Abstract
Whole-genome sequencing has become so affordable that we can now sequence genomes of dozens of individuals of a pathogen population instead of sequencing a single representative isolate. This change of framework enables us to answer questions impossible to address just a few years ago. Recent work on the complex has shown that such genomic approaches have practical implications for disease diagnostics, source attribution, and crop improvement. Most of this work is represented so far by studies on P. syringae pv. aesculi, pv. tomato, pv. actinidiae, and pv. phaseolicola and Pseudomonas cannabina pv. alisalensis. These studies performed comparative genomics and phylogenetic analyses using exclusively populations of P. syringae associated with crops. They thus excluded the vast majority of the P. syringae metapopulation, which is present outside of agricultural fields in habitats associated with the water cycle, leaf litter, and soil, which have been found to be significantly involved in the population dynamics of P. syringae and its evolutionary history. Bulk sequencing of P. syringae strains closely related to pathovar tomato from snowpack and surface water has started to reveal how the genomic diversity of P. syringae in non-agricultural environments relates to that of the epidemic population on crops. This chapter describes how population genomic analyses can take the study of evolution, ecology, disease, and molecular plant–microbe interactions of P. syringae to the next level by integrating all of its diversity and how such studies can be applied to help prevent and/or control future disease outbreaks.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
4.1 Who Is Pseudomonas syringae
Pseudomonas syringae sensu lato is a species that includes a genetically and phenotypically diverse group of Gammaproteobacteria, whose exact taxonomic status is still in flux. This group includes a few dozen causative agents of economically important crop diseases and a large number of genetically distinct strains isolated from wild plants, leaf litter, and compartments of the water cycle (e.g., clouds, precipitation, snowpack, lakes, and rivers) (Morris et al. 2008, 2010, 2013; Monteil et al. 2012). The named species most closely related to P. syringae are P. cichorii (Swingle 1925) and P. viridiflava (Burkholder 1930), which were originally described as species separate from P. syringae sensu stricto (Van Hall 1904) because of distinguishable phenotypic characteristics compared to P. syringae sensu stricto. Interestingly, even considering only crop pathogens, strains related to the P. syringae-type strain could be assigned to nine different species based on DNA similarity (or better dissimilarity). However, since no consistent phenotypic characteristics distinguish these nine groups, most of these groups could not be described as named species and are thus referred to as numbered “genomospecies” 1 through 9 (Gardan et al. 1999). It was recently shown that each genomospecies corresponds to a phylogenetic group (Bull et al. 2011) based on multilocus sequence typing (MLST) (Maiden et al. 1998).
Each genomospecies of P. syringae gathers different crop pathogens that are described based on their host range and the type of disease symptoms they cause. They are referred to as “pathovars” (Dye et al. 1980), and each pathovar is represented by a pathotype strain similar to type strains of named species. However, many genetic lineages of P. syringae have not been assigned to pathovars because either they did not cause disease on any tested plant species (Clarke et al. 2010) or simply because they were isolated from non-plant substrates and have not been tested for host range. They are currently simply assigned to phylogenetic groups based on MLST building on the MLST scheme originally developed by Sarkar and Guttman (2004).
While Chap. 3 describes and compares genomes of relatively distantly related strains belonging to different pathovars—or even different genomospecies—in this chapter, we will focus on how genome sequencing of very closely related strains belonging to the same phylogenetic group—or even the same pathovar—has started to give insight into various aspects of pathogen emergence, evolution, molecular host–microbe interactions, and ecology. We will also show how genome sequencing has the potential to transform plant disease diagnostics and unravel geographic routes of transmission. In the last part of the chapter, we will give our opinion on how we expect population genomics to give new insight into the life history and life cycle of P. syringae as long as we deeply sample from all environments in which P. syringae populations have been found to reside.
4.2 Pseudomonas syringae: One Population or Many?
Because strains that belong to P. syringae could actually be assigned to at least nine different species based on DNA similarity (Gardan et al. 1999), the question is how homogenous these strains are in regard to ecological niches they occupy, their interaction with other organisms, and their population structure. Importantly, is there one P. syringae population or are there many? In other words, does each pathovar represent a separate population or does each genomospecies or phylogenetic group represent a population or is there just one large P. syringae population?
This is not an easy question to answer since it is not even obvious what a bacterial population is. Sexual reproducing organisms belong to the same population if they interbreed. Different populations are isolated from each other, but occasional migration of individuals between populations may occur. In the case of bacteria, the boundaries of populations are not well defined. Although bacteria belonging to the same population can exchange DNA by horizontal gene transfer through site-directed recombination of genomic islands or homologous recombination of any genomic region, the relative contribution of recombination compared to mutation is different in different bacterial species. Absurdly, in bacteria, we may even have species corresponding to a “clonal” population, i.e., a population within which there is no recombination, which takes the original definition of “population” ad absurdum.
After performing a population genetic analysis of a collection of crop pathogens belonging to P. syringae, Sarkar and Guttman (2004) concluded that P. syringae consists of a population, in which members rarely recombine. Whole-genome sequencing (WGS) of multiple isolates of P. syringae pv. aesculi (Pae), the causative agent of bleeding canker of horse chestnut (Green et al. 2010), WGS of multiple isolates of P. syringae pv. tomato (Pto), the pathogen that causes bacterial speck disease around the world (Cai et al. 2011a), WGS of multiple isolates of P. syringae pv. actinidiae (Psa) from kiwifruit plants with bacterial canker disease (Mazzaglia et al. 2012), and WGS of multiple isolates of Pseudomonas cannabina pv. alisalensis (Pal) of diseased crucifers, tomato, or monocots (Sarris et al. 2013), confirmed that each of these pathovars consists of only one or a small number of clonal lineages. Each of these pathogens shows very little genetic variation between isolates. These pathogens could thus be defined as “genetically monomorphic.” “Genetically monomorphic” pathogens were described by Mark Achtman as pathogens with very little genetic variation between isolates, suggesting that these pathogens only recently emerged, in the order of dozens, hundreds, or maybe thousands of years (Achtman 2008).
The interesting question is do genetically monomorphic pathogens represent clonal populations? We do not believe so. Yes, if a pathogen is genetically monomorphic, it has not significantly recombined with other bacteria since its emergence and, in that sense, it appears to have a clonal population structure. However, genetically monomorphic pathogens only very recently emerged. Therefore, it is not appropriate to compare a genetically monomorphic pathogen with a genetically diverse pathogen that emerged much earlier. In the first case, we look at a time scale of a few hundred or thousand years and in the second case at a time scale of a few hundreds of thousands or millions of years. Therefore, genetically monomorphic pathogens may be clonal populations when we choose to look at them individually over a window of hundreds of years, but they may be part of recombining populations if we choose to look at them as a group of populations over a window of hundreds of thousands of years. In fact, a whole-genome phylogeny of isolates belonging to the genetically monomorphic Pto pathogen T1 and the Pto pathogens JL1065 and DC3000 suggests that the ancestors of these lineages recombined (our unpublished data). A very similar picture emerged when constructing a whole-genome phylogeny of Psa (McCann et al. 2013). In fact, the authors inferred that 10 % of each sequenced Psa genome is derived from recombination.
When performing MLST of Pto and of closely related crop pathogens in combination with isolates from compartments of the water cycle, it became evident that ancestors of crop pathogens and ancestors of environmental P. syringae isolates recombined (Monteil et al. 2013). One of the genes that was found to have recently recombined between crop strains and environmental isolates was the fliC gene encoding bacterial flagellin, which has an important role in plant–microbe interactions (Clarke et al. 2013).
Furthermore, since there appears to be relatively little recombination between the different phylogenetic groups of P. syringae based on MLST (Sarkar and Guttman 2004) but considerable recombination within groups (Cai et al. 2011b; Yan et al. 2008), we may conclude that the different phylogenetic groups within P. syringae each represents a separate population and that there is no overall P. syringae population.
Taking all these results together, we propose that the different P. syringae phylogenetic groups/genomospecies each represents separate recombining populations that consist of crop pathogens and related environmental strains. We further believe that genetically monomorphic crop pathogens occasionally emerge from these populations and then acquire an apparent “short-term” clonal population structure while spreading around the world from crop field to crop field [see also Monteil et al. (2013)]. Smith et al. (1993) defined such a combination of recombining and clonal populations as “epidemic population structure.” However, to confirm this hypothesis, a full-fledged population genomic analysis of crop pathogens and environmental isolates will be necessary (see below).
4.3 Geographic Origin of Pseudomonas syringae Crop Pathogens and Their Routes of Transmission
For some fungal and oomycete pathogens, strong evidence has been found for their geographic origin, for example, Phytophthora infestans in the Andes of South America (Gomez-Alpizar et al. 2007; Stukenbrock and McDonald 2008). From South America, P. infestans seems to have migrated to North America and from there to Europe and, finally, to Ireland where it caused the Irish potato famine. Also for some bacterial pathogens, like Xylella fastidiosa subsp. fastidiosa, convincing evidence has been found for an origin in Central America and introduction into the USA has been hypothesized to have occurred with the importation of coffee plants from Costa Rica to California around 1880 (Nunney et al. 2010). Also for Xanthomonas axonopodis pv. manihotis, a South American origin seems likely (Bart et al. 2012). As expected, for some genetically monomorphic bacterial human pathogens, origin and international routes of transmission have been reconstructed in great detail. The most impressive example is the reconstruction of the international spread of Yersinia pestis, the causal agent of plague, out of China based on whole-genome sequences of hundreds of strains (Morelli et al. 2010; Cui et al. 2013). The conceptual basis behind such studies is relatively simple and is mainly based on one population genetic principle: The geographic origin of a species (or a population) is inferred to be where the highest genetic diversity is found (Stukenbrock and McDonald 2008). In fact, members of a population accumulate mutations over time. Therefore, where the pathogen has existed for the longest time, i.e., where it originated, pathogen isolates can be expected to be much more different from each other than in those geographic areas where the pathogen was recently introduced. This is also due to the fact that the genetic diversity in the new area is derived from the usually small number of bacteria that migrated from the geographic origin of the pathogen (for example on a single lot of seeds). This phenomenon is called a “population bottleneck effect.”
Another aspect to consider is that populations at the geographic origin diverged less from their ancestor than the pathogen populations in the geographic areas into which the pathogen was recently introduced. In fact, members of the original population will accumulate mutations, but as long as these mutations do not confer a strong selective advantage, these mutations will stay at a relatively low frequency in this population. In contrast, when a very small number of bacteria of a population are introduced into a new geographic area, the mutations present in these few bacteria will now be present in their entire progeny, that is, in all members of the population in this new geographic area. In other words, the introduced bacteria become the founders of the new population in the new geographic area. This process is known as the “founder effect,” and the genetic differences between the population at the center of origin and the populations in the new geographic area are said to be due to “genetic drift” (instead of selection). Therefore, each time a few members of a population are introduced from one geographic area into the next, the genetic distance from the original population can be assumed to have increased. Importantly, the mutations present in each founder of each geographic transmission are preserved during the following geographic transmissions, making it possible to reconstruct the geographic routes of transmission by correlating phylogeny of isolates with their geographic origin.
Since recent estimates of yearly mutation rates for bacterial human pathogens are in the order of 1–10 mutation per million base pairs per year (for example, Morelli et al. 2010; Nuebel et al. 2010), it is obvious that if we assume a similar mutation rate for plant pathogens, the geographic origin of recently emerged P. syringae crop pathogens can only be reconstructed by sequencing whole genomes. It is thus exciting that sequencing of a large number of genomes has become affordable using next-generation sequencing with prices as low as $100/genome. Therefore, studies of the geographic origin and routes of transmission can now also be performed for P. syringae crop pathogens. However, there is another challenge that needs to be overcome before performing such studies: Populations from different geographic areas can only be compared, and phylogeny can only be correlated with geography (in what is referred to as “phylogeography”) if a sufficient number of isolates are available from each geographic area to be representative of the populations in these areas. Here, we will show how limited availability of isolates has in fact precluded in-depth phylogeographic studies of P. syringae crop pathogens so far.
The first study that compared multiple strains of a recently emerged P. syringae pathogen focused on Pae, the causative agent of bleeding canker disease of European horse chestnut in northwestern Europe (Green et al. 2010). This disease was first described in 2002/2003 and spread from England within a few years all the way to Scotland. The genomes of three British isolates of the pathogen from 2006 and 2008 were found to be almost indistinguishable from each other (3 mutations in 3 million base pairs) but to be significantly different (1,613 mutations in 3 million base pairs) from a P. syringae pathogen that was isolated in 1969 in India and that is the causative agent of a foliar disease of horse chestnut. The bleeding canker disease of horse chestnut has not been observed anywhere else in the world, and the Pae strain from India that only causes foliar symptoms is the closest known relative. Also, no environmental isolate of P. syringae closely related to Pae has yet been identified in Europe. Therefore, the only conclusion that can be made is that the UK outbreak of this disease is due to a single clone that started spreading on horse chestnut very recently. However, nothing can be said about the geographic origin of this clone. Did it preexist in an environmental reservoir, for example, in compartments of the water cycle, or on wild plants in Europe and jumped onto horse chestnut? Or was it introduced from another geographic area where it caused bleeding canker in the past? Maybe the disease has simply not been observed in the geographic area where it first emerged because other factors in that area limit the severity of the disease, for example, unfavorable climate, tolerance of horse chestnut genotypes that grow in that area, and/or absence of insect vectors that appear to contribute to the spread of Pae in the UK (Green et al. 2010).
Bacterial speck disease of tomato is caused by Pto and was first described in the USA by Bryan (1933). Interestingly, reports of the disease popped up around the world in the 1970s (Goode and Sasser 1980). It is not known whether this was due to a real increase in disease incidence or whether it was the awareness of the disease that increased and led to an increase in disease reports. Interestingly, analyzing over 100 Pto isolates from the 1940s until 2008 from around the world, it was found that two separate lineages of Pto (called T1 and JL1065) were common in Europe and North America in the 1970s, while a third lineage (DC3000) was only found in the 1940s in Canada and 1960s in the UK (Cai et al. 2011a) with one more isolate of this lineage from Japan (Sarkar and Guttman 2004). In the 1980s, the T1 lineage almost completely replaced the JL1065 lineage in Europe and North America. The few available Pto isolates from Africa and Australia belonged to either T1 or JL1065, but isolates from South America all belonged to T1. Since the T1 lineage was found on every continent from which isolates were available, any of these continents could be the geographic origin of the T1 lineage. Unfortunately, genomes of only five representative isolates of the T1 lineage from Europe and North America were sequenced, while all other isolates were only analyzed with a small number of genome-derived molecular markers. Moreover, the number of available Pto isolates from outside of Europe and North America was very limited. Therefore, it was impossible to determine which continent had the population with the highest diversity. Interestingly though, three out of eight genotypes of the Pto T1 population in Europe and North America were found to be present in both continents, showing that T1 strains have been exchanged several times between these two continents during recent years. Some of the same genotypes were found also in Australia. However, T1 isolates from Colombia in South America all had at least one mutation absent from all other T1 populations around the world and absent from the other Pto lineages and all related P. syringae pathovars. This suggests that the Pto population in South America may be relatively isolated compared to the Pto population of the Northern Hemisphere.
The mode of intercontinental transfer of Pto is impossible to know at this point. It could be by seed since Pto was found to be transmitted by seed (McCarter et al. 1983), but it could also be atmospheric movement. In fact, strains indistinguishable from Pto DC3000 were isolated in a creek upstream of any agricultural activity in New Zealand (Monteil et al. 2013), suggesting that at least bacteria belonging to the Pto DC3000 lineage can also travel through the water cycle and possibly travel long distance through the atmosphere. Therefore, it is possible that Pto T1 is exchanged between continents via atmospheric movement.
A relatively more detailed phylogeography has been obtained for Psa, the causative agent of bacterial canker disease of kiwifruit. Bacterial canker of kiwifruit was first described in Japan and China in the 1980s and then in Korea in the early 1990s (Scortichini et al. 2012). In 2008, a very severe epidemic started spreading throughout Italy and then the rest of Europe and Turkey. In 2010, the disease was found in New Zealand and in Chile. Three genome-sequenced isolates from Japan and Korea collected during the outbreak in the 1980s and 1990s were found to be clearly distinct from the 2008 epidemic and represent a separate population with strains that differ by more than 2,000 mutations/million base pairs from strains of the 2008 epidemic (Mazzaglia et al. 2012). This high number of mutations could not have accumulated in less than 30 years, and therefore, the Korean and Japanese populations can be excluded as the parent population from which the 2008 population emerged. Two strains from China (isolated from the same location in the same year) were instead found to be highly similar to four strains isolated in Europe with fewer than 2 mutations per million base pairs (of vertically inherited core genome) distinguishing the strains from the two continents (Mazzaglia et al. 2012). Later, a small number of genome sequences of Psa strains from New Zealand and Chile were compared and found to be also extremely closely related to the European and Chinese strains (Butler et al. 2013). Interestingly though, one additional strain from China was found to be more divergent (1 mutations/16,000 base pairs) compared to all 2008 outbreak strains. While many more Psa strains isolated in New Zealand were sequenced later (McCann et al. 2013), the number of strains from China, Europe, and Chile is still very limited and any conclusions made from these data must be considered preliminary and are based on the pure assumption that the isolates from the different countries/continents are representative of the diversity in these countries. This is likely to be the case for Europe, New Zealand, and Chile since the disease emerged so recently in these areas. However, China, Japan, and Korea may harbor many more genotypes than currently known. Nonetheless, the finding that China harbors at least one genotype that is closely related to all the 2008 isolates, but is clearly more divergent, supports the conclusion that China is the geographic origin (Butler et al. 2013). Moreover, there is strong circumstantial evidence pointing to China as geographic origin of the current epidemic: (1) The center of diversification of the kiwifruit genus Actinidia is in China, making it likely that P. syringae strains adapted to Actinidia species possibly coevolving with Actinidia over hundreds of thousands of years; (2) the disease broke out in China years before it broke out in Europe, New Zealand, and Chile; and (3), plant material is known to have been imported from China to Europe and possibly to New Zealand.
The next important question is what were the international routes of transmission of Psa? Since the disease broke out first in Italy and then in New Zealand and isolates from these countries could initially not be distinguished, it was suggested that Italy was the source of the New Zealand outbreak. It was thought that possibly strains from China were imported with contaminated plant material into Italy and from there to New Zealand. However, several results point to a direct import of Psa from China to New Zealand: (1) Some mutations are shared between Chinese and New Zealand isolates but not with European isolates and (2) the same variant of a genomic island is present in one of the sequenced Chinese strains and all of the sequenced New Zealand strains, but this variant of the genomic island is absent in the European strains (Butler et al. 2013). Interestingly, a different Chinese strain has the identical variant of the genomic island present in the European strains, while the Chilean strains carry yet another version of the same island (Butler et al. 2013). This suggests (but does not prove) that the European, the New Zealand, and the Chilean Psa populations are each derived from separate independent importations of Psa from China.
4.4 Comparison of Closely Related Genomes Provides Insight into Molecular Plant: Microbe Interactions for Crop Improvement
Comparison of genomes of closely related strains with different phenotypes is one avenue to identify the genetic differences at the basis of the observed phenotypic differences. In the case of plant pathogens, the most relevant phenotypic differences are differences in host range and differences in virulence. The identification of genes responsible for these differences is facilitated by the fact that many P. syringae genes that determine virulence and, to some degree, host range are already described. It is known that conserved microbial-associated molecular patterns (MAMPs) trigger immunity in most plants but that some MAMP alleles evade recognition. For example, most flagellin alleles trigger a plant immune response, but flagellin of Ralstonia solanacearum and Agrobacterium tumefaciens do not (Pfund et al. 2004; Felix et al. 1999). It is also known that P. syringae translocates via a type III secretion system (T3SS) so-called effector proteins into plant cells that suppress MAMP-triggered immunity in host plants, while some of them trigger an immune response (Effector-triggered immunity (ETI)) in non-host plants (Jones and Dangl 2006).
When comparing two closely related strains, whereby strain A causes disease on a certain plant species and strain B does not cause disease on this plant species, two hypotheses can be experimentally tested: (1) Strain A contains effectors necessary to suppress the immune system of the plant species, and strain B is missing those effectors and (2) strain B contains effectors or MAMP alleles that trigger an immune response in the plant species, and strain A is missing those effectors or has different MAMP alleles. Either hypothesis may be correct, or a combination of both hypotheses may apply. Moreover, other genes necessary for successful invasion of—or interaction with—the host species may be missing in strain B.
Two groups of closely related P. syringae strains have so far been used for this kind of comparison: (1) Pto DC3000 that causes disease on tomato and Arabidopsis thaliana and the closely related Pto T1 strain that causes disease on tomato but not on A. thaliana (Almeida et al. 2009) and (2) P. syringae pv. phaseolicola (Pph) strains that are pathogens of bean (Phaseoulus vulgaris) and mung bean (Vigna radiata) and the soybean pathogen P. syringae pv. glycinea (Pgl) (Baltrus et al. 2012). While there is one known case where deleting a single effector gene (hopQ1) from a strain (Pto DC3000) expanded the host range of the strain to an additional species (Nicotiana benthamiana) (Wei et al. 2007), genome comparisons and follow-up genetic manipulations of Pto T1 and Pto DC3000 and Pph and Pgl showed that host range differences may more often be the result of multiple genes (Sohn et al. 2012; Baltrus et al. 2012).
In the case of Pto T1, it was found that at least two of its effectors, AvrRpt2 and HopAS1, trigger an immune response in A. thaliana (Almeida et al. 2009; Sohn et al. 2012). Deleting these two effectors significantly increased growth of Pto T1 on A. thaliana. However, the double-deletion mutant still grows significantly less than Pto DC3000 and does not cause any disease symptoms at the minimum inoculum dose at which Pto DC3000 causes disease symptoms on A. thaliana (Sohn et al. 2012). Expressing individual Pto DC3000 effectors in the double-deletion mutant only marginally increased growth of Pto T1 in A. thaliana (our unpublished data). Therefore, either multiple Pto DC3000 effectors need to be expressed in the avrRpt2/hopAS1 deletion mutant of Pto T1 at the same time to allow it to reach the virulence of Pto DC3000 or T3SS-independent genes necessary for full virulence on A. thaliana are also missing from Pto T1 but are present in Pto DC3000.
In the case of the Pph and Pgl comparison, expression of the Pgl effectors hopC1 and hopM1 in one of the Pph strains only slightly reduced growth on bean. On the other hand, expression of the Pph effector avrB2 in Pgl only slightly increased growth of Pgl on bean. Therefore, adaptation of Pph to bean appears to be due to multiple effector differences (Baltrus et al. 2012). Only by deleting and expressing multiple effectors in the same strain in order for a Pph strain to acquire the same effector repertoire as the Pgl strain (and/or the Pgl strain to obtain the same effector repertoire of a Pph strain) will it be possible to determine whether effector differences alone can explain the host range differences between these strains. Possibly, allelic differences in effector sequences or expression level differences of effectors also contribute to the observed host range differences. Importantly, since T3SS mutants of the Pph and Pgl strains grew to the same population density, the differences in their host range appear to be limited to effectors and, possibly, allelic differences in the structural components of their T3SSs.
While comparison of Pto T1 with Pto DC3000 and of Pph strains with Pgl revealed differences in effectors, but differences in MAMPs were not noted, comparison of multiple isolates all belonging to the Pto T1 lineage revealed unexpected differences in MAMPs within flagellin (Cai et al. 2011a). First, a group of strains from Colombia in South America had an amino acid substitution in the flagellin epitope flg22, a known MAMP (Felix et al. 1999), although these strains were otherwise indistinguishable from other Pto T1 isolates. Although it was known that different pathogens have different flagellin alleles and different strains of Xanthomonas campestris have different flagellin alleles (Sun et al. 2006), this difference among isolates belonging to the same genetic lineage was surprising. Moreover, all strains from Colombia had a non-synonymous mutation in another region of flagellin downstream of the flg22 epitope and all strains from Europe and North America isolated after 1980 had another non-synonymous mutation only three codons away from it (Cai et al. 2011a). This strongly suggested that the region of flagellin containing these two mutations (called flgII from now on) represented a new MAMP and Pto T1 adapted independently in Colombia and in North America/Europe to tomato by evading recognition of this MAMP through allelic variation. This hypothesis was experimentally confirmed: A 28-amino-acid-long peptide corresponding to the flgII allele typical of European isolates before 1980 triggered a stronger immune response in tomato than peptides corresponding to the two mutated alleles from Colombia and from North America/Europe after 1980 (Cai et al. 2011a). Therefore, comparison of almost identical isolates of the same genetic lineage of pathovar Pto allowed identification of a previously unknown MAMP. Moreover, allelic variation at this MAMP influences bacterial fitness in planta depending on the plant genotype suggesting that different plants have different alleles of the receptor that recognize alleles of flgII with different affinity (Clarke et al. 2013).
The flgII receptor is different from the flg22 receptor and not present outside of the Solanaceae family (Clarke et al. 2013). Therefore, cloning different alleles of the flgII receptor and expression of these receptor alleles in crops that either do not have any native flgII receptor or have a flgII receptor with low affinity for the flgII alleles of their most important pathogens could be used to improve crop disease resistance (or at least significantly decrease pathogen growth) similar to what was proposed in regard to the MAMP receptor elongation factor tu receptor (EFR) (Lacombe et al. 2010).
Identifying differences in effectors or MAMPs is not the only approach to find new targets for crop improvement for disease resistance based on genome comparisons. Identifying the conserved core repertoire of effectors present in every single strain of a pathogen is another promising approach. The underlying hypothesis is that the effectors that are present in every single strain of a pathogen are the effectors that are most important for virulence and that cannot be easily lost by the pathogen without a reduction in virulence. This approach has been proposed for Xanthomonas manihotis (Bart et al. 2012) but can easily be applied to P. syringae pathogens. For example, the genomes of five isolates of P. cannabina pv. alisalensis (Pcal) have been sequenced and the core effector repertoire identified (Sarris et al. 2013). The individual Pcal effectors could be cloned and tested for triggering a defense response in a panel of plant species to identify putative resistance genes to these effectors. The identified resistance genes could then be either bred into Pcal hosts or cloned and transferred into high-yielding commercial Pcal hosts by genetic engineering. The same could be done with the identified core effectors of Psa (McCann et al. 2013).
4.5 Present and Future of Genome-Based Diagnostics and Epidemiology
In plant disease diagnostics today, polymerase chain reaction (PCR) and real-time PCR (RT-PCR) have become routine and complement—or even replace—diagnostic techniques based on phenotype, for example, Biolog® or fatty acid analysis. Some of the PCR and RT-PCR primers have been designed based on individual whole-genome sequences or based on comparison of whole-genome sequences of the pathogen of interest with genome sequences of other related pathogens, for example, for Xanthomonas carotae (Kimbrel et al. 2011) or R. solanacearum race 3 biovar 2 (Guidot et al. 2009). Even more precise diagnostics are possible with markers based on the comparison of genomes of different strains of the same pathogen. For example, Studholme and colleagues have developed simple genotyping assays based on diversity revealed by WGS in Xanthomonas musae (Wasukira et al. 2013) and Balestra et al. (2013) have designed primers that can distinguish between different clones of Psa.
Going one-step further, MLST is starting to be used for precise identification of pathogens. For example, the causative agent of a bacterial leaf spot outbreak of parsley in Ohio (USA) was recently identified as P. syringae pv. coriandricola (Xu and Miller 2013). The use of MLST for P. syringae and other bacterial crop pathogens is possible because a dedicated MLST database was established to simply compare sequences of individual loci with a collection of characterized pathogen strains or to perform a full-fledged MLST study of a pathogen sequencing all loci that were included in published MLST analyses of pathotype strains (Bull et al. 2011; Young et al. 2008).
While MLST has sufficient resolution for precise identification of a P. syringae pathovar, it is not always sufficient for differentiating between different lineages within the same pathovar. For example, MLST can distinguish between the Korean/Japanese population of Psa and the isolates of the 2008 outbreak in Europe, but MLST cannot distinguish between Pae causing bleeding canker of horse chestnut in Europe and the Indian strain causing only leaf spotting (Green et al. 2010). Also, in regard to epidemiology, MLST usually does not provide the resolution to identify the source of a disease outbreak. Therefore, the question is how can WGS be translated into the diagnostic practice to improve strain identification and determine the source of disease outbreaks?
The potential of WGS for improving diagnostics and epidemiology for bacterial human pathogens is very similar to the potential we see for improving diagnostics of plant pathogens. It was recently proposed that WGS could replace multiple separate steps and tests in the diagnosis of human bacterial pathogens and in epidemiological investigations (Didelot et al. 2012). In fact, not only could WGS precisely identify a plant pathogen to the pathovar level or beyond, it could even provide a list of genes present in an outbreak strain encoding antibiotics resistance and thus help choose the best disease control strategy to deploy. Moreover, WGS has the power to give a precise view of the effector repertoire of an outbreak strain and suggest which crop cultivars may be resistant to the outbreak strain. Therefore, WGS could inform growers in regard to the best choice of cultivars that are possibly resistant to a new outbreak strain and that could be planted the following year.
As with MLST, WGS can only be successful if appropriate databases and tools for genome analysis are developed and become accessible in plant disease clinics. All research results in regard to effector repertoires and the corresponding resistance genes known to recognize them need to be included in such a database. Moreover, to identify the source of a disease outbreak, all isolates that are identified by any diagnostic clinic need to be automatically added to a central database. In this way, if a new pathogen or a new pathogen lineage emerges in one geographic area, it will be possible to automatically follow its spread to other areas as long as outbreak isolates are routinely sequenced. Currently, routine genome sequencing is still too expensive in order to implement such a strategy, but we can expect prices to decrease further and we believe that it is only a question of time until such an a strategy will become reality.
While sequencing the genome of isolated bacteria is an effective tool in identification and epidemiology, it would be even better if pathogens could be precisely identified directly from a plant sample without pathogen isolation in a metagenomic approach. This could be done by extracting DNA from a plant sample, or it could be done by extracting RNA from a plant sample. The advantage of RNA over DNA is that by extracting RNA and then reverse transcribing all RNA into DNA, RNA viruses would also be detected. The resolution lost by only sequencing transcribed genes would be minimal for diagnostics and may be negligible for epidemiological purposes since, for example, most of the single-nucleotide polymorphisms (SNPs) that were identified between Pto isolates were intragenic (Cai et al. 2011a). All genes may not be transcribed in planta, but even if only half the genes of a bacterium were transcribed in planta, this would probably still provide the necessary resolution for source identification. Another advantage of RNA sequencing would be that even physiological problems, like mineral deficiencies, might be identifiable if the sample is fresh or flash-frozen in liquid nitrogen immediately after collection. Of course, as for sequencing isolated bacteria directly, such a culture-independent sequencing approach would require an even more comprehensive database and collection of bioinformatics tools to interpret the massive amount of data that would be obtained.
4.6 From Individual Genomes Analysis to Population Genomics: The Next Step to Infer Pseudomonas syringae Crop Adaptation and Evolutionary History
We expect the next key findings in ecology and evolution of P. syringae to be revealed by “population genomics,” an approach that has already demonstrated its power in clinical microbiology. While WGS is extremely useful for disease diagnostics and to determine the molecular determinants of phenotypic traits or to reconstruct international routes of transmission (as described above), it cannot correctly answer these and other more fundamental questions about ecological differentiation or evolutionary history unless these questions are addressed in the context of a population, i.e., applying population genomics. The field of population genomics has existed for dozens of years in its early form whereby genomic divergence within and between populations was assessed using a small number of genomic loci (Nosil and Buerkle 2010). Today, WGS allows extending this approach to whole genomes and thereby dramatically increases the quantity of information that we can analyze and provides access to genomic variation that was previously undetectable. In particular, we can pinpoint all genomic regions that have diverged between individuals and estimate gene flow allowing the inference of population structure, adaptation, and evolutionary history of organisms with an accuracy previously unimaginable (Nosil and Buerkle 2010; Nosil et al. 2009).
When genomes are compared between individuals, the individuals are expected to be representative of the population or metapopulation for which we want to determine genomic variation. Sampling over space, niche, and time is thus the foundation of any population genomic study, because it drives the interpretation of the results we obtain. The less the sample reflects the population, the less accurate our inferences are. This biggest limitation is due to the fact that our observations are always an approximation of the reality depending on the sampling representativeness. Paraphrasing Hunt et al. (2008), “in most ecological sampling, the true habitats or niches are unknown and can only be observed as projections onto the sampling dimensions (‘projected habitats’).” In the case of P. syringae and other plant pathogens, the question is how to get the most representative sample a priori while we do not know the population structure.
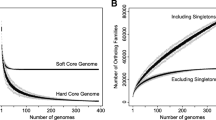
Therefore, carefully choosing the sampling strategy prior to sampling is indispensable and requires considering several factors. The choice of individuals will determine the success of the study in answering the questions we pose. In fact, sampling is what constitutes the difference between studies of population genomics and those of comparative genomics. Comparative genomics provides information about differences in gene content and allelic differences between genomes to test and develop hypotheses on how these differences determine phenotypic differences between organisms. Comparative genomics also permits development of lines of work to investigate ecology and epidemiology of plant pathogens (Sarkar et al. 2006; Potnis et al. 2011; Baltrus et al. 2011; Mann et al. 2013). However, comparative genomics cannot extrapolate the findings to the population scale and cannot distinguish the effect of randomness from real selection by the environment. It is true that when whole-genome sequencing was first applied to epidemiological or evolutionary studies, the sample size was necessarily small because of sequencing costs and time required for gene annotation. Ten genomes were sufficient to obtain first genome-wide phylogenetic trees or to compare genomes of different pathogens or pathogen lineages. However, one individual can never be representative of a population whatever the criterion is (e.g., niche, biology, and lineage). Traits of that individual can be variable within the population, and this variability must be taken into account in the analysis. From a statistical point of view, the higher the number of individuals is, the more powerful the analysis is. However, even considering the low cost of sequencing today, we still cannot sequence as many genomes as we would like to and we still need to make a choice of which strains to sequence. To this end, it is important to select strains from a diverse set of samples that are representative of the environments that the pathogen occupies. In fact, even a large number of strains from a restricted number of samples may not be representative of the pathogen population. Fortunately, sequencing costs are still decreasing and this will allow sequencing more and more strains and make strain selection easier and easier (Didelot et al. 2012).
However, even if we were able to sequence as many strains as we want, choosing the best sampling strategy would still be important. Choice of sampling location, host, environment, or date, the relative number of different samples, and the choice of how many strains to sequence from each sample will always strongly affect how representative the sequenced individuals are of the population. Therefore, to accurately infer the processes that shape genomic variation, samples and strains must be selected properly. Moreover, that choice has to be tailored to the question and to the evolutionary scale at which the effects of the investigated processes are visible. Accordingly, year of isolation, geographic origin, genetic relatedness among individuals, and strain phenotypes do not always have the same importance. For example, when the objective of a study is to investigate early events in ecological differentiation, as in Shapiro et al. (2012), the investigated processes occur over a short time scale in a limited space compared to the evolutionary history of a bacterial species. Therefore, closely related bacteria isolated over a short time and occupying different niches at the same geographic location were chosen. A similar example is the investigation of the microevolution of Staphphyloccoccus aureus within a single host by Young et al. (2012) for which the authors selected dozens of isolates over a 13-month period from the same patient. For studies of biogeography, instead, we need to maximize space and time of sampling. For example, inference of the historical transmission routes of Y. pestis over continents (Morelli et al. 2010; Cui et al. 2013) required this kind of sampling. Finally, when the interest concerns pathogen–host specificity, studies may maximize the number of samples from different hosts (Fitzgerald et al. 2001; Sheppard et al. 2013a, b).
4.7 Pseudomonas syringae in the Footsteps of Human and Animal Bacterial Pathogens: What Have We Learned from Clinical Population Genomic Studies?
Population genomic studies of bacterial human pathogens have already dramatically improved our vision of pathogen ecology and evolution and are revolutionizing medical diagnostics and disease epidemiology. Development of statistical models, databases, and software, like the bacterial isolate genome sequence database (BIGSdb) pipeline (Jolley and Maiden 2010), have made it possible to handle genomic data of hundreds of strains of the same pathogen species. Such tools make it possible, for example, to streamline association-mapping methods to determine the genomic basis of adaptation (Sheppard et al. 2013b). Additionally, several Bayesian modeling approaches have been developed to infer population structure, clonal relationships, and genomic fluxes from large populations and gene sets (Falush et al. 2003; Didelot et al. 2009, 2010; Marttinen et al. 2012; Corander et al. 2008; Shapiro et al. 2012).
Numerous processes associated with genomic divergence between populations and evolutionary history of human pathogens have been revealed, bringing to light new research perspectives. For example, population genomic studies applied to Salmonella enterica have given insights into population structure and the role of recombination far beyond the limitations of classical approaches (den Bakker et al. 2011; Desai et al. 2013; Didelot et al. 2011; Zhou et al. 2013). Classification into serovars and even phylogenies and recombination analysis based on MLST still missed important genetic information (Achtman et al. 2012). However, using a population genomic analysis of 10 % of the core genome of 114 isolates significantly refined our knowledge of the relationships between serovars, identified subpopulations, pinpointed donors and recipients of recombination events, and obtained insight into emergence of genetic lineages and estimated the age of these lineages (Didelot et al. 2011). By a similar approach, Joseph et al. (2012) unraveled the population structure and genomic fluxes within the obligate pathogen Chlamydia trachomatis.
In order to contain disease spread, it is necessary to understand how the bacterial variant that is causing a disease outbreak emerged. It is especially important to assess the importance of species introgression (acquisition of genomic regions from a different bacterial species) and to determine the environment in which it occurred, because this may allow identifying hot spots of bacterial diversification and prevention of future emergence of new variants. Mechanisms of introgression and the regions of the genome that are affected have been recognized and better understood, thanks to population genomics. One of the best examples is the genome-wide introgression that occurred in the zoonotic pathogen Campylobacter coli. Based on MLST data, Sheppard et al. (2011) had previously observed extensive DNA acquisition from another related species Campylobacter jejuni into one lineage of C. coli. Both species cause gastrointestinal symptoms in humans characterized by different host ranges in agricultural and non-agricultural environments. Through WGS of 30 strains of the two species, they were able to determine that gene flow occurred most frequently in those regions of the genome that are most similar between the two species (Sheppard et al. 2013a). These results also suggest that farming has played an important role in the diversification of Campylobacter spp. by enhancing physical opportunities for genetic exchange between the two species. Thus, agriculture appears to provide the conditions that are conducive to the emergence of new adapted hybrids and their proliferation. Sheppard et al. (2013b) then went further in the study of host adaptation applying for the first time whole-genome association mapping to a bacterial model and identified a genomic region significantly associated with isolates from cattle but frequently absent from genomes of strains that infect birds.
The high resolution of the information provided by whole-genome sequences facilitates the study of small genetic changes undetectable with classical genetic approaches. With population genetics, the assumptions we make about gene organization and dynamics in bacterial genomes leave variation at many loci undetected. We can thus not study small microevolutionary events associated with a small numbers of mutations per genome. With population genomics instead, these barriers disappear, permitting the study of microevolutionary events associated with niche adaptation. This can lead to new fundamental knowledge in microbial ecology. Whole-genome comparisons at the population scale have even revealed the processes involved in the early phases of ecological differentiation and speciation of bacteria (Shapiro et al. 2012).
The more we are interested in subtle changes and short time scales, the less powerful classical genetic population analysis is. This is especially well illustrated by the within-host microevolutionary study of methicillin-sensitive Staphylococcus aureus causing fatal bloodstream infections (Young et al. 2012). The genome-wide study of Wilson and his partners (2012) enabled identification of a small number of mutations separating disease-causing variants from commensal bacteria. Following the pathogen population within a patient over a year, they demonstrated that the initial population accumulated 30 SNPs. Just eight of these SNPs affected protein function including a transcriptional regulator and were associated with the emergence of pathogenic variants inside the nasal carrier population. The high resolution of the approach thus brought new insight into the evolutionary dynamics of bacterial pathogens by demonstrating how small evolutionary events can lead to the emergence of virulent variants from nonpathogenic populations within the same host.
These few examples show the many possibilities of how population genomics could be applied to P. syringae and could help answer many unresolved questions about its ecology and evolution. Similar to what was shown for S. aureus, small evolutionary events might occur in planta in a few weeks or months from seed germination to maturity. The microevolutionary dynamics unraveled by population genomics within the plant host could also reveal regulatory and structural changes that lead to pathogenicity from a single cell via mutation or via acquisition or loss of genomic islands as experimentally revealed previously (Pitman et al. 2005; Lovell et al. 2009).
4.8 Looking Beyond Agriculture to Better Infer the Ecology and Evolution of Pseudomonas syringae Crop Pathogens
Previously, we discussed the importance of sampling in population genomic studies. We pointed out how we sample based on our current knowledge of a pathogen’s ecology; in particular, we sample those environments in which we assume the pathogen population to reside. Therefore, our view of the plant–pathogen system is going to determine our sampling strategy and because our sampling strategy will determine the results of the population genomic analysis that we apply, our view of the plant pathogens system will ultimately condition the results we obtain and their interpretation. In plant pathology, the selection of isolates is mostly based on virulence traits, host range, or epidemic history. This is why studies of plant pathogens are often incomplete. Most of the time, strain choice is biased because we impose our current perspective of a plant pathogen’s life history on that choice. Didelot et al. (2011) point out that when only highly virulent isolates are chosen, it is not possible to find the true evolutionary history of a pathogen because we only consider a small portion of the pathogen population and we cannot correctly infer gene flow and population structure.
For most pathogens that can survive outside of their hosts, limiting sampling to hosts is a problem. Sampling from diseased plants is only sufficient when we are interested in the evolution of a clonal lineage or its phylogeography. However, it is not sufficient if we want to understand how the pathogenic lineage relates to the wider diversity and how it originally emerged. The range of ecosystems, where non-obligate plant pathogens can evolve and adapt, have been unexplored and underestimated. A series of investigations of the ecology of P. syringae outside the agricultural context and, more specifically, in alpine ecosystems, have highlighted the various facets of P. syringae lifestyles. P. syringae population dynamics have been found to be inextricably associated with the water cycle (Morris et al. 2008). From the clouds through precipitation to river water, snowpack, and leaf litter, P. syringae is widely present and abundant (Diallo et al. 2012; Monteil et al. 2012; Morris et al. 2008, 2010). Based on these new insights into P. syringae life history, Morris et al. (2013) proposed scenarios of the role of the Earth’s processes in P. syringae ecology and evolutionary history. Evidence was gathered that population dynamics of crop pathogens participate in wider metapopulation dynamics through the global freshwater ecosystem. These environments are not only routes of dissemination for crop pathogens, but they represent reservoirs of genetic diversity and may be motors of diversification. While populations of alpine meadows or surface water are not directly impacted by cropping areas, most of them harbor virulence traits and genes typical of crop pathogens (Monteil et al. 2013). P. syringae studies carried out outside of the agricultural context are still exceptions among bacterial plant pathogens, but clues from others species, such as Erwinia spp. or Pantoea spp., suggest they could have similar life histories (Morris et al. 2009). Phytophthora species are another example of plant pathogens for which recent studies showed the importance of other ecosystems beyond crops and diseased trees. In the past decade, new evidence emerged showing abundance and high diversity of Phytophthora species in forest soils and forest streams. Phytophthora species present in natural habitats could lead to emergence of cryptic species potentially able to cause emerging destructive epidemics (Hansen et al. 2012).
Undoubtedly, determination of genomic fluxes between populations residing in natural environments and those causing disease on crops is indispensable to infer the evolutionary history and adaptation to a pathogenic lifestyle. Recently, a recombination analysis coupled to Bayesian coalescent analysis was performed with P. syringae strains isolated from snowpack and alpine streams that are closely related to the tomato pathogen Pto. This study revealed that ancestors of environmental populations and those of monomorphic crop pathogens recombined (Monteil et al. 2013). Moreover, virulence gene repertoires coding for T3SS effectors in Pto lineages are present in environmental strains. Environmental strains induced slightly less severe symptoms on tomato but had a wider host range. These results suggest that crop pathogens evolved through a small number of evolutionary events from an environmental population of less aggressive ancestors. Environmental pools and crop pathogen populations thus appear to interact, and environmental populations could be a source of novel genes. Similar observations led to the same conclusions for the causal agent of Legionnaire’s disease: Legionella pneumophila. Interestingly, Coscolla and Gonzalez-Candelas (2007) found recombination rates to be high within environmental populations that are abundant in freshwater. When clinical isolates were included in the analysis, conflicting signals within trees based on sequences of individual loci strongly suggested recombination between environmental and clinical isolates (Coscolla and Gonzalez-Candelas 2009).
These observations show how important it is to enlarge the population framework outside of hosts to have a representative sample for population genomic studies. The environment may select genes that can be involved in plant–microbe interaction, virulence, and host range (Morris et al. 2009). In all environments, trophic interactions within the microbial community (e.g., predation/prey relationships, competition for resources, and mutualism) shape bacterial genomes, which act both as sink and donor of genetic information. Genetic exchanges and mutations lead to the formation and selection of new bacterial variants. Since the maintenance of genes in a genome has a cost, the presence of virulence genes in environmental strains suggests that these genes might have another function outside the agricultural context (Martinez 2013). These traits may then become virulence traits if they confer an adaptive advantage for P. syringae fitness in planta. Although this has not formally been demonstrated for any gene in P. syringae, some of the phytotoxins produced by some P. syringae strains (Bender et al. 1999) are known to have antimicrobial activity and may have evolved under selection pressure during competition with other microbes in the environment. Several examples of putative dual-use traits related to pathogenic and environmental fitness have been demonstrated for human pathogens (Morris et al. 2009). Those traits are involved in the formation of biofilms, resistance to predation by nematodes or protists, iron sequestration, oxidation, and resistance to antibiotic compounds. For example, Pseudomonas aeruginosa and Vibrio spp. are abundant in soil and aquatic substrates known to be environmental reservoirs (Vezzulli et al. 2010; Selezska et al. 2012). They possess T3SSs similar to P. syringae to deliver effectors to eukaryotic cells and infect their human hosts. Evidence supports the hypothesis that their T3SSs were originally associated with bacterial survival in soil and water as defense against bacteriovorous amoebae (Matz et al. 2008, 2011). But grazing is not the only selection pressure at the origin of traits involved in virulence. For example, seawater is a reservoir of Vibrio cholerae and Vibrio sp. use the same factors associated with human intestinal colonization for attachment to chitin shells of crustaceans (Vezzulli et al. 2010; Pruzzo et al. 2008). Also, part of the resistome of clinical bacteria is shared with the soil microbiome. A metagenomic approach showed the presence of resistance genes to five classes of antibiotics in nonpathogenic soil-dwelling proteobacteria that were identical to antibiotic resistance genes of human pathogens (e.g., P. aeruginosa, Klebsellia pneumonia, and Acinetobacter baumanii) (Forsberg et al. 2012).
An example of the importance of the non-host environment for an opportunistic human pathogen is Staphylococcus aureus. Genomic diversity, evolutionary relationships, and methicillin resistance trait were investigated by Fitzgerald et al. (2001) on the basis of 36 clinical isolates. The authors showed that the methicillin resistance gene had been horizontally acquired several times independently. Where these events occurred is still unclear, but since Staphylococcus aureus is present in the rhizosphere (Berg et al. 2005), it may very well be that they occurred in the soil.
Therefore, conclusions about the evolutionary pressures shaping pathogen genomes are biased if they are made on the sole basis of pathogenic populations isolated from the host, while the species has a life style that includes non-host environments. Because this is the case for P. syringae, interpretation of genetic changes underlying virulence or host range based on strains isolated only from crop plants might be misleading, like in the studies of Baltrus et al. (2011) or Cai et al. (2011b).
The water cycle does not only consist of water from hydrological networks, but also includes the biosphere and consequently wild plants. P. syringae populations on wild plants or non-diseased crops have usually not been taken into account in population genetics and population genomics studies of P. syringae so far. Yet, P. syringae is an epiphyte and pathogenic lineages can be present in the phyllosphere of non-hosts and hosts at high population density without causing disease (Morris et al. 2008; Hirano and Upper 2000). No population genomic study has yet been performed to determine how genetically diverse epiphytic P. syringae populations are and how different or similar their virulence traits are compared to pathogen populations on diseased hosts. Including wild plants in studies of P. syringae evolution may thus also give new insight into plant adaptation. Indeed, as P. syringae crop pathogens have close relatives in the environment, crop plants have wild relatives outside of cropping areas. We can assume that the pool of wild relatives of crops plays an important role in host adaptation of environmental P. syringae population. The emergence of a P. syringae crop pathogen might possibly involve a progressive increase in fitness on wild relatives and lead to a population adapted to relatives of a certain crop from which a clonal crop pathogen may occasionally emerge, as was suggested for Psa (McCann et al. 2013). Importantly, most of the plant families of agricultural interest are present in alpine ecosystems like Rosaceae, Cruciferae, Fabaceae, Brassicaceae, Fagaceae, or Solanaceae (Sherman et al. 2008; Taberlet et al. 2012). Since Monteil et al. (2012) estimated that about 108 cfu of P. syringae may reside in one square meter of alpine vegetation, wild plants of alpine ecosystems should be included when investigating the adaptation of P. syringae to crop plants.
4.9 Conclusion
So far, population genomic studies based on whole-genome sequences of P. syringae pathogens are relatively few and relatively small in regard to the number of strains that were analyzed. However, because WGS allows us to detect even a single mutation in a whole genome, population genomic approaches show great promise for describing evolutionary processes at a resolution that was inconceivable just a few years ago. Studies of human pathogens have pioneered the field and will facilitate similar studies on bacterial plant pathogens like P. syringae. In the past 10 years, population genomic studies of human pathogens brought new insights into how populations are structured, how genes are exchanged, and how populations adapt to a pathogenic lifestyle. By identifying the processes leading to the emergence of virulent isolates, host specialization, and pathogen movement, we expect to be able to better prevent and control crop diseases in the future. We may even be able to develop advanced warning systems alerting us of new pathogens or pathogen variants allowing more time to develop resistant cultivars.
Sampling remains the most important issue to address in population genomic studies in order to obtain results that reflect the actual pathogen population. Depending on the question, the factors to consider in the choice of strains in regard to time, location, host, and/or environment of isolation will be different. Importantly though, evolutionary history reconstruction of non-obligate pathogens like P. syringae will always need to take into account more than the diversity within the diseased crop host. They need to extend to the pathogen population residing on non-diseased hosts and non-host plants and non-plant environments. We expect that following these guidelines, population genomic studies of P. syringae will significantly deepen and broaden our view of plant pathogen ecology, evolution, epidemiology, and molecular plant–microbe interactions.
Reference
Achtman M (2008) Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiolo 62:53–70
Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, Grp SEMS (2012) Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8(6):e1002776. doi:10.1371/journal.ppat.1002776
Almeida NF, Yan S, Lindeberg M, Studholme DJ, Schneider DJ, Condon B, Liu H, Viana CJ, Warren A, Evans C, Kemen E, Maclean D, Angot A, Martin GB, Jones JD, Collmer A, Setubal JC, Vinatzer BA (2009) A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 22(1):52–62. doi:10.1094/MPMI-22-1-0052
Balestra GM, Taratufolo MC, Vinatzer BA, Mazzaglia A (2013) A multiplex PCR assay for detection of Pseudomonas syringae pv. actinidiae and differentiation of populations with different geographic origin. Plant Dis 97(4):472–478. doi:10.1094/pdis-06-12-0590-re
Baltrus DA, Nishimura MT, Dougherty KM, Biswas S, Mukhtar MS, Vicente J, Holub EB, Dangl JL (2012) The molecular basis of host specialization in bean pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 25(7):877–888. doi:10.1094/mpm1-08-11-0218
Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Shahid Mukhtar M, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7(7):e1002132
Bart R, Cohn M, Kassen A, McCallum EJ, Shybut M, Petriello A, Krasileva K, Dahlbeck D, Medina C, Alicai T, Kumar L, Moreira LM, Rodrigues Neto J, Verdier V, Angelica Santana M, Kositcharoenkul N, Vanderschuren H, Gruissem W, Bernal A, Staskawicz BJ (2012) High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc Natl Acad Sci USA 109(28):E1972–E1979. doi:10.1073/pnas.1208003109
Bender CL, Alarcon-Chaidez F, Gross DC (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63(2):266–292
Berg G, Eberl L, Hartmann A (2005) The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol 7(11):1673–1685. doi:10.1111/j.1462-2920.2005.00891.x
Bryan MK (1933) Bacterial speck of tomatoes. Phytopathology 23:897–904
Bull CT, Clarke CR, Cai R, Vinatzer BA, Jardini TM, Koike ST (2011) Multilocus sequence typing of Pseudomonas syringae sensu lato confirms previously described genomospecies and permits rapid identification of P. syringae pv. coriandricola and P. syringae pv. apii causing bacterial leaf spot on parsley. Phytopathology 101(7):847–858. doi:10.1094/PHYTO-11-10-0318
Butler MI, Stockwell PA, Black MA, Day RC, Lamont IL, Poulter RTM (2013) Pseudomonas syringae pv. actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China. PLoS One 8(2):e57464. doi:10.1371/journal.pone.0057464
Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, Zaccardelli M, Setubal JC, Morales-Lizcano NP, Bernal A, Coaker G, Baker C, Bender CL, Leman S, Vinatzer BA (2011a) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7(8):e1002130. doi:10.1371/journal.ppat.1002130
Cai R, Yan S, Liu H, Leman S, Vinatzer BA (2011b) Reconstructing host range evolution of bacterial plant pathogens using Pseudomonas syringae pv. tomato and its close relatives as a model. Infect Genet Evol 11(7):1738–1751. doi:10.1016/j.meegid.2011.07.012
Clarke CR, Cai R, Studholme DJ, Guttman DS, Vinatzer BA (2010) Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol Plant Microbe Interact 23(2):198–210. doi:10.1094/MPMI-23-2-0198
Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki Y, Martin GB, Leman S, Felix G, Vinatzer BA (2013) Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200(3):847–860. doi:10.1111/nph.12408
Corander J, Marttinen P, Siren J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. Bmc Bioinform 9(1):539. doi:10.1186/1471-2105-9-539
Coscolla M, Gonzalez-Candelas F (2007) Population structure and recombination in environmental isolates of Legionella pneumophila. Environ Microbiol 9(3):643–656. doi:10.1111/j.1462-2920.2006.01184.x
Coscolla M, Gonzalez-Candelas F (2009) Comparison of clinical and environmental samples of Legionella pneumophila at the nucleotide sequence level. Infect Genet Evol 9(5):882–888. doi:10.1016/j.meegid.2009.05.013
Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, Zhang Y, Zheng H, Qin N, Xiao X, Wu M, Wang X, Zhou D, Qi Z, Du Z, Wu H, Yang X, Cao H, Wang H, Wang J, Yao S, Rakin A, Li Y, Falush D, Balloux F, Achtman M, Song Y, Wang J, Yang R (2013) Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA 110(2):577–582. doi:10.1073/pnas.1205750110
den Bakker HC, Switt AIM, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, Wright EM, Fang R, Davis M, Root T, Schoonmaker-Bopp D, Musser KA, Villamil E, Waechter H, Kornstein L, Furtado MR, Wiedmann M (2011) A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp enterica Serovar Montevideo pulsed-field gel electrophoresis type. Appl Environ Microbiol 77(24):8648–8655. doi:10.1128/aem.06538-11
Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Clifton SW, Weinstock GM, McClelland M (2013) Evolutionary genomics of Salmonella enterica subspecies. MBio 4(2): e00579–12. doi:10.1128/mBio.00579-12
Diallo MD, Monteil CL, Vinatzer BA, Clarke CR, Glaux C, Guilbaud C, Desbiez C, Morris CE (2012) Pseudomonas syringae naturally lacking the canonical type III secretion system are ubiquitous in nonagricultural habitats, are phylogenetically diverse and can be pathogenic. ISME J 6(7):1325–1335. doi:10.1038/ismej.2011.202
Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G, Sangal V, Anjum MF, Achtman M, Falush D, Donnelly P (2011) Recombination and population structure in Salmonella enterica. Plos Genet 7(7):e1002191. doi:10.1371/journal.pgen.1002191
Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13(9):601–612. doi:10.1038/nrg3226
Didelot X, Darling A, Falush D (2009) Inferring genomic flux in bacteria. Genome Res 19(2):306–317. doi:10.1101/gr.082263.108
Didelot X, Lawson D, Darling A, Falush D (2010) Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186(4):U1435–U1567. doi:10.1534/genetics.110.120121
Dye DW, Bradbury JF, Goto M, Hayward AC, Lelliott RA, Schroth MN (1980) International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev Plant Pathol 59(4):153–168
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567–1587
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J Cell Mol Biol 18(3):265–276
Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM (2001) Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci USA 98(15):8821–8826. doi:10.1073/pnas.161098098
Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337(6098):1107–1111. doi:10.1126/science.1220761
Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont PAD (1999) DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int J Syst Bacteriol 49:469–478
Gomez-Alpizar L, Carbone I, Ristaino JB (2007) An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proc Natl Acad Sci USA 104(9):3306–3311. doi:10.1073/pnas.0611479104
Goode MJ, Sasser M (1980) Prevention—the key to controlling bacterial spot and bacterial speck of tomato. Plant Dis 64(9):831–834
Green S, Studholme DJ, Laue BE, Dorati F, Lovell H, Arnold D, Cottrell JE, Bridgett S, Blaxter M, Huitema E, Thwaites R, Sharp PM, Jackson RW, Kamoun S (2010) Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum. PLoS One 5(4):e10224. doi:10.1371/journal.pone.0010224
Guidot A, Elbaz M, Carrere S, Siri MI, Pianzzola MJ, Prior P, Boucher C (2009) Specific genes from the potato brown rot strains of Ralstonia solanacearum and their potential use for strain detection. Phytopathology 99(9):1105–1112. doi:10.1094/phyto-99-9-1105
Hansen EM, Reeser PW, Sutton S (2012) Phytophthora beyond agriculture. Annu Rev Phytopathol. doi:10.1146/annurev-phyto-081211-172946
Hirano SS, Upper CD (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64(3):624–653
Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF (2008) Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320(5879):1081–1085. doi:10.1126/science.1157890
Jolley KA, Maiden MCJ (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. Bmc Bioinform 11(1):595. doi:10.1186/1471-2105-11-595
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. doi:10.1038/nature05286
Joseph SJ, Didelot X, Rothschild J, de Vries HJC, Morre SA, Read TD, Dean D (2012) Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol 29(12):3933–3946. doi:10.1093/molbev/mss198
Kimbrel JA, Givan SA, Temple TN, Johnson KB, Chang JH (2011) Genome sequencing and comparative analysis of the carrot bacterial blight pathogen, Xanthomonas hortorum pv. carotae M081, for insights into pathogenicity and applications in molecular diagnostics. Mol Plant Pathol 12(6):580–594. doi:10.1111/j.1364-3703.2010.00694.x
Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz B, Jones JDG, Zipfel C (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotech 28(4):365–369. http://www.nature.com/nbt/journal/v28/n4/abs/nbt.1613.html#supplementary-information
Lovell HC, Mansfield JW, Godfrey SAC, Jackson RW, Hancock JT, Arnold DL (2009) Bacterial evolution by genomic Island transfer occurs via DNA transformation in planta. Curr Biol 19(18):1586–1590. doi:10.1016/j.cub.2009.08.018
Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou JJ, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95(6):3140–3145. doi:10.1073/pnas.95.6.3140
Mann RA, Smits THM, Buhlmann A, Blom J, Goesmann A, Frey JE, Plummer KM, Beer SV, Luck J, Duffy B, Rodoni B (2013) Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PLoS One 8(2):e55644. doi:10.1371/journal.pone.0055644
Martinez JL (2013) Bacterial pathogens: from natural ecosystems to human hosts. Environ Microbiol 15(2):325–333. doi:10.1111/j.1462-2920.2012.02837.x
Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J (2012) Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40(1):e6. doi:10.1093/nar/gkr928
Matz C, Moreno AM, Alhede M, Manefield M, Hauser AR, Givskov M, Kjelleberg S (2008) Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J 2(8):843–852. doi:10.1038/ismej.2008.47
Matz C, Nouri B, McCarter L, Martinez-Urtaza J (2011) Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 6(5):e20275
Mazzaglia A, Studholme DJ, Taratufolo MC, Cai R, Almeida NF, Goodman T, Guttman DS, Vinatzer BA, Balestra GM (2012) Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS One 7(5): e36518. doi:10.1371/journal.pone.0036518
McCann HC, Rikkerink EHA, Bertels F, Fiers M, Lu A, Rees-George J, Andersen MT, Gleave AP, Haubold B, Wohlers MW, Guttman DS, Wang PW, Straub C, Vanneste J, Rainey PB, Templeton MD (2013) Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog 9(7):e1003503. doi:10.1371/journal.ppat.1003503
McCarter SM, Jones JB, Gitaitis RD, Smitley DR (1983) Survival of Pseudomonas syringae pv tomato in association with tomato seed, soil, host tissue, and epiphytic weed hosts in Georgia. Phytopathology 73(10):1393–1398
Monteil CL, Cai R, Liu H, Mechan Llontop ME, Leman S, Studholme DJ, Morris CE, Vinatzer BA (2013) Non-agricultural reservoirs contribute to emergence and evolution of Pseudomonas syringae crop pathogens. New Phytol. doi:10.1111/nph.12316
Monteil CL, Guilbaud C, Glaux C, Lafolie F, Soubeyrand S, Morris CE (2012) Emigration of the plant pathogen Pseudomonas syringae from leaf litter contributes to its population dynamics in alpine snowpack. Environ Microbiol 14(8):2099–2112. doi:10.1111/j.1462-2920.2011.02680.x
Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M (2010) Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 42(12):1140–1143. doi:10.1038/ng.705
Morris CE, Bardin M, Kinkel LL, Moury B, Nicot PC, Sands DC (2009) Expanding the paradigms of plant pathogen life history and evolution of parasitic fitness beyond agricultural boundaries. PLoS Pathog 5(12):e1000693
Morris CE, Monteil CL, Berge O (2013) The life history of Pseudomonas syringae: linking agriculture and Earth system processes. Annu Rev Phytopathol 51:85–104
Morris CE, Sands DC, Vanneste JL, Montarry J, Oakley B, Guilbaud C, Glaux C (2010) Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. mBio 1(3):1–10
Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffiere A, Yan SC, Dominguez H, Thompson BM (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2(3):321–334. doi:10.1038/ismej.2007.113
Nosil P, Buerkle A (2010) Population genomics. Nat Educ Knowl 3(10):8
Nosil P, Funk DJ, Ortiz-Barrientos D (2009) Divergent selection and heterogeneous genomic divergence. Mol Ecol 18(3):375–402. doi:10.1111/j.1365-294X.2008.03946.x
Nuebel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, Zemlickova H, Leblois R, Wirth T, Jombart T, Balloux F, Witte W (2010) A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog 6(4):e1000855. doi:10.1371/journal.ppat.1000855
Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, Moreira L, Ortiz B, Stouthamer R (2010) Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of pierce’s disease of grapevine in the US. PLoS One 5(11):e15488. doi:10.1371/journal.pone.0015488
Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant Microbe Interact 17(6):696–706. doi:10.1094/MPMI.2004.17.6.696
Pitman AR, Jackson RW, Mansfield JW, Kaitell V, Thwaites R, Arnold DL (2005) Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Current biology CB 15(24):2230–2235. doi:10.1016/j.cub.2005.10.074
Potnis N, Krasileva K, Chow V, Almeida NF, Patil PB, Ryan RP, Sharlach M, Behlau F, Dow JM, Momol MT, White FF, Preston JF, Vinatzer BA, Koebnik R, Setubal JC, Norman DJ, Staskawicz BJ, Jones JB (2011) Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12(1):146. doi:10.1186/1471-2164-12-146
Pruzzo C, Vezzulli L, Colwell RR (2008) Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol 10(6):1400–1410
Sarkar SF, Gordon JS, Martin GB, Guttman DS (2006) Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174(2):1041
Sarkar SF, Guttman DS (2004) Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol 70(4):1999–2012. doi:10.1128/aem.70.4.1999-2012.2004
Sarris PF, Trantas EA, Baltrus DA, Bull CT, Wechter WP, Yan S, Ververidis F, Almeida NF, Jones CD, Dangl JL, Panopoulos NJ, Vinatzer BA, Goumas DE (2013) Comparative genomics of multiple strains of Pseudomonas cannabina pv. alisalensis, a potential model pathogen of both Monocots and Dicots. PLoS One 8(3):e59366. doi:10.1371/journal.pone.0059366
Scortichini M, Marcelletti S, Ferrante P, Petriccione M, Firrao G (2012) Pseudomonas syringae pv. actinidiae: a re-emerging, multi-faceted, pandemic pathogen. Mol Plant Pathol 13(7):631–640. doi:10.1111/j.1364-3703.2012.00788.x
Selezska K, Kazmierczak M, Müskeb M, Garbe J, Schobert M, Häussler S, Wiehlmann L, Rhode C, Sikorski J (2012) Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Env Microbiol 14(8):1952–1967
Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, Polz MF, Alm EJ (2012) Population genomics of early events in the ecological differentiation of bacteria. Science 336(6077):48–51. doi:10.1126/science.1218198
Sheppard SK, Colles FM, McCarthy ND, Strachan NJC, Ogden ID, Forbes KJ, Dallas JF, Maiden MCJ (2011) Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol 20(16):3484–3490. doi:10.1111/j.1365-294X.2011.05179.x
Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJC, Ogden ID, Forbes K, French NP, Carter P, Miller WG, McCarthy ND, Owen R, Litrup E, Egholm M, Affourtit JP, Bentley SD, Parkhill J, Maiden MCJ, Falush D (2013a) Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22(4):1051–1064. doi:10.1111/mec.12162
Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MCJ, Parkhill J, Falush D (2013b) Genome-wide association study identifies vitamin B-5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci USA 110(29):11923–11927
Sherman R, Mullen R, Haomin L, Zhendong F, Yi W (2008) Spatial patterns of plant diversity and communities in Alpine ecosystems of the Hengduan Mountains, Northwest Yunnan China. J Plant Ecol 1(2):117–136. doi:10.1093/jpe/rtn012
Smith JM, Smith NH, Orourke M, Spratt BG (1993) How clonal bacteria are? Proc Natl Acad Sci USA 90(10):4384–4388. doi:10.1073/pnas.90.10.4384
Sohn KH, Saucet SB, Clarke CR, Vinatzer BA, O’Brien HE, Guttman DS, Jones JDG (2012) Hop AS1 recognition significantly contributes to Arabidopsis non-host resistance to Pseudomonas syringae pathogens. New Phytol 193(1):58–66. doi:10.1111/j.1469-8137.2011.03950.x
Stukenbrock EH, McDonald BA (2008) The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 46:75–100. doi:10.1146/annurev.phyto.010708.154114
Sun WX, Dunning FM, Pfund C, Weingarten R, Bent AF (2006) Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18(3):764–779. doi:10.1105/tpc.105.037648
Taberlet P, Zimmermann NE, Englisch T, Tribsch A, Holderegger R, Alvarez N, Niklfeld H, Coldea G, Mirek Z, Moilanen A, Ahlmer W, Marsan PA, Bona E, Bovio M, Choler P, Cieślak E, Colli L, Cristea V, Dalmas J-P, Frajman B, Garraud L, Gaudeul M, Gielly L, Gutermann W, Jogan N, Kagalo AA, Korbecka G, Küpfer P, Lequette B, Letz DR, Manel S, Mansion G, Marhold K, Martini F, Negrini R, Niño F, Paun O, Pellecchia M, Perico G, Piękoś-Mirkowa H, Prosser F, Puşcaş M, Ronikier M, Scheuerer M, Schneeweiss GM, Schönswetter P, Schratt-Ehrendorfer L, Schüpfer F, Selvaggi A, Steinmann K, Thiel-Egenter C, van Loo M, Winkler M, Wohlgemuth T, Wraber T, Gugerli F, IntraBioDiv C (2012) Genetic diversity in widespread species is not congruent with species richness in alpine plant communities. Ecol Lett 15(12):1439–1448. doi:10.1111/ele.12004
Vezzulli L, Pruzzo C, Huq A, Colwell RR (2010) Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ Microbiol Rep 2(1):27–33. doi:10.1111/j.1758-2229.2009.00128.x
Wasukira A, Tayebwa J, Thwaites R, Paszkiewicz K, Aritua V, Kubiriba J, Smith J, Grant M, Studholme DJ (2013) Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pv. musacearum. Genes. doi:10.3390/genes3030361
Wei C-F, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin N-C, Martin GB, Huang H-C, Collmer A (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51(1):32–46. doi:10.1111/j.1365-313X.2007.03126.x
Xu X, Miller SA (2013) First report of bacterial leaf spot of parsley caused by Pseudomonas syringae pv. coriandricola in Ohio. Plant Dis 97(7):988. doi:10.1094/pdis-12-12-1181-pdn
Yan S, Liu H, Mohr TJ, Jenrette J, Chiodini R, Zaccardelli M, Setubal JC, Vinatzer BA (2008) Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Appl Environ Microbiol 74(10):3171–3181. doi:10.1128/aem.00180-08
Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, Iqbal Z, Rimmer AJ, Cule M, Ip CLC, Didelot X, Harding RM, Donnelly P, Peto TE, Crook DW, Bowden R, Wilson DJ (2012) Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci USA 109(12):4550–4555. doi:10.1073/pnas.1113219109
Young JM, Park DC, Shearman HM, Fargier E (2008) A multilocus sequence analysis of the genus Xanthomonas. Syst Appl Microbiol 31(5):366–377. doi:10.1016/j.syapm.2008.06.004
Zhou Z, McCann A, Litrup E, Murphy R, Cormican M, Fanning S, Brown D, Guttman DS, Brisse S, Achtman M (2013) Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet 9(4):e1003471. doi:10.1371/journal.pgen.1003471
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Vinatzer, B.A., Monteil, C.L. (2014). Pseudomonas syringae Genomics: From Comparative Genomics of Individual Crop Pathogen Strains Toward Population Genomics. In: Gross, D., Lichens-Park, A., Kole, C. (eds) Genomics of Plant-Associated Bacteria. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-55378-3_4
Download citation
DOI: https://doi.org/10.1007/978-3-642-55378-3_4
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-55377-6
Online ISBN: 978-3-642-55378-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)