Abstract
Cooperativity and allostery within catalytically active protein complexes are important concepts for control of phenotypic outcome within a living system. In this chapter, the putative molecular determinants for cooperativity and allostery of a multi-subunit antibiotic efflux pump complex are discussed. While resisting multiple antibiotic stresses, Gram-negative bacteria deploy a network of single and multicomponent drug efflux pumps to reduce the concentration of the drugs in the cytoplasm and periplasm. The dual membrane setup imposes a particular challenge for transport of drugs from the cytoplasm to the medium, and current hypothesis describes a multistep transport path of drugs across the inner and outer membrane by the action of different drug efflux pumps. Drug transport from the cytoplasm to the periplasm is catalyzed by single component drug efflux systems belonging to the ABC-transporter superfamily, the Major Facilitator Superfamily (MFS), The Multi Antimicrobial Extrusion (MATE) Family, and the Small Multidrug Resistance (SMR) family. Transport from the periplasm across the outer membrane is catalyzed by a tripartite transport complex consisting of an inner membrane Resistance-Nodulation-cell Division (RND) transporter, an adaptor protein, and an outer membrane channel. Cooperative effects occur at multiple levels within this three component RND system: Anticipated allostery for binding of ligands (drugs and H+) at the level of the protomer, interdependence of the protomers within the trimer, and cooperative effects between the three components within the entire transport complex. The molecular determinants of multiple substrates binding at different sites within the inner membrane transporter and its coupling to H+ binding and transport are being described here for the paradigm tripartite transport machinery AcrAB-TolC from Escherichia coli. High resolution structures of the three components, the anticipated three component setup, and a multitude of biophysical and biochemical data are combined to address the overall molecular understanding of secondary H+/drug antiport. With focus on the inner membrane RND component, drug and H+ binding and transport cooperativity are exemplified by exploiting a mechanism based on binding change and the strict coupling to the influx of H+.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Multidrug efflux
- Functional rotation
- Cooperativity
- Tripartite RND systems
- Proton translocation
- Drug/H+ antiporter
- Multiple drug binding
- Conformational changes
- Transporter states
- Membrane transport
9.1 Introduction
Multidrug resistance by bacteria is an increasing concern as the arsenal of antibiotics used to treat infections has been described as being increasingly ineffective (Bush et al. 2011). Bacteria defend themselves against stress by deployment of resistance mechanisms which are either constitutively present by moonlighting activities of systems with different physiological functions or induced upon contact with stress signals. For antibiotic stress, common defense mechanisms are modification of the antibiotic by hydrolysis, phosphorylation, acetylation, or adenylation of the specific drugs, sequestering of the toxic compound by proteins with nonessential function in the cell, or modification of the antibiotic target by mutation or methylation (Walsh 2000). These resistance mechanisms can be combined to render the bacteria resistant against multiple antibiotics. Since entry of the toxic compound into the cells is the first event to reach the target(s) inside the cells, the first line of defense for many bacteria is to prevent entry of the drug via the membrane. This is accomplished by transport proteins which extrude the antibiotics from within the cell or cell membrane towards the outside. For Gram-negative bacteria, containing an inner and out membrane, the downregulation of the expression of porins, often a preferred route for drugs to enter the periplasmic space between the two membranes, is another effective means to resist the incoming threat (Davin-Regli et al. 2008). Gram-negative cells face an interesting ambiguity in relation to drug permeation, since the outer membrane presents on the one hand a protective barrier for drugs entering the periplasmic space, but on the other hand, drugs expelled out of the cytoplasm across the inner membrane accumulate in the limited space of the periplasm leading to very high concentrations in this compartment and therefore limited effectiveness of inner membrane transporters to clean the cytoplasm due to the reflux of drugs along the inward gradient. Past research has shown that Gram-negative bacteria install transport complexes which span the inner and outer membrane as to avoid drug accumulation in the periplasmic space (Nikaido and Pagès 2012). The best characterized complexes are the tripartite systems build up out of an inner membrane transporter, connected to an outer membrane channel and stabilized by a periplasmic adaptor molecule. The action of these systems prevents drug accumulation in the periplasm, thereby avoiding large gradients across the inner-membrane and/or lethal concentrations of drugs inside the periplasm (e.g., of β-lactam antibiotics).
Infections by (multidrug-resistant) Gram-negative bacteria (e.g., Escherichia coli, Acinetobacter baumanii, and Pseudomonas aeruginosa) became in the last decades a major health issue (Vila and Martínez 2008). The additional outer membrane of Gram-negative bacteria is contributing to a reduced permeability for drugs into the bacterial cytosol. The resistance phenotype in Gram-negative bacteria appears to be largely connected with the expression of efflux pump genes. In E. coli, for example, 20 different membrane transporters were identified to be involved in mediating antibiotic resistances (Nishino and Yamaguchi 2001; Sulavik et al. 2001). In general, there are five superfamilies of membrane-embedded transporters that are involved in antibiotic resistances and are summarized in Table 9.1.
Many clinical isolates of multidrug resistant pathogens exhibit overexpression of endogenous membrane transporters, particularly, members of the Resistance-Nodulation-cell Division (RND) superfamily (Piddock 2006). For example, overexpression of the RND transporter genes acrB (E. coli) or mexB (P. aeruginosa) is sufficient to render bacterial cells resistant against a huge variety of structurally unrelated molecules, including antibiotics (Nikaido 1998). Disruption of these genes from the clinical isolates chromosome renders these cells susceptible for many antibiotics, indicating the particular importance of these efflux pumps (Piddock 2006).
The tripartite system, consisting of AcrA, AcrB, and TolC (AcrAB-TolC), was shown to play a central role in generating a drug resistant phenotype (Nishino and Yamaguchi 2001). Consequently, acrB-deficient (or acrA respectively tolC-deficient) E. coli mutants were rendered largely susceptible to a wide variety of antibiotics, disinfectants, and dyes (see also Chaps. 10 and 11). The deletion of other transporter genes has, relative to the acrB-deficient E. coli mutant, a smaller impact on the resistance phenotype. Tal and Schuldiner (Tal and Schuldiner 2009) revealed by combinatorial knock-outs of emrE, mdfA, and acrB in E. coli that drug efflux is most likely organized in a coordinated network, where several transporters with overlapping substrate spectra are transporting drugs from the cytosol into the periplasm from where these are transported by AcrAB-TolC across the outer membrane (Fig. 9.1). This cooperative behavior between single component efflux transporters and tripartite efflux systems has been reported also earlier on in the case of resistance against tetracycline and chloramphenicol (de Cristóbal et al. 2006; Lee et al. 2000).
Efflux-mediated resistance is most likely organized in a coordinated network. Transporters of the inner membrane like EmrE and MdfA are transporting substrates from the cytosol into the periplasm. Drugs from the periplasm are then transported by AcrAB-TolC across the outer membrane [figure taken from Tal and Schuldiner (2009)]
The AcrAB-TolC three-component system has been studied most intensively on a structural and functional level, and considerable research effort has also been focused on other tripartite RND efflux systems, like MexAB-OprM from P. aeruginosa as well (Nikaido and Takatsuka 2009). X-ray crystallographic studies have been conducted on several RND systems, which are, apart from the systems mentioned above, CusCBA from E. coli (Su et al. 2011a), and the SecDF protein export machinery from Thermus thermophilus (Tsukazaki et al. 2011). We will mainly focus on the work on E. coli AcrAB-TolC with references to the results obtained from homolog systems. We will shortly introduce AcrA and TolC and the current knowledge on tripartite assembly, while most of this chapter will focus on the functional and structural analysis including allosteric and cooperative activities displayed by the trimeric RND inner membrane antiporter AcrB.
9.1.1 The Membrane Fusion Protein AcrA
AcrA is a member of the membrane fusion protein (MFP) family, and its gene is cotranscribed in E. coli with acrB (Ma et al. 1995). Like all subunits of the tripartite system, AcrA is indispensable for the AcrAB-TolC-mediated resistance. AcrA is postulated to support interactions between AcrB and TolC in a scaffold-like manner (Tikhonova et al. 2011). However, recent studies indicate alternative functions for AcrA besides its adaptor function and might include transduction of conformational changes between TolC and AcrB, as well as playing a role in substrate specificity, most likely caused by an indirect effect via AcrB (Weeks et al. 2010; Zgurskaya et al. 2009).
AcrA is a periplasmic soluble protein, which is secreted across the inner membrane via a signal peptide-mediated export mechanism (Kawabe et al. 2000). Upon cleavage of the N-terminal signal peptide, the newly N-terminally exposed cysteine residue at position 25 is palmitoylated and anchors AcrA to the inner membrane. Palmitoylation appears not to be essential for the function of AcrA within the tripartite system (Zgurskaya and Nikaido 1999), but surface plasmon resonance (SPR) spectroscopy experiments indicated that the lipidation of AcrA is important for AcrA-dimerization, which resulted in increased affinity for AcrB in comparison to the AcrA monomer (Tikhonova et al. 2011).
The structure of AcrA can be described by three globular domains (membrane proximal domain, β-barrel domain, and lipoyl domain) and a helical domain (α-hairpin domain) (Fig. 9.2); a molecular setup shared by the structurally elucidated close homologues MexA (Higgins et al. 2004; Akama et al. 2004) and CusB (Su et al. 2009).
Structures of MexA (AcrA) and TolC. (a) Structure of MexA (pdb entry 2V4D, homolog of AcrA) in cartoon representation. MexA consists of 4 domains, where the lipoyl domain (yellow), the β-barrel domain (magenta), and the membrane proximal domain (green) are globular. The α-hairpin domain consists of two α-helices (blue). The general structures of MexA and AcrA are highly congruent to each other, whereas the α-hairpin domain of MexA is by 7 amino acids shorter than AcrA. (b, c) Structure of TolC (pdb entry 2VDE) in cartoon representation. The monomers are color-coded in blue, yellow, and red, respectively. (b) View on TolC parallel to the membrane plane. (c) View from the periplasmic space on TolC in its open state (Koronakis et al. 2000)
9.1.2 The Outer Membrane Factor TolC
TolC (tolerance to colicin E1) of E. coli is a member of the outer membrane factor (OMF) family of membrane proteins and encoded by a single gene at a location on the chromosome unrelated to the acrAB operon. Like other members of the OMF family, TolC is embedded in the outer membrane of Gram-negative bacteria and serves many tripartite systems in E. coli including AcrA-AcrB-TolC (Symmons et al. 2009), EmrA-EmrB-TolC (Tanabe et al. 2009), HlyB-HlyD-TolC (Balakrishnan et al. 2001), and MacA-MacB-TolC (Lu and Zgurskaya 2012).
Trimeric TolC (Koronakis et al. 2000) contains an outer membrane embedded 12-sheeted β-barrel domain, a cylinder-like α-helical domain protruding 100 Å into the periplasm, and an equatorial domain consisting of laterally positioned short N- and C-terminal helices at the equator of the α-helical cylinder (Fig. 9.2). TolC crystal structures in the closed (Koronakis et al. 2000), open (Bavro et al. 2008) and the transition state (Pei and Hinchliffe 2011) demonstrated that the opening and closing of the TolC α-helical cylinder is accomplished by an iris-like movement, reminiscent of aperture opening/closing of optical devices like cameras or microscopes.
9.1.3 Assembly of RND-MFP-OMF Tripartite Systems
The assembly of the RND-MFP-OMF tripartite system is a heavily researched field and a critically discussed topic in the field of efflux transporters. Due to the distribution of RND and OMF proteins in two distinct membranes, the technical challenges to obtain structural and in vitro data are unparalleled. Nevertheless, several biochemical approaches using appropriate members of RND-MFP-OMF tripartite systems were able to indicate interactions of the single components, whereas the MFP component seems to play a central role in the assembly of tripartite systems. For example, the MFP component MexA was successfully co-purified with OprM (OMF) in the absence of MexB (RND) and also within the complete MexA-MexB-OprM tripartite system in pull down experiments (Mokhonov et al. 2004). MexA, furthermore, co-purified with OprM, whereas MexB was not able to be co-purified as bipartite complex, neither as MexB-OprM nor as MexA-MexB complex. On the other hand, spatial proximity of AcrB (RND) with AcrA (MFP) (Tikhonova and Zgurskaya 2004; Touzé et al. 2004; Zgurskaya and Nikaido 2000) and AcrB with TolC (OMF) (Tamura et al. 2005) is supported by in vivo cross-linking experiments using dithiosuccinimidylpropionate, which indicates interactions between these components. A recent study using SPR spectroscopy and size exclusion chromatography showed that N-terminal lipidation of AcrA was essential for dimerization. Lipidated and dimeric AcrA, on its part, exhibited a higher binding affinity to AcrB in comparison to unlipidated monomeric AcrA (Tikhonova et al. 2011). The affinity of the AcrA dimer was apparently pH-dependent, where experiments at pH 6 were able to detect interactions of the AcrA dimer with AcrB whereas no interactions could be detected at pH 7.5. Particularly, the stoichiometry of the tripartite system is an intensively debated aspect. So far, ratios of 4:1:1 (Akama et al. 2004), 3:1:1 (Higgins et al. 2004), 2:1:1 (Akama et al. 2004; Su et al. 2011b), and 1:1:1 (Symmons et al. 2009; Xu et al. 2011) for MFP-RND-OMF systems were postulated.
9.2 The Structure of the Resistance-Nodulation-Cell Division Pump AcrB
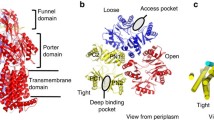
The gene locus of acrB (Acriflavin resistance protein B) was initially identified by (Nakamura et al. (1978). Deletion of acrB rendered E. coli cells highly susceptible to acriflavine. AcrB is a member of the hydrophobe/amphiphile efflux-1 family (HAE-1, transporter class number (TC#) 2.A.6.2) within the RND superfamily (TC#: 2.A.6) characterized by its typical repeated topology of 1 + 5 transmembrane (TM) helices connected by extensive soluble loops (in Gram-negative bacteria located in the periplasm) between TM helices TM1 and TM2 and TM helices TM7 and TM8, respectively (Fujihira et al. 2002; Murakami et al. 2002). AcrB functions within the tripartite AcrAB-TolC setup and extrudes antibiotics from the periplasmic side over the outer membrane. This process is energized by the proton motive force across the inner membrane. While drug binding/specificity and transport have been shown to be located at the periplasmic loops (Elkins and Nikaido 2002; Mao et al. 2002), proton translocation occurs at the transmembrane domain (TMD) of AcrB (Seeger et al. 2009; Takatsuka and Nikaido 2006). Energy transduction and drug binding/transport are therefore spatially separated, which is atypical for secondary transporters but common for primary transporters like ABC-transporters or F1Fo ATP-synthases. The substrate specificity spectrum of AcrB (within the AcrAB-TolC complex) includes structurally diverse molecules ranging from organic solvents, dyes, detergents, and antibiotics including hydrophobic β-lactams, but excluding the more hydrophilic aminoglycosides [references in Pos (2009)]. Crystallization and initial X-ray crystal diffraction of AcrB was first reported in 2002 by Pos and Diederichs (2002), and its crystal structure was first solved in 2002 by Murakami et al. (2002) in a R32 crystal lattice at 3.5 Å resolution (pdb entry 1IWG). This trimeric AcrB structure exhibited a threefold symmetry axis perpendicular to the membrane plane and indicated homotrimeric assembly, where all protomers adopted identical conformation. The general structure of AcrB can be subdivided into three major domains: (1) the transmembrane domain, (2) the porter domain, and (3) the TolC-docking domain (Fig. 9.3).
Side view parallel to the membrane plane onto the symmetric AcrB structure [pdb entry 1IWG (Murakami et al. 2002)] in cartoon representation. The structure can be subdivided in a TolC docking domain, a porter domain and a transmembrane domain. The sizes of the domains are indicated. (Center and Right) 120° rotation side views in surface representation. In the right representation, the front protomer is displayed in transparent yellow. Trimeric AcrB contains two large cavities: the “funnel” in the TolC docking domain and a most likely lipid-filled “central cavity” in the TM domain. The central cavity is connected to the periplasm by “vestibules” at the interfaces of the protomers
In the TM domain of each protomer, TM helices TM4 and TM10 are positioned in the topological core surrounded by the other 10 TM helices, whereas TM1 and TM7 are positioned in a more peripheral location (Fig. 9.4). Site-directed mutagenesis studies identified a functional H+-translocation triad of D407 and D408 positioned on TM4 and at equiplanar level K940 on TM10, as well as the additional essential residue R971 on TM11 (Murakami et al. 2002; Seeger et al. 2009; Su et al. 2006; Takatsuka and Nikaido 2006).
Overview of the AcrB structure. Top panel (Center): Side view parallel to the membrane plane of the symmetric AcrB structure (pdb entry 1IWG) in surface representation. A large and deep cavity, denoted as the “cleft,” (dotted ellipse) is visible between subdomains PC1 and PC2. (Right) View onto the porter domain from the periplasm in cartoon representation. The porter domain of AcrB is organized into PN1, PN2, PC1, and PC2 subdomains. The clefts between the PC1 and PC2 subdomains are indicated by open ellipses. The TolC docking domain is not shown in this view. (Left) View onto the TolC docking domain is shown in transparent cartoon representation. The protruding loops connecting the protomers are highlighted by transparent spheres. The Nα2 helices (pore helices) constituting the pore-like structure in the center of the AcrB trimer are displayed as opaque cartoons. Lower panel (Left): Topological view from the periplasm onto the transmembrane domain of the symmetric AcrB structure in cartoon representation. The lipid-filled central cavity is indicated by the closed circle, as well as the symmetry operator (triangle). (Center) Detailed view of one protomeric TM domain. TM helices are numbered TM1-TM12. TM4 and TM10 are encircled by the other helices, whereas TM1 and TM7 are located in a more peripheral position. Residues essential for AcrB function and most likely involved in proton translocation are shown in sticks (C: green, O: red, N: blue). (Right) Side view (90° rotation view of center figure) of one AcrB protomer with residues essential for proton translocation
The periplasmic porter domain is subdivided in four subdomains: PN1, PN2, PC1, and PC2 (Fig. 9.4). The coupling of the subdomains can be described by their primary sequence proximity (PN1 with PN2 and PC1 with PC2) and by structural proximity of PN1 with PC2 and PN2 with PC1. The PN1/PC2 and PN2/PC1 units are in addition coupled by a shared β-sheet. The PN2, PC1, and PC2 subdomains encircle the PN1 subdomain and are laterally exposed to the periplasm, with a large solvent accessible opening between PC1 and PC2, denoted as “cleft.” The PN1 subdomain is located at the interior of the AcrB trimer and exposes a short helix into the center of the trimer, denoted as Nα2 or pore helix. The pore helices of all three protomers are forming a closed pore-like structure (Fig. 9.4). These helices form strong non-covalent contacts and are considered a prime interface for the conformational communication between the protomers. The porter domain is connected at its outer membrane-proximal side to the TolC docking domain. There is experimental evidence that this domain provides the interaction site of AcrB with TolC (Tamura et al. 2005). A long loop is protruding from the TolC-docking domain of one protomer to the adjacent protomer. Studies using laser induced liquid beam ion desorption mass spectrometry (LILBID MS) and phenotypic analysis showed the essential role for this loop in the assembly of AcrB protomers to form a stable trimer (Brandstatter et al. 2011; Yu et al. 2011).
The AcrB structure reveals two large cavities (Fig. 9.4): a large central cavity, open to the cytoplasm, and a funnel-like cavity, open to the periplasm. The most likely lipid-filled “central cavity” is situated in the center of the TM domains of all three protomers and extends into the periplasmic domain. The “funnel”-like opening in the TolC-docking domain of AcrB on its part widens towards TolC and is most likely part of the AcrB exit from where substrates are further transported into the TolC channel. The “central cavity” and “funnel” are separated from each other by the pore helices. The “central cavity,” with a 30 Å diameter, is connected to the periplasm by “vestibules.” This might provide pathways for drugs tunneling along the membrane surface below the interface of PN1 and PC2 of one protomer and PN2 and PC1 of the adjacent protomer.
9.3 Functional Rotation Mechanism, Peristalsis, and Cooperativity During Catalysis
In 2006 and 2007, asymmetric crystal structures were solved independently by three groups (Murakami et al. 2006; Seeger et al. 2006; Sennhauser et al. 2007). The asymmetry was defined by the lack of the threefold symmetry in these structures, with each protomer within the homotrimer adopting a distinct conformation (see also Chap. 4). In 2009, an X-ray structure of MexB from P. aeruginosa (a close homolog of AcrB) was reported, showing a similar asymmetric composition (Sennhauser et al. 2009). The three conformations found in these asymmetric trimers were designated Loose (L), Tight (T), and Open (O) (Seeger et al. 2006), or, Access, Binding, and Extrusion (Murakami et al. 2006), respectively. It has been proposed that the different conformations represent consecutive states in a catalytic cycle for drug export defining a “functional rotation” mechanism in analogy to the binding change mechanism of the F1Fo ATP-synthase (Boyer 1997). In this model, protomers cycle consecutively through all conformations, i.e., from L to T to O back to L. In general this would suggest that, since all protomers in the trimer adopt the next conformation in this cycle, the overall trimeric composition remains “LTO” at any time of the cycle. The prerequisite conformational flexibility and interdependent conformational changes of the single protomers within the trimer were further biochemically analyzed by engineered disulfide bonds (Seeger et al. 2008; Takatsuka and Nikaido 2007). The cross-linking data showed reporting of the conformational states as predicted by the X-ray structure. Interestingly, the amount of cross-linking expected for the L and T conformational states was far beyond the theoretical one-third maximum of cross-linking expected within one trimer (data show approximately 41 % crosslinking for L or T conformations), suggesting more than one L or T protomer within the trimer. This might indicate structural flexibility of the protomers within the trimer and presence of more than one L or T state at any time of the conformational cycle, e.g., LLT or LTT (Pos 2009). Cross-linked AcrB variants with anticipated conformational restriction (i.e., cross-links in regions with large conformational changes deduced from the X-ray structure) showed strongly reduced activities. Upon reduction of the Cys-disulfide bonds, activity was recovered to a large extent, showing proper folding of the cross-linked AcrB trimers and that loss of activity was due to the conformational restraints caused by the disulfide bridges (Seeger et al. 2008).
Drug transport is proposed to start in the L-protomer, where substrates can enter the protein through the large periplasmic cleft. Indeed, using computational tools (Petrek et al. 2007) a tunnel (Tunnel 2) was detected with its lateral entrance at the cleft in the L protomer about 15 Å above the membrane plane, possibly representing an entrance site for substrates from the periplasm (Fig. 9.5). The conformation of the protomers of the symmetric structure, where the large cleft initially was identified, resembles the L protomer in the asymmetric structure. This symmetric “all-Loose” (LLL) state is suggested to be a “resting state,” where all protomers are in a state to recruit substrates (Pos 2009; Su et al. 2006).
(Left) Side view parallel to the membrane plane of asymmetric AcrB [pdb entry 4DX5 (Eicher et al. 2012)]. The protomers are shown in blue (L), yellow (T), and red (O). Drug transport is anticipated to start in the L conformation where Tunnel 2 connects the internal part of AcrB with the periplasm. The entrance of Tunnel 2 is located at the periplasmic “cleft,” approximately 15 Å above the membrane plane. After conformational transition from L to T, an additional second tunnel (Tunnel 1) is present. This tunnel connects the deep binding pocket with the periplasmic end of a groove present between TM8 and TM9. In the O protomer, Tunnels 1 and 2 disappear due to closure of the cleft by movements of the PC2 subdomain. In this conformation, Tunnel 3 is generated, connecting the now closed deep binding pocket with the “funnel” leading to the TolC channel. (Right) Top view onto the membrane plane of asymmetric AcrB (pdb entry 4DX5) in cartoon representation. The protomers are shown in blue (L), yellow (T), and red (O). Residues involved in binding of minocycline and doxorubicin in the deep binding pocket, as well as the phenylalanine-rich cluster, are represented as spheres. Detailed pictures of the deep binding pocket reveal that the deep binding pocket is closed in the L (bottom left inset) and the O (top left inset) protomer. Binding of minocycline (transparent spheres; C: green, O: red, H: grey) is prevented by sterical clashes as shown. In contrast, the deep binding pocket is widened in the T protomer (bottom right inset), which enables binding of minocycline (indicated sphere; C: green, O: red, H: grey). Residues involved in binding of minocycline (sphere representation) and doxorubicin (not shown) in the deep binding pocket, as well as the phenylalanine cluster, are shown in stick representation in the top right inset
During the transition from L to T, substrate is transported into a hydrophobic pocket, designated “deep binding pocket” (Fig. 9.5, right) (Eicher et al. 2012). In several co-crystal structures, electron densities in the T protomer deep binding pocket were assigned to minocycline or doxorubicin (Eicher et al. 2012; Murakami et al. 2006; Nakashima et al. 2011). Several residues within the deep binding pocket are involved in interaction with the substrates in the deep binding pocket, including a most prominent phenylalanine cluster (F136 + F178 + F610 + F615 + F617 + F628) (Bohnert et al. 2008). Different residues are involved in the binding of minocycline (N274 + G179 + I277 + A279 + V612 + N176) or doxorubicin (Q176 + F615 + F617 + S134 + F178 + G179 + I277 + L177). The opening of the deep binding pocket during the L to T transition entails motions within the PN2/PC1 unit (RMSD ~2.3), whereas the PN1/PC2 unit remains largely rigid (RMSD < 0.5) (Schulz et al. 2011). A second tunnel (Fig. 9.5, “Tunnel 1”) was identified connecting the deep binding pocket with an entrance at the periplasmic end of the TM8/9 interface at the height of the outer leaflet of the inner membrane (Sennhauser et al. 2007). Very recently, computational free energy analysis and corresponding mutagenesis experiments (Yao et al. 2013) indicated that based on the physiochemical properties the different drug substrates preferred either Tunnel 2 or Tunnel 1. Relatively, low molecular weight hydrophobic and lipophilic substrates are proposed to be preferably taken up via Tunnel 1, whereas larger and less hydrophobic substrates are entering AcrB via Tunnel 2. Moreover, an additional pathway was identified, which could not be identified using the X-ray structures.
In analogy to the binding change mechanism of the F1Fo ATP-synthase (Boyer 1997), binding of substrates in the T conformation is proposed to be prerequisite for the energy-dependent T to O transition step (Pos 2009). Large movements within the porter domain are apparent during the T to O transition on the periplasmic side of AcrB, where the cleft closes from a width of over ~27 Å in L and T to 18 Å in the O protomer (distances from K708 to A654) and thereby close the entrance of tunnel 2. The flexibility of the cleft was confirmed to be essential for functionality of AcrB by disulfide crosslinking experiments (Takatsuka and Nikaido 2007). The O protomer is proposed to represent the conformation where substrates exit through the AcrB funnel region to TolC. Husain and Nikaido (2010) recently reported that cysteines introduced in the vicinity of the cleft (N274C or E273C) were able to covalently link the maleimide-labeled AcrB substrate Bodipy-FL, indicating that this area of the cleft is part of the drug pathway. Additionally, computational calculations indicated a movement of doxorubicin by 8 Å from the deep binding pocket through tunnel 3 during the T to O transition. This study showed that the porter domain of AcrB exhibits a bottom-to-top zipper-like closing motion (Schulz et al. 2010; Vargiu et al. 2011), strongly supporting the peristaltic efflux pump model proposed by Seeger et al. (2006). After the release of substrates to TolC, conformational transition from O back to L is the anticipated final step of one catalytic cycle.
9.4 Proton Translocation and Energy Coupling in the AcrB Protomer
The TM domain of the AcrB protomer is organized in two repeats consisting of Repeat 1 (R1: TM1 + TM3–TM6) and Repeat 2 (R2: TM7 + TM9–TM12). In addition, elongated TM helices, TM2 and TM8, connect the PN2/PC1 unit and the PN1/PC2 unit with R1 and R2, respectively. These helices are proposed to be coupling units for the energy transduction from the TM domain to the substrate transporting porter domain. In fact, TM8 undergoes large structural rearrangements, where the periplasmic N-terminal end of the TM8 is in a coiled state in L, a helix-coil-helix state in T, and exhibits a continuous helix state in the O protomer (Murakami et al. 2006; Seeger et al. 2006; Sennhauser et al. 2007).
The 1.9 Å resolution AcrB/minocycline (pdb entry 4DX5) co-crystal structure reveals the importance of interactions along the interface of R1 and R2 for proton translocation (Eicher et al. 2012). Here two distinct states can be described: an “engaged” state in the L and T protomers in contrast to a “disengaged” state in the O protomer. In the L and T states, residues of the proton translocation triad D407, D408 (TM4), and K940 (TM10) interact directly with each other, i.e., K940 from R2 is sandwiched between D407 and D408 on TM 4 from R1 by ionic interactions (Fig. 9.6, “L”). Additional interactions across R1 and R2 are described, amongst others, by the interaction of D407 of R1 with T978 (TM11) of R2. Further interactions within one repeat are described for example between D408 and S481 (TM6). All interactions from the engaged state in the T conformation are detached in the O conformation, the “disengaged” state. D407, for example, rotates into its own repeat R1 and interacts with the backbone oxygen of G403, while D408 reorients to interact with the backbone of L442 (TM5). Also, K940 of R2 turns away from D407 and D408 and interacts in the disengaged state with N941 (TM10) and T978 (TM11) of R1.
(a) Schematic representation of the catalytic cycle of drug/H+ antiport by the AcrB protomer. A substrate molecule (S) accesses the AcrB L conformer (blue). Substrate binding induces a conformational change from L to T involving by TM2. In the T conformation (yellow), the transmembrane domain is accessible for protons from the periplasm. Protonation of the D407 and D408 residues of the proton translocation triad results in a conformational change from T to O mediated by TM8. In analogy to catalytic cycle of the F1Fo ATP-synthase, the energy consuming step is proposed to be the release of substrate molecules from the porter domain towards the TolC channel. Release of substrate triggers the reverse of the protomer to the L conformation releasing the bound protons into the cytoplasm. (b) Detailed view on proton translocation residues within the TMD of the AcrB protomer. In the L (blue) and T (yellow) conformation, proton translocation triad residues exhibit an “engaged” state, where K940 is sandwiched between D407 and D408. All three residues are charged in this “engaged” state. Substrate binding in the porter domain of the L protomer triggers the L to T transition based on structural rearrangements in the TMD. In the T conformation, residues D407, D408, and K940 of the proton translocation triad are connected to the periplasm by a continuous cluster of water molecules leading to a stepwise protonation. This triggers a substantial reorientation of the TMD helices towards the protonated “disengaged” state of the O conformer (red). Bound protons on D407 and D408 are released towards the cytoplasm during the O to L transition recovering the engaged state of the proton translocation triad (TDD TolC docking domain, TM2 transmembrane helix 2, TM8 transmembrane helix 8, TMD transmembrane domain)
Several observations point towards the central role of the T conformation in the proton translocation. In the T conformation, all triad residues D407/D408/K940 are postulated to be charged. In the high resolution AcrB/minocycline structure, a network of water molecules, exclusively present in the T state, almost continuously connects the periplasm with the titratable residues within the proton translocation triad (Fig. 9.6, “T”) (Eicher et al. 2012). Computational simulations in the group of Kandt independently suggested a major role of the water molecule network in the TMD for proton translocation (Fischer and Kandt 2011). We hypothesize that D407 accepts, in a stepwise manner, two periplasmic protons during the T to O transition, where one proton is further transported to D408. Therefore, D407 and D408 are both protonated in the O conformation (Fig. 9.6, “O”). This postulate is supported by the observation that K940H and K940R variants of AcrB are largely active in transport (Seeger et al. 2009). In contrast, K940A and K940M variants of AcrB were inactive. R971 on TM11 is also predicted to be charged in all three states and interacts with the triad in the T monomer by the coordination of a water cluster residing between D407 and R971. Protonation of D407 and D408 during the T to O transition converts the net charge of the triad from −1 to +1. This most likely weakens the interaction between R971 and the triad. Reorientation of the R971 side chain in the O conformation to neighboring side chains (e.g., F948 in TM10) and surrounding cytoplasmic bulk water might be a consequence of the electrostatic change in the triad. As a consequence the interactions, which were observable in the T conformation, are disengaged in the O conformation. The actual release of the bound protons on D407 and D408 in the O conformation is proposed to occur during the O to L transition (Fig. 9.6, “O → L”), coinciding with the release of drugs in the porter domain of AcrB towards TolC. The release of protons to the cytoplasm during the O to L transition causes the TM domain to revert to its engaged state including reorientation of D407, D408, K940, and R971 (Fig. 9.6).
More support of a directional cycling in the drug/H+ antiport catalysis and interdependence of the protomers within the RND trimer comes from mutational studies with (1) a covalent trimer of AcrB (Takatsuka and Nikaido 2009), (2) monomeric AcrB (Brandstatter et al. 2011; Yu et al. 2011), and (3) the heterotrimeric MdtBC RND homolog from E. coli (Kim and Nikaido 2012; Kim et al. 2010).
Ad (1) The acrB gene was triplicated in-frame as to express a single giant covalently linked trimeric AcrB by the Nikaido lab. This setup has the advantage that substitutions at sites essential for function (i.e., in the proton binding/translocation sites D407 and/or D408) can be exclusively introduced in one protomer of the trimer, whereas the other two protomers remain wild type. The experiments gave rise to some background activity due to formation of truncation products (AcrB monomers) able to form non-covalent and active AcrB trimers, but in principle very clearly indicated that inactivation of one of the protomers in the trimer results in a nonfunctional transporter. This observation disfavors a transport mechanism with independent stochastic cycling of the protomers.
Ad (2) If drug efflux activity is dependent on strictly coupled conformational cycling within the AcrB trimer, monomeric AcrB would be inactive. Two cases of monomeric AcrB production have been reported. In one case, the intermonomeric loop (Brandstatter et al. 2011) (Fig. 9.4) was genetically deleted and gave rise to monomeric AcrB which could be purified from membrane preparations in dodecyl-β-maltoside (DDM) and consisted of mainly not only monomers but also a species eluting within a retention volume alike to wild-type trimeric AcrB as shown by a size-exclusion chromatography. Interestingly, some residual activity could be measured for E. coli ΔacrB cells expressing the deleted-loop acrB gene mutant for some of the substrates tested. This might indicate that deleted-loop monomeric variants retain some ability to form functional trimeric AcrB or that residual activity is caused by monomeric species. Another interesting monomeric variant was created in the Wei Lab containing a P223G substitution [within the (tip) intermonomeric loop] (Yu et al. 2011). Here again, the activity was severely reduced, but some residual activity was maintained for some of the substrates. Via BN-PAGE it was shown that the P223G variant was monomeric; however, using Cys-crosslinking of the loop with the PC1 domain [shown previously to have no effect on the activity of trimeric AcrB (Seeger et al. 2008)], much of the activity could be recovered, showing the structural integrity of the monomeric species and the capability to assemble into a trimeric setup for function. The situation for the AcrB trimer appears to be different from another trimeric transporter, BetP. In the case of this betaine/Na+ symporter, the monomeric species still transports its substrate efficiently; however, the observed regulation by hyperosmotic shock in case of BetP trimer, is absent (Perez et al. 2011). It appears therefore that trimerization for AcrB and BetP might serve different regulatory roles, the former showing catalytic cooperativity and the latter regulatory cooperativity.
Ad (3) The E. coli heterotrimeric RND transporter homolog MdtBC offers a unique experimental flexibility which has been used by the Nikaido lab to address the interdependence of the protomers within the MdtB2C trimer (Kim and Nikaido 2012; Kim et al. 2010). Purified native MdtBC showed a clear MdtB2C complex formation. As with the in-gene triple fusions of acrB, combinations of mdtB and mdtC were fused to obtain trimers with different MdtB and/or MdtC combinations, showing that an Nterminus-BCB-Cterminus construct was most active. Within this heterotrimeric complex, MdtB and MdtC appeared to have different and complementing roles concerning the energy transduction (H+ binding/transport) and substrate binding/transport. MdtB with one of the D410A, D411A, K933A, R964A, or T971A substitutions (within the TMD proton translocation site, homologous to the D407, D408, K940, R971, and T978 residues in AcrB) yielded an inactive MdtB2C transporter trimer, but these substitutions in MdtC (D401A, D402A, K919A, R950A, or T957A) did not severely affect activity of the trimer. Interestingly, in MdtB2C mutants with an D410A substitution in MdtB appeared to influence the conformation of MdtC and its ability to accommodate substrates, since labeling with the MdtBC substrate Fluorescein-5-maleimide in MdtB_D410A_MdtC_F610C (or F598C, both located in the deep binding pocket region of MdtC) was enhanced compared to labeling of MdtB_wt_MdtC_F610A (or F598C). On the other hand, Cys-substitutions in the periplasmic deep binding pocket of MdtC affected the cloxacillin efflux activity substantially, whereas the Cys-subtitutions in homologous positions in MdtB did in general not affect the efflux activity. Moreover, the deep binding pocket Cys-substituted residues were labeled effectively in MdtC, whereas in MdtB at homolog positions only 10–20 % (two exceptions of 50 %) of the MdtC labeling efficiency could be achieved. These data indicate a clear separation of the functional roles of MdtB (energy transduction) and MdtC (drug binding/transport) within the MdtB2C heterotrimer and also points towards a clear interdependence of these subunits. It also shows that energy is not only transduced within the protomers as depicted in Fig. 9.6 for AcrB but also transduced interprotomerically, explaining the activity of the heterotrimer containing H+-binding-deficient MdtC. Conformational cycling and interprotomeric energy transduction therefore appear to be of intrinsic importance to the postulated functional rotation within the MdtBC transporter. Cross-linking studies within the MdtB or MdtC subunits might be needed to indicate whether conformational flexibility within the periplasmic domain is necessary for the cycling function of the trimer.
The interdependence of the protomers within the trimeric RND pump is recently also further substantiated by a fascinating observation: For the AcrAB-TolC system, very strong cooperative kinetics with Hill coefficients of 4–5 were measured by the Nikaido lab (Lim and Nikaido 2010; Nagano and Nikaido 2009) for cephalosporins (with low apparent affinity to AcrB, but not for nitrocefin, a cephalosporin with high affinity) extrusion, which might indicate bi-site activation as postulated (Pos 2009).
This observation immediately raises many interesting questions, e.g., the physiological role for cooperativity of this pump system but also on the determinants for cooperativity within the AcrAB-TolC complex. Cooperative effects can be anticipated on different levels within the complex: e.g., each protomer appears to be able to bind multiple drugs, and binding of one drug might be directly [by providing direct interaction interfaces (homotropic or heterotropic)] or indirectly (by allosteric effects within the protomer) influencing subsequent drug binding. Further cooperative effects can be anticipated by protomers within the trimer, where binding of drug(s) to one monomer influences binding/activity of the neighboring protomer as seems to be the case for the MdtB2C system. Another speculation concerns the role of the large (lipid-filled) cavity formed by the TM domains of the protomers. Activity might be controlled via the concentration of membrane-soluble drugs residing in the cavity area, by, e.g., changing lateral pressure compared to exclusively lipid-filled conditions and therefore stimulating the turnover rates of the trimeric complex. Another regulatory role might be given for the lipid composition, where specific lipids might bind preferably to (a) specific conformational state(s) of the AcrB trimer. BetP, e.g., is also trimeric, but no cooperative transport behavior can be shown for the transport for its substrate betaine. But interdependence has been shown by heterotropic allostery depending on osmotic stress (e.g., internal K+ concentration) (Ressl et al. 2009), as well as a periplasmic ionic network involved in crosstalk between the monomers affecting turnover rates (Gärtner et al. 2011). The concerted conformational changes in case of trimeric AcrB resembles more catalytic cooperativity as shown for the F1Fo-ATP-synthase (Boyer 1997) rather than the regulatory cooperativity seen for, e.g., the ACTase (Lipscomb and Kantrowitz 2011). Further cooperativity might involve the membrane adaptor component AcrA. Although its role in drug binding is not yet completely clear, shared (allosteric) binding sites with AcrB might exist. Its homolog CusB from the CusCBA Ag+ and Cu+ transport system has been shown to bind monovalent cations independent from the RND component (Bagai et al. 2007). AcrA might furthermore be involved in conformational energy conservation by, e.g., conserving energy in conformational strain delivered by the proton motive force via the RND AcrB and being released at a subsequent step, e.g., the O to L transition. The interaction between each AcrB monomer and its affiliated AcrA molecule has been investigated very recently (Symmons et al. 2009) and might suggest that during functional rotation of AcrB, conformational changes are transmitted towards AcrA. Rather than just being a rigid adapter, AcrA might actively transduce energy generated by AcrB and induce subtle peristaltic motions in the TolC channel. Similar to the tunnels observed in a single AcrB protomer during functional rotation (Murakami et al. 2006; Seeger et al. 2006; Sennhauser et al. 2007), TolC is possibly not a rigid hollow cylinder but changes its diameter by conformational change of each monomer, thereby pushing substrate unidirectionally from the closing aperture into the media (Pos 2009; Vaccaro et al. 2006, 2008).
9.5 The Role of a Switch Loop in Multiple Drug Binding in the Access and Deep Binding Pockets
Recent AcrB X-ray structures by two groups (Eicher et al. 2012; Nakashima et al. 2011) have substantiated multiple drug binding within the AcrB trimer as was postulated by the functional rotation hypothesis. The PC1/PC2 cleft in the L protomer of AcrB (Fig. 9.4) was proposed to function as a putative initial binding site for substrates (Murakami et al. 2006; Pos 2009; Seeger et al. 2006). A 2.25 Å resolution AcrB/doxorubicin co-crystal structure (pdb entry 4DX7) solved in our lab (Eicher et al. 2012) indicated simultaneous binding of a doxorubicin molecule in the deep binding pocket of the T protomer, a twofold-symmetric sandwich of stacked doxorubicin molecules (Fig. 9.7), and is likely to represent a structural indication for an access site for drugs into AcrB. This site was designated as the “access pocket.”
Binding of doxorubicin to the access pocket of the L protomer. (a) Close-up view of the access pocket with two doxorubicin molecules in stick representation (cyan: carbon, red: oxygen, blue: nitrogen) and 2Fo–Fc electron density (2.25 Å resolution) displayed as green mesh after refinement and contoured at 1σ. Side chain residues within 3.5 Å distance from the doxorubicin molecules are shown in stick representation (blue: carbon, red: oxygen, light blue: nitrogen, yellow: sulfur). Residues identified as part of the drug export pathway by Bodypy-FL maleimide labeling (Husain and Nikaido 2010) are depicted as magenta sticks (magenta: carbon, red: oxygen, blue: nitrogen) as well as alanine-substituted residues (F617A, F666A, R717A), causing reduced E. coli resistance towards drugs. (b) Side view of the asymmetric AcrB trimer in cartoon representation (L in blue, T in yellow, O in red) with two doxorubicin molecules bound to the access or one doxorubicin molecule bound to the deep binding pocket in sphere representation (cyan or orange, respectively: carbon, red: oxygen, blue: nitrogen)
The two doxorubicin molecules are tightly interacting with each other by stacking interactions of their anthracyclic rings. The stacked dimer itself is predominantly coordinated by hydrophobic interactions in the access pocket. In vivo substrate cross-linking experiments support the crystallographic observation that substrates bind to the access pocket (Fig. 9.7). Cysteine substitutions of those residues interacting with the doxorubicin dimer were also strongly cross-linked by the maleimide-labeled AcrB-substrate Bodipy-FL (Husain and Nikaido 2010). Strong cross-linking efficiencies indicate that these residues are part of the substrate pathway and accessible for substrate molecules under in vivo conditions. Moreover, AcrB variants F617A, F666A, and R717A conferred significantly decreased resistances against various compounds (Bohnert et al. 2008; Yu et al. 2005) (Fig. 9.7) and are therefore indicative to play an important role in AcrB-mediated transport of substrates.
Most strikingly, high molecular mass drugs rifampicin and erythromycin were identified in X-ray structures solved in the Yamaguchi lab in 2011 (Nakashima et al. 2011) within the L protomers of asymmetric AcrB co-crystal structures [pdb entries 3AOB (resolution = 3.35 Å), 3AOC (resolution = 3.35 Å) and 3AOD (resolution = 3.3 Å)] (Fig. 9.8). One of the structures showed the remarkable co-crystallization of rifampicin and minocycline in the L and T protomers, respectively, within the same AcrB trimer, indicating true “multiple drug” binding. Superimpositions of these structures with the 2.25 Å resolution doxorubicin co-crystal structure [pdb entry 4DX7, also containing drugs in both access and deep binding pockets (Eicher et al. 2012)] indicated that the position of rifampicin was largely congruent with the position of the doxorubicin dimer in 4DX7 (Fig. 9.8, D), whereas erythromycin was positioned more towards the deep binding pocket relative to the location of the doxorubicin dimer. The structures might indicate the intermediate phases of drug transport through the AcrB porter domain (Fig. 9.8). Of essential importance for the binding of substrates is a large loop (AcrB amino acids 613–623) of the PC1 subdomain protruding into the core of the porter domain of each AcrB protomer and is noticeably positioned between access and deep binding pocket (Fig. 9.9). This loop was designated as the “switch loop” and is partially defining the deep binding pocket as well as the access binding pocket. The switch loop adopts a specific conformation in the L protomer of the AcrB/doxorubicin co-crystal structure (pdb entry 4DX7) and interacts with the doxorubicin dimer in the access pocket.
Detailed view on the local structure of the porter domain of L protomers represented as transparent cartoons of the AcrB/substrate co-crystal structures (a) pdb entry 3AOC (erythromycin bound) and (b) pdb entry 3AOD (rifampicin and minocycline bound) solved by Nakashima et al. (2011). (a) Erythromycin is represented in green sticks. Residues interacting with erythromycin are shown in dark green sticks. (b) Rifampicin is displayed in cyan sticks. All residues involved in binding of rifampicn are shown in dark cyan sticks. (c) Porter domain of pdb entry 3AOC represented as transparent green cartoon. Erythromycin (green spheres) within pdb entry 3AOC is substantially displaced from the superimposed doxorubicin dimer (transparent blue spheres) identified in pdb entry 4DX7 (Eicher et al. 2012) and is nearer to the theoretical position of the deep binding pocket (presented by yellow spheres). (d) Porter domain of pdb entry 3AOD represented as transparent cyan cartoon. Rifampicin (cyan spheres) within pdb entry 3AOD is almost similarly positioned within the access pocket as the superimposed doxorubicin dimer (transparent blue spheres) identified in pdb entry 4DX7
Detailed view on the switch loop in cartoon representation of AcrB L monomers with a doxorubicin dimer from pdb entry 4DX7 (a, blue sticks), rifampicin from pdb entry 3AOD (b, cyan sticks) and erythromycin from 3AOC (c, green sticks) bound to the access pocket. The observed switch loop conformations correlate with the different substrates used for co-crystallization and might indicate that the switch loop conformation is substrate induced. (d) All switch loop conformations of the L monomers differ significantly from the switch loop conformation seen in the T monomer of pdb entry 4DX5 (AcrB with minocycline bound to only the deep binding pocket). In all pictures the switch loops of the respective other structures are displayed as transparent cartoons to demonstrate the reorientation of switch loop. Side chain residues are displayed in stick representation and substrate molecules as stick and transparent spheres to demonstrate the van der Waals volumes
A single amino acid exchange of G616N within the switch loop of AcrB was able to inhibit the efflux activity mainly for rather large substrates, like macrolide antibiotics (Eicher et al. 2012). Nevertheless, the transport of another high molecular mass drug, novobiocin, was not affected by the G616 substitution. Moreover, resistance against small molecular mass drugs ciprofloxacin, linezolid, and TPP+ was affected as well by the G616N substitution. Clearly, other physicochemical parameters of drug molecules, besides the molecular mass, are causing the rather puzzling phenotype. The crystal structure of AcrB G616N revealed an alternative switch-loop conformation in the L protomer of AcrB compared to the wild-type conformation. This altered switch-loop conformation would prevent or inhibit binding of a doxorubicin dimer in the access pocket by sterical clash. The multiple distinct L-switch loop conformations, observed in various crystal structures, indicate that the access pocket might be a highly flexible binding pocket, where the switch loop acts as an adaptor module. The putatively flexible adaptor module like the switch loop appears to be used to exclusively alter the shape of the binding pocket without altering the global protein conformation. This “adaptor-mediated binding mechanism” might be a general feature of multidrug recognition in membrane transporters and can either function according to an induced fit or conformational selection mechanism.
Combining the results obtained from the structural and functional data described above presents the tripartite drug efflux proteins as highly dynamic entities with allosteric and cooperative properties. Gaining deeper insight into the underlying mechanisms of substrate specificity of AcrB and the initial hypothesis of peristaltic motion driving drug efflux (Seeger et al. 2006) is in need of in vitro (reconstitution) assays (for AcrAB-TolC or any of the homologue tripartite systems) and time-resolved structural analysis. Moreover, the assignment of the physiological function of many of these tripartite pumps will help to understand why these machineries are highly cooperative and in need of allosteric regulation.
References
Akama H, Matsuura T, Kashiwagi S et al (2004) Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem 279:25939–25942, doi: 10.1074/jbc.C400164200 C400164200 [pii]
Bagai I, Liu W, Rensing C et al (2007) Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem 282:35695–702. doi:10.1074/jbc.M703937200
Balakrishnan L, Hughes C, Koronakis V (2001) Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol 313:501–510, doi: 10.1006/jmbi.2001.5038 S0022-2836(01)95038-7 [pii]
Bavro VN, Pietras Z, Furnham N et al (2008) Assembly and channel opening in a bacterial drug efflux machine. Mol Cell 30:114–21. doi:10.1016/j.molcel.2008.02.015
Bohnert JA, Schuster S, Seeger MA et al (2008) Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol 190:8225–8229
Boyer PD (1997) The ATP synthase–a splendid molecular machine. Annu Rev Biochem 66:717–749
Brandstatter L, Sokolova L, Eicher T et al (2011) Analysis of AcrB and AcrB/DARPin ligand complexes by LILBID MS. Biochim Biophys Acta 1808:2189–96. doi:10.1016/j.bbamem.2011.05.009
Bush K, Courvalin P, Dantas G et al (2011) Tackling antibiotic resistance. Nat Rev Microbiol 9:894–6. doi:10.1038/nrmicro2693
Davin-Regli A, Bolla J-M, James CE et al (2008) Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets 9:750–9
de Cristóbal RE, Vincent PA, Salomón RA (2006) Multidrug resistance pump AcrAB-TolC is required for high-level, Tet(A)-mediated tetracycline resistance in Escherichia coli. J Antimicrob Chemother 58:31–6. doi:10.1093/jac/dkl172
Eicher T, Cha HJ, Seeger MA et al (2012) Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692, doi: 1114944109 [pii] 10.1073/pnas.1114944109
Elkins CA, Nikaido H (2002) Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J Bacteriol 184:6490–6498. doi:10.1128/JB.184.23.6490
Fischer N, Kandt C (2011) Three ways in, one way out: water dynamics in the trans-membrane domains of the inner membrane translocase AcrB. Proteins 79:2871–2885. doi:10.1002/prot.23122
Fujihira E, Tamura N, Yamaguchi A (2002) Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J Biochem 131:145–51
Gärtner RM, Perez C, Koshy C, Ziegler C (2011) Role of bundle helices in a regulatory crosstalk in the trimeric betaine transporter BetP. J Mol Biol 414:327–36. doi:10.1016/j.jmb.2011.10.013
Higgins MK, Bokma E, Koronakis E et al (2004) Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci U S A 101:9994–9999, doi: 10.1073/pnas.0400375101
Husain F, Nikaido H (2010) Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol 78:320–330. doi:10.1111/j.1365-2958.2010.07330.x
Kawabe T, Fujihira E, Yamaguchi A (2000) Molecular construction of a multidrug exporter system, AcrAB: molecular interaction between AcrA and AcrB, and cleavage of the N-terminal signal sequence of AcrA. J Biochem 128:195–200
Kim H-S, Nikaido H (2012) Different functions of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB(2)C complex of Escherichia coli. Biochemistry 51:4188–97. doi:10.1021/bi300379y
Kim HS, Nagore D, Nikaido H (2010) Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J Bacteriol 192:1377–1386, doi: JB.01448-09 [pii] 10.1128/JB.01448-09
Koronakis V, Sharff A, Koronakis E et al (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914–919. doi:10.1038/35016007
Lee A, Mao W, Warren M (2000) Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182(11):3142–3150, doi: 10.1128/JB.182.11.3142-3150.2000
Lim SP, Nikaido H (2010) Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother 54:1800–1806, doi: AAC.01714-09 [pii] 10.1128/AAC.01714-09
Lipscomb W, Kantrowitz E (2011) Structure and mechanisms of Escherichia coli aspartate transcarbamoylase. Acc Chem Res 45:444–53. doi:10.1021/ar200166p
Lu S, Zgurskaya HI (2012) Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter. Mol Microbiol 86:1132–43. doi:10.1111/mmi.12046
Ma D, Cook DN, Alberti M et al (1995) Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55
Mao W, Warren MS, Black DS et al (2002) On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol Microbiol 46:889–901
Mokhonov VVV, Mokhonova EIEI, Akama H, Nakae T (2004) Role of the membrane fusion protein in the assembly of resistance-nodulation-cell division multidrug efflux pump in Pseudomonas aeruginosa. Biochem Biophys Res Commun 322:483–489. doi:10.1016/j.bbrc.2004.07.140
Murakami S, Nakashima R, Yamashita E, Yamaguchi A (2002) Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–93. doi:10.1038/nature01050
Murakami S, Nakashima R, Yamashita E et al (2006) Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–9. doi:10.1038/nature05076
Nagano K, Nikaido H (2009) Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A 106:5854–8. doi:10.1073/pnas.0901695106
Nakamura H, Hachiya N, Tojo T (1978) Second acriflavine sensitivity mutation, acrB, in Escherichia coli K-12. J Bacteriol 134(3):1184–1187
Nakashima R, Sakurai K, Yamasaki S et al (2011) Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480:565–569. doi:10.1038/nature10641
Nikaido H (1998) Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis 27(Suppl 1):S32–41
Nikaido H, Pagès J-M (2012) Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–63. doi:10.1111/j.1574-6976.2011.00290.x
Nikaido H, Takatsuka Y (2009) Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781, doi: S1570-9639(08)00334-8 [pii]10.1016/j.bbapap.2008.10.004
Nishino K, Yamaguchi A (2001) Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol 183:5803–5812. doi:10.1128/JB.183.20.5803
Pei X, Hinchliffe P (2011) Structures of sequential open states in a symmetrical opening transition of the TolC exit duct. Proc Natl Acad Sci U S A 108(5):2112–2117, doi: 10.1073/pnas.1012588108/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1012588108
Perez C, Khafizov K, Forrest LR et al (2011) The role of trimerization in the osmoregulated betaine transporter BetP. EMBO Rep 12:804–10. doi:10.1038/embor.2011.102
Petrek M, Kosinova P, Koca J, Otyepka M (2007) MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15:1357–1363
Piddock LJ (2006) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi:10.1128/CMR.19.2.382
Pos KM (2009) Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–93. doi:10.1016/j.bbapap.2008.12.015
Pos KM, Diederichs K (2002) Purification, crystallization and preliminary diffraction studies of AcrB, an inner-membrane multi-drug efflux protein. Acta Crystallogr D Biol Crystallogr 58:1865–1867
Ressl S, Terwisscha van Scheltinga AC, Vonrhein C et al (2009) Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature 458:47–52. doi:10.1038/nature07819
Saier MH, Tran CV, Barabote RD (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34:D181–6. doi:10.1093/nar/gkj001
Schulz R, Vargiu AV, Collu F et al (2010) Functional rotation of the transporter AcrB: insights into drug extrusion from simulations. PLoS Comput Biol 6:e1000806. doi:10.1371/journal.pcbi.1000806
Schulz R, Vargiu AV, Ruggerone P et al (2011) Role of water during the extrusion of substrates by the efflux transporter AcrB. J Phys Chem B 115:8278–8287. doi:10.1021/jp200996x
Seeger MA, Schiefner A, Eicher T et al (2006) Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298. doi:10.1126/science.1131542
Seeger MA, von Ballmoos C, Eicher T et al (2008) Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol 15:199–205
Seeger MA, von Ballmoos C, Verrey F, Pos KM (2009) Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48:5801–5812. doi:10.1021/bi900446j
Sennhauser G, Amstutz P, Briand C et al (2007) Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol 5:e7, doi: 06-PLBI-RA-1517R2 [pii] 10.1371/journal.pbio.0050007
Sennhauser G, Bukowska MA, Briand C et al (2009) Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol 389:134–145, doi: S0022-2836(09)00401-X [pii] 10.1016/j.jmb.2009.04.001
Su CC, Li M, Gu R et al (2006) Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol 188:7290–7296, doi: 188/20/7290 [pii] 10.1128/JB.00684-06
Su CC, Yang F, Long F et al (2009) Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol 393:342–355, doi: S0022-2836(09)01025-0 [pii] 10.1016/j.jmb.2009.08.029
Su C-C, Long F, Zimmermann MT et al (2011a) Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–62. doi:10.1038/nature09743
Su CC, Long F, Zimmermann MT et al (2011b) Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562, doi: nature09743 [pii] 10.1038/nature09743
Sulavik MC, Houseweart C, Cramer C et al (2001) Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother 45:1126–1136
Symmons MF, Bokma E, Koronakis E et al (2009) The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A 106:7173–7178, doi: 0900693106 [pii] 10.1073/pnas.0900693106
Takatsuka Y, Nikaido H (2006) Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol 188:7284–7289, doi: 188/20/7284 [pii] 10.1128/JB.00683-06
Takatsuka Y, Nikaido H (2007) Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol 189:8677–8684, JB.01127-07 [pii] 10.1128/JB.01127-07
Takatsuka Y, Nikaido H (2009) Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol 191:1729–1737, JB.01441-08 [pii] 10.1128/JB.01441-08
Tal N, Schuldiner S (2009) A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A 106:9051–9056, doi: 0902400106 [pii] 10.1073/pnas.0902400106
Tamura N, Murakami S, Oyama Y et al (2005) Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 44:11115–11121
Tanabe M, Szakonyi G, Brown KA et al (2009) The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem Biophys Res Commun 380:338–42. doi:10.1016/j.bbrc.2009.01.081
Tikhonova EB, Zgurskaya HI (2004) AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem 279:32116–32124. doi:10.1074/jbc.M402230200
Tikhonova EB, Yamada Y, Zgurskaya HI (2011) Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem Biol 18:454–463, doi: S1074-5521(11)00090-1 [pii] 10.1016/j.chembiol.2011.02.011
Touzé T, Eswaran J, Bokma E et al (2004) Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol 53:697–706. doi:10.1111/j.1365-2958.2004.04158.x
Tsukazaki T, Mori H, Echizen Y et al (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474:235–8. doi:10.1038/nature09980
Vaccaro L, Koronakis V, Sansom MS (2006) Flexibility in a drug transport accessory protein: molecular dynamics simulations of MexA. Biophys J 91:558–564
Vaccaro L, Scott KA, Sansom MS (2008) Gating at both ends and breathing in the middle: conformational dynamics of TolC. Biophys J 95:5681–5691, doi: S0006-3495(08)81985-6 [pii] 10.1529/biophysj.108.136028
Vargiu AV, Collu F, Schulz R et al (2011) Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J Am Chem Soc 133:10704–10707. doi:10.1021/ja202666x
Vila J, Martínez JL (2008) Clinical impact of the over-expression of efflux pump in nonfermentative Gram-negative bacilli, development of efflux pump inhibitors. Curr Drug Targets 9:797–807
Walsh C (2000) Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775–81. doi:10.1038/35021219
Weeks JW, Celaya-Kolb T, Pecora S, Misra R (2010) AcrA suppressor alterations reverse the drug hypersensitivity phenotype of a TolC mutant by inducing TolC aperture opening. Mol Microbiol 75:1468–83. doi:10.1111/j.1365-2958.2010.07068.x
Xu Y, Lee M, Moeller A et al (2011) Funnel-like hexameric assembly of the periplasmic adapter protein in the tripartite multidrug efflux pump in gram-negative bacteria. J Biol Chem 286:17910–20. doi:10.1074/jbc.M111.238535
Yao X-Q, Kimura N, Murakami S, Takada S (2013) Drug uptake pathways of multidrug transporter AcrB studied by molecular simulations and site-directed mutagenesis experiments. J Am Chem Soc. doi:10.1021/ja310548h
Yu EW, Aires JR, Mcdermott G, Nikaido H (2005) A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol 187:6804–6815, doi:187/19/6804 [pii]10.1128/JB.187.19.6804-6815.2005
Yu L, Lu W, Wei Y (2011) AcrB trimer stability and efflux activity, insight from mutagenesis studies. PLoS One 6:e28390. doi:10.1371/journal.pone.0028390
Zgurskaya HI, Nikaido H (1999) AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol 285:409–420, doi: 10.1006/jmbi.1998.2313
Zgurskaya HI, Nikaido H (2000) Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol 182:4264–4267
Zgurskaya HI, Yamada Y, Tikhonova EB et al (2009) Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta 1794:794–807, doi: S1570-9639(08)00335-X [pii]10.1016/j.bbapap.2008.10.010
Acknowledgments
The work of the Pos lab presented in this chapter was supported by the Swiss National Foundation, the German Research Foundation (SFB 807, Transport and Communication across Biological Membranes), the DFG-EXC115 (Cluster of Excellence Macromolecular Complexes at the Goethe-University Frankfurt), the Innovative Medicine Initiative (IMI), Project TRANSLOCATION (http://www.imi.europa.eu/content/translocation) and by grants from Europe Aspire and Human Frontier Science Program.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Cha, Hj., Pos, K.M. (2014). Cooperative Transport Mechanism and Proton-Coupling in the Multidrug Efflux Transporter Complex ArcAB-TolC. In: Krämer, R., Ziegler, C. (eds) Membrane Transport Mechanism. Springer Series in Biophysics, vol 17. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-53839-1_9
Download citation
DOI: https://doi.org/10.1007/978-3-642-53839-1_9
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-53838-4
Online ISBN: 978-3-642-53839-1
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)