] (Fig. 3.2). For boranophosphate, wild-type T7 RNA polymerase is good enough for the transcription. The initial purpose was to deliver aptamers for boron neutron capture therapy (BNCT), where boron-10 was used as a non-radioactive isotope that has a high propensity to capture slow neutrons to emit high-energy charge particles and result in the cell death. And aptamer would be a perfect probe to deliver this boron-10 to the cancer target. This concept was preliminarily demonstrated by performing a SELEX against ATP [61], with 5′-α-P-borano G or U in the library.
Phosphorothioate internucleotide linkage was utilized to generate nuclease resistance preferred to DNAse. Although the chiral property of phosphorothioate linkage is usually neglected and both diastereomers are used in the phosphoramidite chemistry synthesis, stereoregular phosphorothioate are usually prepared using α-thio-dNTPs in Taq polymerase amplification or T7 RNA polymerase transcription [35]. Examples include cytokine TGF-β1-targeted RNA aptamers [62] and Venezuelan equine encephalitis virus capsid protein [63].
3.2.1.3 Artificial Ribose
An alternative method is to use artificial ribose, to prevent aptamer from nuclease degradation. One way is to replace 4′-oxygen by sulfur on ribose (Fig. 3.2). 4′-thio UTP and CTP were used in library for SELEX to generate aptamers for thrombin [64]. It was claimed that by adding the 4′-thio, besides enhancement of the nuclease resistance by about 50 times, the base pair strength was found to be increased too.
Another heroic work was the generation of Spiegelmers done by Furste and his coworkers [45, 65, 66] (Fig. 3.2). The basic design of the work was based on the fact that all natural enzymes, no matter a polymerase or a nuclease, are all in l-amino acids and born to fit d-nucleosides. In this case, theoretically, an l-oligonucleotide should escape from enzymatic recognition and subsequent degradation. However, because of the same reason those l-oligonucleotides will not be accepted by polymerase so that it cannot be copied in the in vitro selection process. A compromised method was carried out by these smart scientists (Fig. 3.3). Instead of select against natural target, they used the mirror-image target and the natural oligonucleotides in the SELEX process. As long as the natural oligonucleotides targeting, mirror-image target was successfully obtained at the end of SELEX, an l-oligonucleotide would certainly binds to the natural target. And an unnatural enantiomer of aptamer, named Spiegelmer (from German word ‘Spiegel,’ means mirror), will thus be generated. Based on this technology, a number of spiegelmers have been selected [65–68].
Schematic of spiegelmer technology: mirror-image in vitro selection. Adapted from Eulberg and Klussmann [45]
3.2.2 Appending Functionality on Aptamers
On above, we discussed about modifications on (deoxy) ribose-sugar backbone and their applications in SELEX. However, the motivations were more on to enhance the in vivo stability, while contribute little on increasing diversity. The essential shortage of nucleic acid as a binding probe lies on its lack of diversified functional groups and thus relatively weak binding capability after combinatorial assembly of them. While the biding sites of aptamers are usually believed to be the unpaired nucleobases (compared with side chains of peptides). Introducing functional groups direct on nucleobases are, therefore, crucial to enlarge the diversity of library. However, as mentioned before, adding functional groups onto nucleic acid has to follow the principle, that is, this addition cannot interfere with the approach and functionalization of polymerase. This limitation largely reduces the possibility of introducing more functional groups. So far, the most permissive sites for modifications are 5-position on U/T, and a variety of functional groups have been successfully appended (Fig. 3.4).
3.2.2.1 Hydrophobic Groups
The earliest attempt using this strategy was done by Latham and his colleagues [69] (Fig. 3.4a). In their design, a 5-pentynyl-modified dU was used to displace dT in the randomized region of DNA library and this library was used to successfully generate aptamers against thrombin. Vent DNA polymerase was used in their experiment. The survivors at the end of the selection were cloned and sequenced using Sequenase to obtain the specific sequences of candidate aptamers. Because functional group added on C5 cite of pyrimidine, which does not interfere the function of polymerase, the PCR and sequencing were not influenced too much by the modification. Despite that this example was the first attempt to prove the feasibility of introducing a new group onto C5 site of pyrimidine, the affinity of their selected aptamers is not superior of their previous selected aptamers using natural DNA library [70].
3.2.2.2 Positively Charged Groups
Sooner after the success of introducing hydrophobic, another type of functional groups which is lack in natural nucleic acid, cationic group, was also being introduced into SELEX process [71]. In this work, Benner and his coworkers added an aminopropynyl group onto 5 sites of deoxyuridine (dJ, Fig. 3.4b), and as an example, the modified nucleotide were incorporated into library and performed a SELEX against ATP molecules. Vent DNA polymerase was used for PCR in this experiment. It was revealed that the selection yielded aptamer having sequences differing from the one generated from standard selection using natural nucleotides. However, convergence including both binding to two ATP molecules and very similar affinity were discovered. This work confirmed the capability to introduce functionality onto natural nucleic acid and use it for in vitro selection.
Another practice was carried out by Szostak and McLaughlin groups several years later [72]. In this work, a 5-(3-aminopropyl)uridine analogue was incorporated into a degenerate RNA library by enzymatic polymerization (UNH 2 , Fig. 3.4b). Again, this in vitro selection generated an aptamer binding to ATP molecules. And it was observed that the modified RNA can interact with the triphosphate group of ATP, which was not found in the case of natural RNA aptamer. This could further prove the strength of positive charged group.
This line of research continues to develop, and new positive charged nucleobase analogous kept emerging. Sawai and his colleagues successfully incorporated a 1,6-diaminohecyl-N-5-carbamoyl methyl deoxyuridine (dT HM, Fig. 3.4b) into DNA library to perform SELEX to generated aptamers against viral infection related cell surface component sialyllactose [73] as well as (R)-thalidomide [74]. Recently, Perrin group has expanded introduction of cationic groups onto other standard nucleobases, and those modified nucleotides are being applied in in vitro selection of DNAzyme [27, 75] (dU aa TP, dU ga TP, Fig. 3.4b).
3.2.2.3 Mimic of Amino Acid Side Chains
The idea of adding functional groups was initially motivated by presented diversity of amino acids. Recently, this strategy has been propelled one step further by endowing nucleic acids with protein-like properties via functional groups mimicking amino acid side chains [29]. Eaton and coworkers introduced an amide linkage at the 5 position so that six different side chains could be armed on deoxyuridine, including 5-isobutylaminocarbonyl-dU (iBudU), 5-benzylaminocarbonyl-dU (BndU), 5-histaminocarbonyl-dU (HisdU), 5-naphthylmethylaminocarbonyl-dU (NapdU), 5-tryptaminocarbonyl-dU (TrpdU), and 5-pyridylmethylaminocarbonyl-dU (PrdU) (Fig. 3.4c). Besides mimicking amino acids, other advantages of introducing amide linkage are to restrict rotation of the linkage bond and to provide with extra hydrogen bond acceptors and donors. After screening, all these modified deoxyuridines contained DNA were found to be fully replicated by D.Vent and KOD XL polymerase, which set the foundation for their usage in in vitro selection. Several of these modified deoxyuridines were incorporated into DNA library to perform in vitro selection against tumor necrosis factor receptor superfamily member 9 (TNFRSF9), which used to be considered as a challenging target. BndU and TrpdU incorporated selection generated aptamers having affinity much higher than natural DNA library where deoxythymidine replacing modified deoxyuridines [29].
This approach is now being commercially utilized in SomaLogic, Inc. to generate slow off-rate modified aptamers (SOMAmers), which could significantly increase the number of addressable targets of human proteins as well as the binding affinity [30]. Since the incorporation of four modified nucleotides, BndU, NapdU, TrpdU, and iBudU, into SELEX experiments, out of over 1,000 different proteins, the overall success rate rose from below 30 % to around 84 %, with aptamers having satisfactory affinities (Kd < 30 nM) and specificity. Exploiting those SOMAmers, an aptamer-based multiplexed proteomic technology for biomarker discovery, as well as for diagnostic and clinical applications, has been established in their company [28, 31].
3.2.3 Expanded Genetic Alphabets in Aptamer Selection
The effort of adding functional groups has been proved to be successful and productive. All those functional groups added into in vitro selection process indeed help generate aptamers with improved properties and fulfilled the goal in each initial design. However, there is another desire of human for nucleic acid which cannot be covered simply by introducing functional groups, that is, to increase the information density. It would be easier to understand if we transfer this description to chemical terminology. The desired larger chemical diversity could be achieved by introducing functional groups. However, the structural diversity, especially reflected in folding patterns, will not be increased by grafting stuff on one of the four basic building blocks. It is also highly possible that these nucleotides armed with functional groups will be folded into duplex structure and thus losing function, as stem region is usually believed not involved in the binding site. While adding additional nucleotides, also called expanded genetic alphabets, beyond A/G/C/T(U) will give much higher folding diversity and immediately minimize nonfunctioning folds that compete with the folds that have the desired function. Besides, those expanded genetic systems were usually designed to possess modifiable sites, which allow them to potentially carry functional groups if needed. Therefore, the two strategies could be perfectly combined helped by chemical synthesis method.
However, to create those expanded genetic alphabets is not as easy as to picture it. A successful design of additional nucleotide has to incorporate considerations from broad areas. At first, the additional nucleotide must be stable. Considering the nature of conjugated structures and multiple protonizable and oxidizable sites in aromatic rings, to design a nucleotide stable under different circumstances is definitely not an easy thing to do. Actually most of the expanded genetic systems developed so far have gone through repeating process of modify–test–modify to optimize the most stable structures [76]. Secondly, the designed nucleotides have to be able to be compatible with the enzyme system. Unlike simply adding functional group to replicable uredines, the new designed nucleotides have to be capable of being replicated by polymerase or being transcribed by transcriptase in the evolution process, which of course require not a single, but a pair of complementary nucleotides to be added at the same time. No doubt it took even more effort of scientists, again, to repeat the process of modify–test–modify to cater those enzymes which have been evolved for billions of years to accept only natural nucleotides [77, 78].
It is indeed the essence of synthetic biology, a discipline in which people use tool of synthetic chemistry to modify biological molecules to manipulate the biological systems, more importantly, in which process to understand deeper about biological systems [39].
3.2.3.1 Developed Artificial Genetic Systems
As mentioned above, it is extremely hard to build an artificial genetic system. It has been almost a quarter century since this idea been proposed; however, only several groups in the world have successfully developed artificial genetic systems [34, 77, 78]. Among them, only two examples of in vitro evolution have ever been reported [33, 79].
In 1989, Benner’s lab designed a pair of isomers of guanine and cytidine, named isoG and isoC (Fig. 3.5a), with nonstandard hydrogen-bonding patterns but similar geometry with Watson–Crick pairs [80]. And very soon, another pair (Χ and K, Fig. 3.5b) was reported too [37]. Although some shortcomings were found on these pairs, for example, lack of electron pairs in minor groove which were believed to be the polymerase recognition site and keto–enol tautomerization which could induce mismatching isoG with T(U), these pioneering studies inspired the subsequent studies to a great extent. And after 25 years of effort to keep improving, Benner’s group have now developed a whole system containing up to 12 different nucleotide ‘letters’ pair via six distinguishable hydrogen-bonding patterns [39, 76]. We will further introduce more details about this system in next section.
In 1998, Kool’s group proposed their classic non-hydrogen-bonded base pairs. Instead of hydrogen-bonding, they exploited shape analogues of the natural bases, Z(Q) and F (Fig. 3.5c, f), to be incorporated into the replicable nucleic acids [81, 82]. It was the first time researchers realized that the hydrogen bonding between nucleobases is not an absolutely requirement for replication. The role of shape complementarity and hydrophobicity in nucleic acid stability and replication has been illustrated in these works. The absence of hydrogen bonding will inevitably reduce the stability between DNA duplex; however, the pairing between artificial bases is still more stable than mispairing with natural bases; thus, thermal selectivity could be achieved. Similarly, this work inspired a lot of subsequent researches. After all, the strategy of rearrangement of hydrogen bonds between nucleobases could easily reach its limit, that is, maximum 12 bases could be conceived following this rule, and not all of them could be replicated efficiently by enzymes [39].
Hirao, Yokoyama, and their coworkers started from the nonstandard hydrogen-bonding strategy at first, developing x and y (Fig. 3.5d) as a complementary and replicable pair [83]. A serial of modifications and accommodations were conducted in order to increase the duplex stability and incorporation efficiency, generating base pair of s and z (Fig. 3.5e), however end up without satisfactory results [84, 85]. Then, they shifted attention to the strategy of exploiting hydrophobicity and stacking interaction, by designing new base (Pa, Fig. 3.5f) to be paired with Q base developed by Kool’s group [86]. And coincidently, they found Pa could be paired with s also, which pair has higher efficiency compared to the precedent s-z pair [87, 88]. After another several years of unremitting efforts, the structures were optimized to generate the pair of Ds and Px (Fig. 3.5g), which was reported that they could be incorporated by Deep Vent DNA polymerase (exo+) with selectivity of over 99.9 % [89].
Romesburg and colleagues also developed their expanded genetic systems. In 1999, they reported the hydrophobic self-paired PICS base (Fig. 3.5h), which was believed to be as stable as G:C pair in duplex DNA [90, 91]. Similarly, due to their hydrophobic stacking ability, this self-paired base could be replicated by KF exo− polymerase with reasonable efficiency. Another four self-paired bases (3MN [91, 92], SNICS [93], 7AI [94], and 3FB [95], Fig. 3.5i–l) were also developed with incorporation efficiency to a certain extent.
Then, they shifted the way of thinking, and a combinatorial chemistry method was utilized to screen nucleobase pairs for efficient replication [96]. Consequently, following the rule of hydrophobicity, more than 60 base analogues were synthesized and screened for KF exo− extension. A pair of nucleobases, SICS and MMO2 (Fig. 3.5m), who exhibited the best efficiency and selectivity, was successfully obtained. On basis of structure of MMO2, the 5SICS-NaM and 5SICS-DMD (Fig. 3.5n, o) pairs were finally developed [97, 98]. In a newly published work, they applied the 5SICS-NaM pair into the E.coli, proving that the replication could be efficiently achieved in biological system [42].
3.2.3.2 First Example of In Vitro Selection Using Expanded Genetic Alphabets
It was not until 2013 did researches first report utilizing expanded genetic systems for in vitro selection [33]. Hirao and his colleagues have incorporated his dDs deoxynucleoside into the DNA library and performed the SELEX experiment to generated aptamers targeting VEGF-165 and IFN-γ with better affinity than what people developed using natural RNA.
Due to the lack of deep sequencing technology for their artificial nucleobase, there were some modifications on most of these SELEX protocol (Fig. 3.6). At first, for each randomized region, only 1-3 Ds could be introduced, and it has to be on some predetermined position, and a short tag sequence will be appended. Secondly, the Ds has to be allocated several nucleotides away instead of consecutively or closely in consideration of polymerization efficiency. Thirdly, in the PCR amplification, besides natural dNTPs and dDsTP, triphosphate of Px pairing with Ds also needs to be added into the reaction to amplify the Ds contained template. And finally, after enrichment of those Ds contained DNA, a replacement PCR needed to be run to switch all Ds into either A or T, and after deep sequencing the Ds contained sequences will be inferred based on the tag information.
Schematic of in vitro selection using Ds as the fifth nucleotide. Adapted from Kimoto et al. [33]
In spite of its success as the first ever use expanded genetic alphabets to do in vitro selection, this method still have lots of room to be improved. As mentioned before, to successfully exploit an unnatural DNA into SELEX experiment, a delicate deep sequencing method should have been established beforehand. To put the artificial base on predetermined position will undoubtedly reduce the randomization effect of the library and thus make the mimicking evolution process less powerful. Besides, only adding one nucleobase into the library cannot help much about the increase of the information density, it is more like an extension of those adding functional group strategies.
However, this example still shows us a lot of potential and sheds light on the future of applying expanded genetic alphabets. After all, the survivors after over 100 round of PCR can still maintain the expanded genetic alphabets. And the high affinity brought by introducing the well-designed fifth ‘letter’ largely demonstrated what expanded genetic systems can do and how to do it. This work proved the power of synthetic biology and also contributed hugely on the development of SELEX technique.
3.3 AEGIS System and Application in Cell-SELEX
Hirao and his colleagues’ work is exciting and it is definitely a milestone in the development of expanded genetic systems. However, they can hardly be called as a complete SELEX. The deep sequencing technology needs to be well developed; the Ds and/or Px have to be proved to be sustained in closer distance in the evolution process; and only if both complementary bases are added into library could it truly improve SELEX, in terms of increasing information density and chemical diversity.
To date, there is only one pair of expanded nucleotides toward which a deep sequencing method has been reported, along with the solid synthesis technology and PCR amplification technology. This pair is two members of AEGIS developed by Benner and his coworkers [39, 76]. And our collaborative team has been utilizing this system in our cell-SELEX practice and generated a panel of DNA aptamers binding to several kinds of cancer cells. In this section, we will specifically introduce AEGIS system and its application in cell-SELEX. One of our recent work will be presented as an example.
3.3.1 Artificial Expanded Genetic Information Systems
The development of AEGIS was inspired by recognizing that two natural nucleobase pairs (G:C and A:T or U) have not fully exploited all possible hydrogen-bonding patterns (Fig. 3.7). To rearrange hydrogen bond donors and acceptors between nucleotide bases with the help of synthetic chemistry can increase the number of independently replicable nucleosides, from 4 to at most 12 [76]. The hydrogen-bonding pattern assigned to each nucleobase pair is unique and distinguishable that allow the 12 bases to form a system to support the basic DNA properties, that is, to be complemented, to be copied, and, more importantly, to be evolved. At the same time by introducing those much of unnatural nucleotides, the information density will be increased dramatically [39]. We mentioned in last section that the isoG and isoC pair, as well as Χ and K pair developed by Benner group are the prototypes of the current members of AEGIS system. And no doubtfully this system will be kept improving to meet the potential different requirements. As it developed, now the second generation of AEGIS system has overcome these potential tautomerization problems and can carry additional functional groups on those unnatural nucleotides (R group shown in Fig. 3.7). This is to combine the strengths of strategy of adding functionality and the strategy of introducing new nucleotides. AEGIS are now being applied as orthogonal binding elements for human disease diagnosis, including FDA-approved assays for HIV, hepatitis B, and hepatitis C viruses [99].
Among the AEGIS system members, a pair of nucleotides, (2-amino-8-(1′-β-D-2-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)one, trivially known as P, and 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone, trivially known as Z) [100, 101] (Fig. 3.7), were found to be highly suitable for being incorporated into cell-SELEX experiment.
3.3.2 AEGIS Cell-SELEX
The confidence of incorporating Z and P into cell-SELEX experiment attributes to the successful development of a series of molecular biology technologies: (1) synthesis of DNA libraries containing Z and P nucleotides together with four natural nucleotides [100], (2) highly efficient PCR amplification of DNA sequences containing Z and P [102], and (3) deep sequencing of DNA molecule survivors after the evolution process [103]. As mentioned in previous sections, these three key technologies determine whether a designed expanded genetic system can be used for in vitro evolution or not. Based on those fundamental technologies, we have tried multiple AEGIS cell-SELEX on different target cells. In this book, we will cite one of these examples to illustrate the SELEX process [79].
3.3.2.1 Design and Synthesis of AEGIS DNA Primers and Library
The purpose of cell-AEGIS-SELEX is to screen six-letter DNA sequences capable of recognizing and binding to the target cells in the natural state. To achieve this goal, two 16-mer primers were designed with the following principles: unlikely to form intramolecular hairpin structure, similar melting temperature (Tm) between two primers, and unlikely to form neither self-dimer nor hetero-dimer. No artificial bases were included in the primer part. The forward primer was labeled with Fluorescein isothiocyanate (FITC) at the 5′ end, and the reverse primer was labeled with biotin at the 5′ end.
A GACTZP DNA library having a 20-nucleotide random region flanked by two primer binding segments (each 16 nt) was prepared by solid-phase phosphoramidite DNA synthesis. Each of the 20 randomized sites was synthesized to have all six (GACTZP) phosphoramidites in equal amounts (Table 3.1). The presence of Z and P in the random region was confirmed by digestion of the GACTZP library, with the nucleotide fragments being quantitated by HPLC.
We could do some simple calculations to show the diversity increased by adding new nucleotides. If only four natural nucleotides were involved in 20-nt-long randomized region, the number of all possible sequence would be 420 (1.099 × 1012). If two more nucleotides added, this number will increases to 620 (3.656 × 1015), which is 3,000 times more possibilities. When we use longer randomized region, this ratio could be even larger.
3.3.2.2 AEGIS Cell-SELEX Generates GACTZP Aptamers
The procedure for AEGIS cell-SELEX is shown schematically in Fig. 3.8. As a proof of concept, we did not add the counter-selection in this experiment. The aptamers were expected to be generated faster but with less specificity. A breast cancer cell line MDA-MB-231 (from ATCC) was used as the target cell for the positive selection.
Schematic of AEGIS cell-SELEX process. Reprinted from Ref. [79] by permission of PNAS
In the in vitro selection experiment, an aliquot of the six-letter ssDNA library (20 nmol) was first denatured by heating at 85 °C (different from standard SELEX using 95 °C, to prevent unnatural nucleotides from damage). It was then ‘snap cooled’ to force the DNA sequences to form their kinetically most accessible secondary structures, incubated with target cells and those binding sequences will be extracted and amplified. These steps followed the protocol of standard cell-SELEX [104].
The enriched GACTZP were amplified by six-nucleotide PCR using Taq polymerase [102, 103] (Table 3.2). Note that we used much larger amount of Taq polymerase and much longer elongation time compared to what were used in standard PCR protocol. The polymerase will amplify those standard DNA nucleotides much faster than unnatural ones, larger concentration of polymerase, and longer elongation time will allow polymerase to sufficiently amplify unnatural DNA.
The rest of the selection process is similar to what we used in the standard protocol. A total of 12 rounds of selection were performed. The stringency of the selection was increased in later rounds by decreasing the number cells and the incubation times. Starting from the 9th round, the progress of the selection was monitored by monitoring the ability of the FITC labeled library to fluorescently label individual cells (Fig. 3.9).
Monitoring the progress of GACTZP AEGIS cell–SELEX using flow cytometer. Reprinted from Ref. [79] by permission of PNAS
It is interesting to find out that the fluorescence intensity keep increasing from rounds 9 to 11 rounds, however back shift in round 12. We assume the reason is dZ-and dP-contained DNA were kept losing in the PCR process, although they were supposed to be enriched in the selection process. The selection was then stopped at round 12, and the library from round 11 was prepared for deep sequencing.
3.3.2.3 Deep Sequencing of GACTZP Survivors Using NextGen Sequencing Technology
One of the most advantages that AEGIS has over other artificial genetic system is the well-developed deep sequencing technology. The emerging next-generation sequencing technology provided the chance to identify every single sequence in the enriched pool, avoiding miss any potential aptamer candidate and also avoiding the complicated cloning operation. It is indeed a huge improvement to develop corresponding deep sequencing technology for Z and P pair, which distinguishes it from others to be used in the in vitro selection process.
We will introduce here briefly about this method. It wisely exploits the strategy that the Z and P could be directionally controlled to be converted into corresponding natural bases [103]. After conversion, the common deep sequencing methods could be used for the sequencing. Then, the ancestral sequences will be inferred based on the conversion information used. To assure the accuracy of the sequencing, two of the conversion protocols were established and they are usually conducted in parallel (Fig. 3.9). Specifically, in the first protocol, sites holding Z and P nucleotides in the GACTZP survivors were converted predominantly into sites holding C and G nucleotides, respectively; less than 15 % were other nucleotides. Under the second conversion protocol, sites holding Z were converted to sites holding a mixture of C and T, with their ratio lying between 60:40 and 40:60, depending on the sequence surrounding that site. Sites holding P is converted to a mixture of G and A with roughly the same range of ratios, again depending on the sequence context surrounding that site.
In this specific SELEX, survivors enriched after 12 rounds of AEGIS-SELEX were divided into two equal portions. These were separately converted by barcoded copying into standard DNA using two conversion protocols. Following conversion, two barcoded samples were combined and submitted for Ion Torrent ‘next generation’ sequencing. A software was coded to cluster sequenced DNA and infer the ancestral sequences contained Z and/or P based on the information of conversion strategy. The clustered sequences obtained under the first conversion conditions (Z–C and P–G) serve as reference for the clustered sequences obtained under the second conversion conditions. Sites where C and T were found in approximately equal amounts after conversion under the second conditions were assigned as Z in their ‘parent.’ Sites where G and A were found in approximately equal amounts after conversion under the second conditions were assigned as P in their ‘parent.’ The inferred ancestral sequences were aligned to identify candidate aptamers.
Honestly, this is not a straightforward method as it took complicated steps to do these conversions and even need the help of computer software to run the inference. However, using this deep sequencing method does help to truly apply expanded genetic systems in the selection process. Compared to Hirao’s work mentioned in previous section, now the artificial bases do not have to be put in predetermined positions. This allowed the maximum chemical diversity to be achieved, as well as the pairing of nucleotides to form more complex secondary structures.
3.3.2.4 Characterization of Selected Aptamer
In this specific work, according to the alignment result, the most enriched sequence has around 30 % of populations in the whole pool (ZAP-2012: 5′-TCC CGA GTG ACG CAG CCC CCG GZG GGA TTP ATC GGT GGA CAC GGT GGC TGA C-3′). And this very sequence has very strong binding signal on flow cytometry as well as satisfactory affinity according to the low disassociation constant. As expected, it does not have very good specificity as only positive selection was conducted.
In this aptamer we selected, only one Z and one P are contained. Given the six nucleotides were synthesized equally in amount in the randomized region, it might arouse the skepticism that Z and P play less important role in the binding process. However, if think about over 200 rounds of PCR performed in the selection process, using the polymerase which has been evolved for billions of years to accept natural nucleobases, this results should be quite gratifying. Those DNA containing only GCAT will be enriched much faster than DNA containing full six letters. And Z and P tend to keep being lost in the amplification process, resulting in the rare Z and P present in the winning aptamers.
To show that the AEGIS nucleotides were essential for binding in aptamers, we tried to synthesize those analogs where Z and P are replaced by natural nucleotides. It turned out that in all cases, the binding abilities were largely diminished (Fig. 3.10). We still do not fully understand the role that Z and P are playing in the aptamer binding process; however, at least we have the clue that they are indispensable (Fig. 3.11).
Binding of analogs of aptamer ZAP-2012 with Z and P replaced by standard nucleotides. Reprinted from Ref. [79] by permission of PNAS
3.3.2.5 Other SELEXs Using AEGIS Nucleotides
This work is the first example of AEGIS cell-SELEX. It is actually first experiment ever to perform a complete in vitro selection using artificial bases exactly following the principle of SELEX. Even though the aptamer we obtained from this work might not have broad application due to its lack of specificity, it proved the concept. We have used this strategy to successfully overcome some very challenging targets. For example, we generated aptamers binding to liver cancer cells, lung cancer cells, and several proteins (data not published). Unlike in this work, only one aptamer carrying only one Z and one P is recovered; in other cases, we have obtained those aptamers containing 1–4 Z and/or P, with some of them very close or even adjacent to each other. This method is now becoming a routine method in aptamer generation and could be applied for a wider range of targets.
3.4 Perspective
Modified library and corresponding SELEX have been utilized effectively to generate aptamers with high affinity and specificity for almost two decades. The natural shortages of natural DNA have been largely covered by chemical method. And aptamers with endowed stability, functionality, and diversity are kept being generated using those modified libraries. With all these successful examples of modification achieved by our brilliant scientists, it seems researchers have sufficient reasons to be proud of what we have done. However, our understanding of aptamers and their interactions with their targets are far away from maturation.
Conversely, the studies of unnatural DNA and its application are still on the stage of infant. One can imagine the final goal of researchers is to find a way to generate the aptamers ‘on demand.’ That means, whenever people need an aptamers binding to whatever target, there is always a method to generate such aptamers in a quick and efficient, maybe also economic way. Those targets could include tumor sample of individual patient, a mutant Ebola virus, or a new emerged water pollutant. Of course, those aptamers on demand cannot be designed, at least not before human fully understanding every single detail of biological systems. The in vitro selection might be still the only method to generate those aptamers in a long period in future.
The direction and requirements for the development of SELEX are clear. Whether those requirements can be fully fulfilled by effort of chemists is still worth exploring. On one hand, we keep trying to improve our unnatural DNA to accommodate the practical requirement. On the other hand, which is perhaps more important, in the process of improving and testing our modification, the understanding of biology is accumulating. This is the true genius of synthetic biology, which is a subject that requires understanding of biology to synthetically improve or even mimic it and at the same time using synthetic method to understand more about biology [39, 105].
Particularly in the synthetic of unnatural DNA, what we need is larger diversity, more efficient enzyme incorporation, and detail understanding of mechanism of how these unnatural DNAs interact with targets. Obviously, these future goals have to be achieved by involvement of not only organic chemist, but also collaboration of molecular biologists, physical chemists, microbiologist, and a lot of specialist from all other areas. Only if all of these happen, could the aptamers truly rival antibodies and serve better for human community.
References
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818–822. doi:10.1038/346818a0
Robertson DL, Joyce GF (1990) Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344(6265):467–468. doi:10.1038/344467a0
Tolle F, Mayer G (2013) Dressed for success—applying chemistry to modulate aptamer functionality. Chem Sci 4(1):60–67. doi:10.1039/C2sc21510a
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45(9):1628–1650
Famulok M (1999) Oligonucleotide aptamers that recognize small molecules. Curr Opin Struct Biol 9(3):324–329. doi:10.1016/S0959-440x(99)80043-8
Gopinath SC (2007) Methods developed for SELEX. Anal Bioanal Chem 387(1):171–182. doi:10.1007/s00216-006-0826-2
Sun W, Du L, Li M (2010) Aptamer-based carbohydrate recognition. Curr Pharm Des 16(20):2269–2278
Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W (2006) Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA 103(32):11838–11843. doi:10.1073/pnas.0602615103
Sefah K, Bae KM, Phillips JA, Siemann DW, Su Z, McClellan S, Vieweg J, Tan W (2013) Cell-based selection provides novel molecular probes for cancer stem cells. Int J Cancer 132(11):2578–2588. doi:10.1002/ijc.27936
Van Simaeys D, Turek D, Champanhac C, Vaizer J, Sefah K, Zhen J, Sutphen R, Tan W (2014) Identification of cell membrane protein stress-induced phosphoprotein 1 as a potential ovarian cancer biomarker using aptamers selected by cell systematic evolution of ligands by exponential enrichment. Anal Chem 86(9):4521–4527. doi:10.1021/ac500466x
Jimenez E, Sefah K, Lopez-Colon D, Van Simaeys D, Chen HW, Tockman MS, Tan W (2012) Generation of lung adenocarcinoma DNA aptamers for cancer studies. PLoS ONE 7(10):e46222. doi:10.1371/journal.pone.0046222
Mitchell P, Annemans L, White R, Gallagher M, Thomas S (2011) Cost effectiveness of treatments for wet age-related macular degeneration. Pharmacoeconomics 29(2):107–131. doi:10.2165/11585520-000000000-00000
Hamula CLA, Guthrie JW, Zhang HQ, Li XF, Le XC (2006) Selection and analytical applications of aptamers. Trac-Trends Anal Chem 25(7):681–691. doi:10.1016/j.trac.2006.05.007
Li N, Ebright JN, Stovall GM, Chen X, Nguyen HH, Singh A, Syrett A, Ellington AD (2009) Technical and biological issues relevant to cell typing with aptamers. J Proteome Res 8(5):2438–2448. doi:10.1021/Pr801048z
Proske D, Blank M, Buhmann R, Resch A (2005) Aptamers—basic research, drug development, and clinical applications. Appl Microbiol Biotechnol 69(4):367–374. doi:10.1007/s00253-005-0193-5
King DJ, Ventura DA, Brasier AR, Gorenstein DG (1998) Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry 37(47):16489–16493. doi:10.1021/bi981780f
Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J (1998) Locked Nucleic Acids (LNA): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54(14):3607–3630. doi:10.1016/S0040-4020(98)00094-5
Koshkin AA, Rajwanshi VK, Wengel J (1998) Novel convenient syntheses of LNA [2.2.1]bicyclo nucleosides. Tetrahedron Lett 39(24):4381–4384. doi:10.1016/S0040-4039(98)00706-0
Wang RW, Zhu GZ, Mei L, Xie Y, Ma HB, Ye M, Qing FL, Tan WH (2014) Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J Am Chem Soc 136(7):2731–2734. doi:10.1021/Ja4117395
Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W (2007) Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol Cell Proteomics 6(12):2230–2238. doi:10.1074/mcp.M700026-MCP200
Keefe AD, Pai S, Ellington A (2010) Aptamers as therapeutics. Nat Rev Drug Discovery 9(7):537–550. doi:10.1038/nrd3141
Ng EW, Shima DT, Calias P, Cunningham ET Jr, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discovery 5(2):123–132. doi:10.1038/nrd1955
Jellinek D, Green LS, Bell C, Janjic N (1994) Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry 33(34):10450–10456
Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N (1998) 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 273(32):20556–20567
Carrigan MA, Ricardo A, Ang DN, Benner SA (2004) Quantitative analysis of a RNA-cleaving DNA catalyst obtained via in vitro selection. Biochemistry 43(36):11446–11459. doi:10.1021/Bi049898l
Hollenstein M, Hipolito CJ, Lam CH, Perrin DM (2009) A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M-2). Nucleic Acids Res 37(5):1638–1649. doi:10.1093/Nar/Gkn1070
Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Kim N, Saccomano NA, Wilcox SK, Zichi D, Sanders GM (2011) From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE 6(10):e26332. doi:10.1371/journal.pone.0026332
Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton BE (2010) Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc 132(12):4141–4151. doi:10.1021/ja908035g
Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic J, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D (2010) Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5(12):e15004. doi:10.1371/journal.pone.0015004
Zichi D, Eaton B, Singer B, Gold L (2008) Proteomics and diagnostics: let’s get specific, again. Curr Opin Chem Biol 12(1):78–85. doi:10.1016/j.cbpa.2008.01.016
Kimoto M, Cox RS 3rd, Hirao I (2011) Unnatural base pair systems for sensing and diagnostic applications. Expert Rev Mol Diagn 11(3):321–331. doi:10.1586/erm.11.5
Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I (2013) Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol 31(5):453–457. doi:10.1038/nbt.2556
Henry AA, Romesberg FE (2003) Beyond A, C, G and T: augmenting nature’s alphabet. Curr Opin Chem Biol 7(6):727–733
Keefe AD, Cload ST (2008) SELEX with modified nucleotides. Curr Opin Chem Biol 12(4):448–456. doi:10.1016/j.cbpa.2008.06.028
Kuwahara M, Sugimoto N (2010) Molecular evolution of functional nucleic acids with chemical modifications. Molecules 15(8):5423–5444. doi:10.3390/molecules15085423
Piccirilli JA, Krauch T, Moroney SE, Benner SA (1990) Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature 343(6253):33–37. doi:10.1038/343033a0
Benner SA, Allemann RK, Ellington AD, Ge L, Glasfeld A, Leanz GF, Krauch T, MacPherson LJ, Moroney S, Piccirilli JA et al (1987) Natural selection, protein engineering, and the last riboorganism: rational model building in biochemistry. Cold Spring Harb Symp Quant Biol 52:53–63
Benner SA, Yang ZY, Chen F (2011) Synthetic biology, tinkering biology, and artificial biology: what are we learning? C R Chim 14(4):372–387. doi:10.1016/j.crci.2010.06.013
Moran S, Ren RX, Kool ET (1997) A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA 94(20):10506–10511
Moran S, Ren RX, Rumney S, Kool ET (1997) Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J Am Chem Soc 119(8):2056–2057. doi:10.1021/ja963718g
Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR Jr, Romesberg FE (2014) A semi-synthetic organism with an expanded genetic alphabet. Nature 509(7500):385–388. doi:10.1038/nature13314
Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A (2012) KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry. Nat Chem Biol 8(7):612–614. doi:10.1038/Nchembio.966
Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD (2005) Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol 12(1):25–33. doi:10.1016/j.chembiol.2004.10.017
Eulberg D, Klussmann S (2003) Spiegelmers: biostable aptamers. ChemBioChem 4(10):979–983. doi:10.1002/cbic.200300663
Lin Y, Qiu Q, Gill SC, Jayasena SD (1994) Modified RNA sequence pools for in-vitro selection. Nucleic Acids Res 22(24):5229–5234. doi:10.1093/nar/22.24.5229
Huang Y, Eckstein F, Padilla R, Sousa R (1997) Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry 36(27):8231–8242. doi:10.1021/bi962674l
Burmeister PE, Wang C, Killough JR, Lewis SD, Horwitz LR, Ferguson A, Thompson KM, Pendergrast PS, McCauley TG, Kurz M, Diener J, Cload ST, Wilson C, Keefe AD (2006) 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 16(4):337–351. doi:10.1089/oli.2006.16.337
Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DP, Abesinghe P, Pieken WA et al (1995) Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34(36):11363–11372
O’Connell D, Koenig A, Jennings S, Hicke B, Han HL, Fitzwater T, Chang YF, Varki N, Parma D, Varki A (1996) Calcium-dependent oligonucleotide antagonists specific for L-selectin. Proc Natl Acad Sci USA 93(12):5883–5887
Pagratis NC, Bell C, Chang YF, Jennings S, Fitzwater T, Jellinek D, Dang C (1997) Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat Biotechnol 15(1):68–73. doi:10.1038/nbt0197-68
Lin Y, Nieuwlandt D, Magallanez A, Feistner B, Jayasena SD (1996) High-affinity and specific recognition of human thyroid stimulating hormone (hTSH) by in vitro-selected 2′-amino-modified RNA. Nucleic Acids Res 24(17):3407–3414
Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D (1996) High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J Immunol 157(1):221–230
Kubik MF, Bell C, Fitzwater T, Watson SR, Tasset DM (1997) Isolation and characterization of 2′-fluoro-, 2′-amino-, and 2′-fluoro-/amino-modified RNA ligands to human IFN-gamma that inhibit receptor binding. J Immunol 159(1):259–267
Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA (2002) RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419(6902):90–94. doi:10.1038/nature00963
Biesecker G, Dihel L, Enney K, Bendele RA (1999) Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 42(1–3):219–230
White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B (2001) Generation of species cross-reactive aptamers using “toggle” SELEX. Mol Ther J Am Soc Genet Ther 4(6):567–573. doi:10.1006/mthe.2001.0495
Davis KA, Lin Y, Abrams B, Jayasena SD (1998) Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res 26(17):3915–3924
Richardson FC, Zhang C, Lehrman SR, Koc H, Swenberg JA, Richardson KA, Bendele RA (2002) Quantification of 2′-fluoro-2′-deoxyuridine and 2′-fluoro-2′-deoxycytidine in DNA and RNA isolated from rats and woodchucks using LC/MS/MS. Chem Res Toxicol 15(7):922–926
Richardson FC, Tennant BC, Meyer DJ, Richardson KA, Mann PC, McGinty GR, Wolf JL, Zack PM, Bendele RA (1999) An evaluation of the toxicities of 2′-fluorouridine and 2′-fluorocytidine-HCl in F344 rats and woodchucks (Marmota monax). Toxicol Pathol 27(6):607–617
Lato SM, Ozerova ND, He K, Sergueeva Z, Shaw BR, Burke DH (2002) Boron-containing aptamers to ATP. Nucleic Acids Res 30(6):1401–1407
Kang J, Lee MS, Copland JA 3rd, Luxon BA, Gorenstein DG (2008) Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-beta1 protein. Bioorg Med Chem Lett 18(6):1835–1839. doi:10.1016/j.bmcl.2008.02.023
Kang J, Lee MS, Watowich SJ, Gorenstein DG (2007) Combinatorial selection of a RNA thioaptamer that binds to venezuelan equine encephalitis virus capsid protein. FEBS Lett 581(13):2497–2502. doi:10.1016/j.febslet.2007.04.072
Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, Matsuda A (2005) New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res 33(9):2942–2951. doi:10.1093/nar/gki578
Klussmann S, Nolte A, Bald R, Erdmann VA, Furste JP (1996) Mirror-image RNA that binds D-adenosine. Nat Biotechnol 14(9):1112–1115. doi:10.1038/nbt0996-1112
Nolte A, Klussmann S, Bald R, Erdmann VA, Furste JP (1996) Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat Biotechnol 14(9):1116–1119. doi:10.1038/nbt0996-1116
Szeitner Z, Lautner G, Nagy SK, Gyurcsanyi RE, Meszaros T (2014) A rational approach for generating cardiac troponin I selective spiegelmers. Chem Commun 50(51):6801–6804. doi:10.1039/c4cc00447g
Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S (2002) GnRH binding RNA and DNA spiegelmers: a novel approach toward GnRH antagonism. Chem Biol 9(3):351–359
Latham JA, Johnson R, Toole JJ (1994) The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res 22(14):2817–2822
Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ (1992) Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355(6360):564–566. doi:10.1038/355564a0
Battersby TR, Ang DN, Burgstaller P, Jurczyk SC, Bowser MT, Buchanan DD, Kennedy RT, Benner SA (1999) Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J Am Chem Soc 121(42):9781–9789
Vaish NK, Larralde R, Fraley AW, Szostak JW, McLaughlin LW (2003) A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 42(29):8842–8851. doi:10.1021/bi027354i
Masud MM, Kuwahara M, Ozaki H, Sawai H (2004) Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg Med Chem 12(5):1111–1120. doi:10.1016/j.bmc.2003.12.009
Shoji A, Kuwahara M, Ozaki H, Sawai H (2007) Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J Am Chem Soc 129(5):1456–1464. doi:10.1021/ja067098n
Hollenstein M, Hipolito CJ, Lam CH, Perrin DM (2013) Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb Sci 15(4):174–182. doi:10.1021/co3001378
Benner SA (2004) Understanding nucleic acids using synthetic chemistry. Acc Chem Res 37(10):784–797. doi:10.1021/ar040004z
Hirao I, Kimoto M, Yamashige R (2012) Natural versus artificial creation of base pairs in DNA: origin of nucleobases from the perspectives of unnatural base pair studies. Acc Chem Res 45(12):2055–2065. doi:10.1021/ar200257x
Hirao I, Kimoto M (2012) Unnatural base pair systems toward the expansion of the genetic alphabet in the central dogma. Proc Jpn Acad Ser B Phys Biol Sci 88(7):345–367
Sefah K, Yang Z, Bradley KM, Hoshika S, Jimenez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, Tan W, Benner SA (2014) In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci USA 111(4):1449–1454. doi:10.1073/pnas.1311778111
Switzer C, Moroney SE, Benner SA (1989) Enzymatic incorporation of a new base pair into DNA and RNA. J Am Chem Soc 111(21):8322–8323. doi:10.1021/Ja00203a067
Morales JC, Kool ET (1998) Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat Struct Biol 5(11):950–954. doi:10.1038/2925
Morales JC, Kool ET (1999) Minor groove interactions between polymerase and DNA: more essential to replication than Watson-Crick hydrogen bonds? J Am Chem Soc 121(10):2323–2324. doi:10.1021/ja983502+
Ohtsuki T, Kimoto M, Ishikawa M, Mitsui T, Hirao I, Yokoyama S (2001) Unnatural base pairs for specific transcription. Proc Natl Acad Sci USA 98(9):4922–4925. doi:10.1073/pnas.091532698
Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, Kodama K, Yokogawa T, Nishikawa K, Yokoyama S (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nat Biotechnol 20(2):177–182. doi:10.1038/nbt0202-177
Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S (2004) A two-unnatural-base-pair system toward the expansion of the genetic code. J Am Chem Soc 126(41):13298–13305. doi:10.1021/ja047201d
Mitsui T, Kitamura A, Kimoto M, To T, Sato A, Hirao I, Yokoyama S (2003) An unnatural hydrophobic base pair with shape complementarity between pyrrole-2-carbaldehyde and 9-methylimidazo[(4,5)-b]pyridine. J Am Chem Soc 125(18):5298–5307. doi:10.1021/ja028806h
Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I (2007) Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res 35(16):5360–5369. doi:10.1093/nar/gkm508
Hikida Y, Kimoto M, Yokoyama S, Hirao I (2010) Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat Protoc 5(7):1312–1323. doi:10.1038/nprot.2010.77
Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I (2009) An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res 37(2):e14. doi:10.1093/nar/gkn956
McMinn DL, Ogawa AK, Wu YQ, Liu JQ, Schultz PG, Romesberg FE (1999) Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-fairing hydrophobic base. J Am Chem Soc 121(49):11585–11586. doi:10.1021/Ja9925150
Ogawa AK, Wu YQ, McMinn DL, Liu JQ, Schultz PG, Romesberg FE (2000) Efforts toward the expansion of the genetic alphabet: Information storage and replication with unnatural hydrophobic base pairs. J Am Chem Soc 122(14):3274–3287. doi:10.1021/Ja9940064
Ogawa AK, Wu YQ, Berger M, Schultz PG, Romesberg FE (2000) Rational design of an unnatural base pair with increased kinetic selectivity. J Am Chem Soc 122(36):8803–8804. doi:10.1021/Ja001450u
Yu C, Henry AA, Romesberg FE, Schultz PG (2002) Polymerase recognition of unnatural base pairs. Angew Chem 41(20):3841–3844. doi:10.1002/1521-3773(20021018)41:20<3841:AID-ANIE3841>3.0.CO;2-Q
Wu YQ, Ogawa AK, Berger M, McMinn DL, Schultz PG, Romesberg FE (2000) Efforts toward expansion of the genetic alphabet: optimization of interbase hydrophobic interactions. J Am Chem Soc 122(32):7621–7632. doi:10.1021/Ja0009931
Matsuda S, Leconte AM, Romesberg FE (2007) Minor groove hydrogen bonds and the replication of unnatural base pairs. J Am Chem Soc 129(17):5551–5557. doi:10.1021/ja068282b
Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE (2008) Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J Am Chem Soc 130(7):2336–2343. doi:10.1021/ja078223d
Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE (2009) PCR with an expanded genetic alphabet. J Am Chem Soc 131(41):14620–14621. doi:10.1021/ja906186f
Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE (2010) Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chemistry 16(42):12650–12659. doi:10.1002/chem.201000959
Arens MQ, Buller RS, Rankin A, Mason S, Whetsell A, Agapov E, Lee WM, Storch GA (2010) Comparison of the eragen multi-code respiratory virus panel with conventional viral testing and real-time multiplex PCR assays for detection of respiratory viruses. J Clin Microbiol 48(7):2387–2395. doi:10.1128/JCM.00220-10
Yang ZY, Hutter D, Sheng PP, Sismour AM, Benner SA (2006) Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res 34(21):6095–6101. doi:10.1093/Nar/Gkl633
Yang ZY, Sismour AM, Sheng PP, Puskar NL, Benner SA (2007) Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res 35(13):4238–4249. doi:10.1093/Nar/Gkm395
Yang Z, Chen F, Chamberlin SG, Benner SA (2010) Expanded genetic alphabets in the polymerase chain reaction. Angew Chem 49(1):177–180. doi:10.1002/anie.200905173
Yang Z, Chen F, Alvarado JB, Benner SA (2011) Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J Am Chem Soc 133(38):15105–15112. doi:10.1021/ja204910n
Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W (2010) Development of DNA aptamers using cell-SELEX. Nat Protoc 5(6):1169–1185. doi:10.1038/nprot.2010.66
Benner SA, Sismour AM (2005) Synthetic biology. Nat Rev Genet 6(7):533–543. doi:10.1038/nrg1637
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Zhang, L. (2015). Unnatural Nucleic Acids for Aptamer Selection. In: Tan, W., Fang, X. (eds) Aptamers Selected by Cell-SELEX for Theranostics. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-46226-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-662-46226-3_3
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-46225-6
Online ISBN: 978-3-662-46226-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)